Abstract
Free full text

A critical role for the loop region of the basic helix–loop–helix/leucine zipper protein Mlx in DNA binding and glucose-regulated transcription
Abstract
The carbohydrate response element (ChoRE) is a cis-acting sequence found in the promoters of genes induced transcriptionally by glucose. The ChoRE is composed of two E box-like motifs that are separated by 5 bp and is recognized by two basic helix–loop–helix/leucine zipper (bHLH/LZ) proteins, ChREBP and Mlx, which heterodimerize to bind DNA. In this study, we demonstrate that two ChREBP/Mlx heterodimers interact to stabilize binding to the tandem E box-like motifs in the ChoRE. Based on a model structure that we generated of ChREBP/Mlx bound to the ChoRE, we hypothesized that intermolecular interactions between residues within the Mlx loop regions of adjacent heterodimers are responsible for stabilizing the complex. We tested this hypothesis by preparing Mlx variants in which the loop region was replaced with that of another family member or mutated at several key residues. These Mlx variants retained their ability to bind to a single perfect E-box motif as a heterodimer with ChREBP, but no longer bound to the ChoRE nor supported glucose responsive activity. In summary, our results support a model in which the loop regions of Mlx play an important functional role in mediating the coordinate binding of ChREBP/Mlx heterodimers to the ChoRE.
INTRODUCTION
In mammals, when carbohydrate intake exceeds the immediate energy needs of the organism, the excess is converted into glycogen and triglycerides. Triglycerides are predominantly synthesized in the liver by de novo lipogenesis and in turn, exported in the form of VLDL particles to adipose and other tissues. Whereas triglycerides serve as an important energy source during fasting, their excess contributes to obesity, which is associated with type 2 diabetes and the metabolic syndrome. High carbohydrate intake stimulates the transcription of most genes in the de novo lipogenesis pathway, including enzymes involved in glycolysis [glucokinase and pyruvate kinase (PK)], in fatty acid synthesis [acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS)] and in NADPH production (malic enzyme) (1–3). Two signaling pathways control the transcription of lipogenic enzymes: the well known insulin signaling pathway and a less well characterized glucose signaling pathway (4). For example, full induction of ACC and FAS mRNA levels requires both glucose metabolism and insulin (5,6). A cis-acting sequence that activates the transcription of several lipogenic enzyme genes in response to glucose has been identified. This sequence, designated as the carbohydrate response element (ChoRE), consists of two E box-like motifs related to the consensus sequence CACGTG that are separated by 5 bp (5,7–10). The two E box sequences are usually not exact fits to the CACGTG consensus, most often matching at four or five positions to the 6 bp consensus. However, the spacing between them within the ChoRE is critical for the transcriptional response to glucose (9). All naturally occurring ChoREs analyzed to date have a 5 bp spacing between their two E box motifs. Mutations that change this spacing to 6 bp result in a strongly impaired glucose response, whereas mutations to 4 bp result in a complete loss of the response (9). This stringent spacing requirement suggests that proteins binding to the two E box motifs might interact sterically.
The presence of E box motifs in the ChoRE implicates members of the basic helix–loop–helix/leucine zipper (bHLH/LZ) protein family as transcription factors in regulating glucose-responsive genes (11–13). Proteins of this family always bind to the E box sequence as dimers or higher order oligomers. The contiguous HLH and LZ motifs mediate protein dimerization (12,13). Structurally, the second helix of the HLH motif and the leucine zipper domain form a continuous α-helical segment. The basic region projects from the first helix of the HLH motif and makes base-specific contacts with the major groove of the DNA helix. Dimerization orients the basic regions to enable favorable interactions with DNA. The loop region between the two helices of the HLH motif serves as a linker that tethers the interacting helices and may also stabilize DNA binding by making non-base specific contacts with the minor groove of the DNA helix.
The bHLH/LZ protein ChREBP was purified by DNA affinity chromatography using the PK ChoRE and demonstrated to function as a transcription factor involved in regulating glucose-responsive lipogenic enzyme genes (14–16). Several lines of evidence suggested that ChREBP requires another bHLH/LZ protein, Mlx, to exert its function. In particular, Mlx is required for ChREBP binding to the ChoRE and its activation of ChoRE-containing promoters (17). Transcriptional complexes formed on ChoREs contain both ChREBP and Mlx proteins. We recently reported that overexpressing a dominant negative form of Mlx in hepatocytes (in which both wild-type Mlx and ChREBP are present) abolishes the glucose response of several lipogenic enzyme genes and that this response can be rescued by either wild-type Mlx or ChREBP (18). Together, this evidence implicates Mlx as an obligatory partner of ChREBP in regulating glucose-responsive genes.
Here, we investigate the mechanism by which ChREBP and Mlx collaborate to activate glucose-responsive genes. We found that ChREBP/Mlx binding to the ChoRE requires the Mlx, but not the ChREBP, loop region, a requirement that does not apply when ChREBP/Mlx binds to a single perfect E box sequence. Furthermore, the ChREBP/Mlx/ChoRE complex migrates significantly more slowly than the ChREBP/Mlx/E-box complex in an electrophoretic mobility shift assay (EMSA), suggesting its significantly larger size. Based on these findings, we present a model structure in which two ChREBP/Mlx heterodimers interact with each other to stabilize binding to the tandem E box-like sites in the ChoRE. We demonstrate the functional importance of our binding data in an experiment that reveals the Mlx loop region to be essential for the ability of ChREBP/Mlx to support a glucose response.
MATERIALS AND METHODS
Primary hepatocyte culture and transfection
Male Harlan Sprague–Dawley rats (200–300 g) were fed ad libitum, and primary hepatocytes were isolated by the collagenase perfusion method (19). After a 3 h attachment to 35 mm Primaria plates, cells were transduced with adenovirus for 2 h in Williams' E medium containing 5.5 mM glucose, 23 mM HEPES, 0.01 μM dexamethasone, 2 mM glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 26 mM sodium bicarbonate and 0.1 U/ml insulin. Cells were then transfected using F1 reagent (Targeting Systems) with a mixture of a firefly luciferase reporter driven by an ACC ChoRE-containing promoter region (5) and an internal control plasmid encoding Renilla luciferase (pRL-CMV, Promega). In some experiments, expression plasmids for wild-type or variant Mlx or ChREBP (17,20) were cotransfected with the reporter plasmid mixture. Four h after transfection, Matrigel (Collaborative Biomedical Products) was added at 667 μg/35 mm plate and cells were maintained in low glucose medium for 16 h. Cells were then cultured in either low or high (27.5 mM) glucose medium for 24 h. Hepatocytes were harvested using the Passive Lysis Buffer (Promega). Luciferase assays were performed using the Dual-Luciferase Assay System kit (Promega). Results of luciferase assays are expressed as relative firefly light units/Renilla light units.
293 cell culture and transfections
Experiments were performed as described with minor modification (17). Human embryonic kidney 293 cells were maintained in DMEM supplemented with 5% fetal bovine serum. For whole cell extracts used in EMSAs, 293 cells were transfected in 100 mm plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Fifteen μg each of plasmids expressing ChREBP and Mlx were cotransfected. Whole cell lysates were prepared from cells 40 h post-transfection using the ‘whole cell extract’ protocol in the Nuclear Extract kit from ActiveMotif.
Plasmids and mutagenesis
All mutations of Mlx and ChREBP were created using the QuickChange™ site-directed mutagenesis kit (Stratagene). The dominant negative form of ChREBP was created by inserting a double mutation (R673A/R674G) into the basic region of the bHLH/LZ domain. This mutant was then used to prepare an adenoviral vector as described previously for dominant negative Mlx (18). Mutated coding regions were confirmed by DNA sequencing.
Electrophoretic mobility shift assay
EMSAs were conducted as described (21). A typical reaction contained 100 000 c.p.m. of 32P radio-labeled oligonucleotide and 5 μg of whole cell extract from 293 cells. After incubation with oligonucleotide for 30 min at room temperature, samples were loaded and separated on a 4.5% non-denaturing polyacrylamide gel. Results were obtained by PhosphorImager analysis.
Immunoblot analysis
Proteins obtained from the whole cell extract protocol were boiled in reducing buffer and separated on a 10% SDS–PAGE. Proteins were electro-transferred onto an Immobilon-P PVDF membrane (Millipore). Mlx and Mlx variants were detected with the M2 mouse monoclonal anti-HA antibody (Sigma) and ChREBP and ChREBP variants were detected either by anti-ChREBP (Santa Cruz Biotechnology) or anti-Flag antibody (Sigma). Signals were detected using the ECL western blot detection kit (Amersham Biosciences) with anti-mouse horseradish peroxidase-conjugated IgG as the secondary antibody (Santa Cruz Biotechnology).
Molecular modeling
A crystal structure is available for the bHLH/LZ domain from the homodimeric transcription factor Max complexed to an E box DNA sequence (13). We used the deposited coordinates of this structure to generate models of ChREBP/Mlx bound to the ChoRE sequence in the following manner. From within InsightII (Accelrys, Inc.), the Max homodimer complexed to the E-box sequence was duplicated, and the new complex ligated to the original to generate two Max dimers bound to an extended sequence containing two E-boxes: the ChoRE sequence. Homology-based modeling was used to generate coordinates for two ChREBP/Mlx heterodimers based on the Max homodimers. Each of these structures was superimposed onto a Max homodimer, which was then deleted. Impact (Schrodinger, LLC) was used to properly minimize the final structure, which was achieved by using 1000 steps of steepest descent minimization followed by another 1000 steps of the conjugate gradient minimization. The loop regions of ChREPB and Mlx were further refined by subjecting them to 20 ps of molecular dynamics with 1 fs time steps at 298 K. During the molecular dynamics simulation, the protein helices and entire DNA molecule was held rigid. The oplsaa 2005 forcefield was used along with the generalized born solvent model during the energy minimization and molecular dynamics simulation.
RESULTS
Two ChREBP/Mlx heterodimers bind coordinately to tandem E boxes in the ChoRE
All ChoREs studied to date contain two E box-like motifs separated by 5 bp (22). Previous results indicate that ChREBP and Mlx bind the ChoRE as a complex requiring both proteins (17). Interestingly, ChREBP and Mlx also bind as a complex to oligonucleotides containing only a single E box, as long as this site contains a perfect CACGTG consensus sequence (17,23). This complex migrates more rapidly in an EMSA than that formed on several ChoREs (17). We surmised that each of the E box-like motifs of the ChoRE binds to a ChREBP/Mlx heterodimer, which would cause such complexes to migrate more slowly in an EMSA compared to a single ChREBP/Mlx heterodimer formed with only one E box motif. To test whether binding of ChREBP/Mlx to the ChoRE is dependent on interactions between heterodimers bound at the tandemly arranged E box-like motifs, we performed an EMSA experiment with various amounts of overexpressed ChREBP/Mlx proteins and either an oligonucleotide containing a single perfect E box motif (WBSCR14) or the PK ChoRE oligonucleotide with two degenerate E boxes (Figure 1). Lanes 1 and 8 are the negative controls of mock-transfected 293 whole cell extract corresponding to the highest amount of protein used in the titration. All bands in these lanes represent background binding, as ChREBP and Mlx are expressed at negligible levels in 293 cells. The remaining lanes are loaded with increasing amounts of whole cell extract from 293 cells co-expressing a S196A/T666A double mutant ChREBP and wild-type Mlx. The double mutant ChREBP was used here to enhance the binding of ChREBP/Mlx to the ChoRE sequence (18,24). As reported previously (17), a single complex was observed on both PK ChoRE and WBSCR14 oligonucleotides. However, the PK ChoRE complex migrated more slowly than the WBSCR14 complex. Only the slower migrating complex appeared on the PK ChoRE even at the lowest amount of ChREBP/Mlx protein. If the binding of ChREBP/Mlx heterodimers to the two E box-like motifs were independent of each other, then at low ratios of protein to DNA, the faster migrating population should appear. Therefore, we hypothesized that two ChREBP/Mlx heterodimers are required to stabilize the binding of Mlx/ChREBP to the tandemly arranged E box-like motifs of the ChoRE.
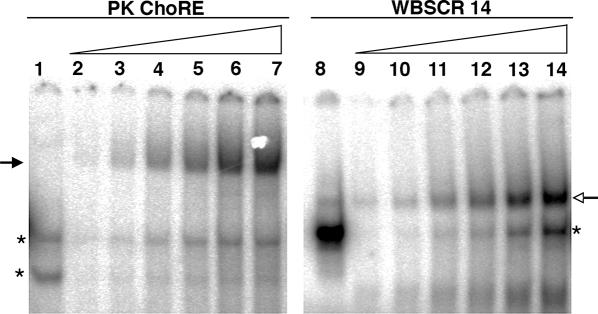
Comparison of binding of ChREBP/Mlx to oligonucleotides containing a single E box or the PK ChoRE. EMSA was performed with a probe containing the ChoRE of the PK gene, which consists of two E box-like motifs (5), or a probe containing only one perfect E box sequence (WBSCR14) (23). Sequences of these oligonucleotides are shown in Figure 2. Lanes 1 and 8 are controls with 5 μg of mock-transfected 293 whole cell extract. The remaining lanes are loaded with increasing amounts (0.5–5 μg) of whole cell extract from 293 cells co-expressing ChREBP and Mlx. The solid arrow indicates the slower migrating ChREBP/Mlx complex. The open arrow indicates the faster migrating ChREBP/Mlx complex. Asterisks denote background binding.
To test our hypothesis, we performed an experiment in which we individually mutated each E box-like motif in the PK ChoRE (Figure 2). Introduction of a 2 bp mutation into either of the PK ChoRE E box-like motifs resulted in the loss of the more slowly migrating ChREBP/Mlx complex. Importantly, no detectable formation of the more quickly migrating, heterodimer complex was observed, indicating that the single E box-like motif is not sufficient for recognition by the ChREBP/Mlx heterodimer (compare lanes 4 and 6 with lane 2). On the other hand, if we altered these two mutant PK ChoREs so that the remaining E box-like sequence consisted of a perfect CACGTG motif, we found that the more quickly migrating band, representing a single heterodimer of ChREBP/Mlx bound to one perfect E box, appeared (lanes 10 and 12). This new complex migrated identically with the ChREBP/Mlx heterodimer band observed on the WBSCR14 probe (lane 8). The appearance of the faster migrating band with these mutant oligonucleotides indicates that two ChREBP/Mlx heterodimers are not required for binding when a perfect E box site is present. In addition, we found that these mutant PK probes retained formation of the slower migrating complex. We suggest that retention of the slower migrating band arises from the presence of one strong E box site that can cooperatively stabilize binding to the mutant E box motif. These data support the conclusion that binding to naturally occurring ChoRE elements requires the coordinate action of two heterodimers. One mechanism by which such tandem binding could overcome the weak recognition of each E box-like motif is through the DNA promoting favorable protein–protein interactions between the two heterodimers. To test the plausibility of such protein–protein interactions, we generated a structural model of the ChREBP/Mlx dimers bound to the ChoRE.
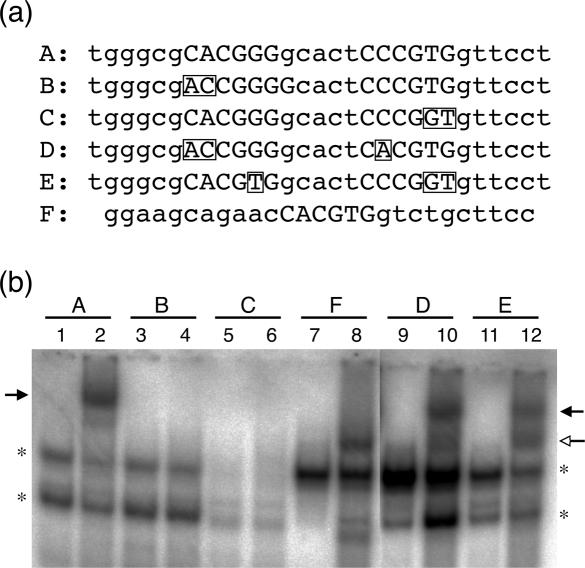
Mutational analysis of ChREBP/Mlx binding to the PK ChoRE. (a) Sequences of probes used to analyze ChREBP/Mlx binding. Probe A is the wild-type PK ChoRE and probe F is the synthetic WBSCR14 probe. Mutations from the wild-type PK ChoRE in probes B through E are shown in rectangles. The E box-like sequence of the PK probe and the corresponding region of mutant probes are capitalized, as is the single E box of the WBSCR14 probe. (b) EMSA was performed with probes shown in part (a). All odd-numbered lanes contain 5 μg of mock-transfected 293 whole cell extract and even-numbered lanes contain 5 μg of whole cell extract from 293 cells transfected with ChREBP and Mlx. The solid arrow indicates the slower migrating ChREBP/Mlx complex, and the open arrow the faster migrating complex. Asterisks denote background binding.
A model of the bHLH/LZ domain of ChREBP/Mlx bound to the ChoRE
We used the structure of the bHLH/LZ domain of the homodimeric transcription factor Max complexed to an E box DNA sequence (13) to generate models of ChREBP/Mlx bound to the ChoRE sequence (Figure 3). The models illustrate that it is sterically feasible for two ChREBP/Mlx heterodimers to bind simultaneously to two E boxes separated by 5 bp and that the intervening distance allows each of the dimers to bind to the same face of the DNA helix. Three possible binding arrangements exist and differ by which loops are positioned over the 5 bp intervening sequence. The ChREBP loop is significantly shorter than that of Mlx (Figure 4a) and the positioning of the ChREBP loop from each heterodimer over the intervening sequence does not result in direct intermolecular contacts. Mlx has a relatively long loop of 15 residues compared to that of ChREBP and MondoA, a paralog of ChREBP, which contain an 11- and 7-residue loops, respectively (25). In addition, the Mlx loop contains large hydrophobic residues, which suggests the possibility of hydrophobic protein–protein interactions between the two DNA-bound heterodimers. To test the likelihood of interactions between residues within a Mlx loop region with those of a neighboring heterodimer, we performed molecular dynamics simulations on these regions as the protein helices and entire DNA molecule was held rigid. Intriguingly, when the Mlx loop regions are interior, numerous favorable hydrophobic interactions are formed, as are salt bridges involving Q160 (Figure 3a). In these models, DNA acts as a scaffold to promote favorable interactions between residues within the loop region. In contrast, no such interactions are observed between Mlx and the shorter ChREBP loop region (Figure 3b).
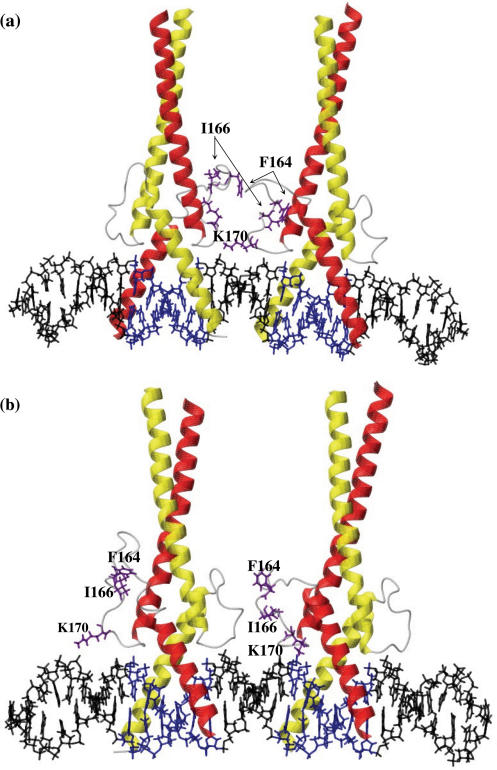
Model of the bHLH/LZ region of two ChREBP/Mlx heterodimers binding to the ChoRE. The models were generated by applying energy minimization and molecular dynamics (for the loop regions) on homology-based structures. The helical regions of Mlx and ChREBP are indicated in red and yellow, respectively, whereas the E boxes and flanking DNA are highlighted in blue and black. The three residues that were hypothesized and confirmed to contribute to ChoRE binding are highlighted in purple: F164, I166 and K170. Molecular modeling was performed on complexes in which the Mlx loop of one heterodimer partner is oriented toward the Mlx loop of the other partner (a) or toward the ChREBP loop of the other partner (b).
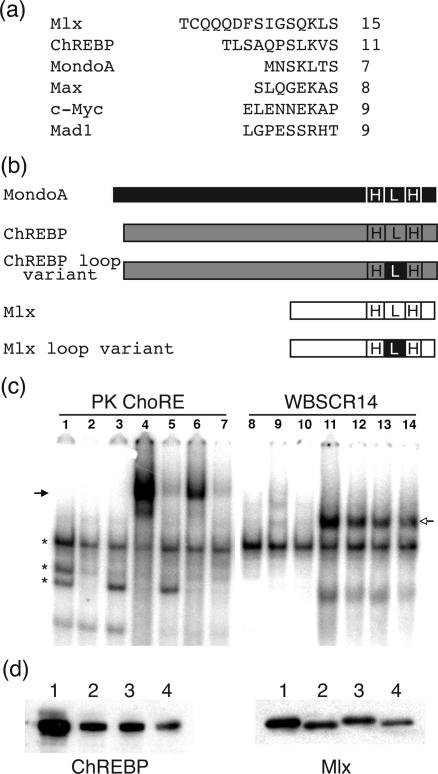
The Mlx loop region is critical for ChoRE binding. (a) Comparison of loop regions of various bHLH/LZ proteins. Sequences of the loop regions of several representative bHLH/LZ proteins are shown. The number of residues in the loop region is indicated. Mlx has a significantly longer loop domain than most other bHLH/LZ proteins, allowing it to potentially interact across the interface between heterodimer pairs. (b) Construction of ChREBP and Mlx loop variants. ChREBP loop variant: the loop region of ChREBP was substituted with the MondoA loop region; Mlx loop variant: the loop region of Mlx was substituted with the MondoA loop region. (c) EMSAs were performed with an oligonucleotide containing the PK ChoRE or the WBSCR14 probe and 5 μg of 293 cell extract. Lanes 1 and 8 are 293 mock-transfected whole cell extract. The other lanes contain extract from 293 cells transfected with the following expression plasmids: lanes 2 and 9, ChREBP loop variant alone; lanes 3 and 10, Mlx loop variant alone; lanes 4 and 11, ChREBP and Mlx; lanes 5 and 12, ChREBP and Mlx loop variant; lanes 6 and 13, ChREBP loop variant and Mlx; lanes 7 and 14, ChREBP loop variant and Mlx loop variant. The solid arrow indicates the slower migrating and the open arrow indicates the faster migrating ChREBP/Mlx complexes. Asterisks denote background binding. (d) Protein levels of ChREBP and Mlx in 293 cell extracts. Loading is as follows: lane 1, ChREBP and Mlx; lane 2, ChREBP and Mlx loop variant; lane 3, ChREBP loop variant and Mlx; lane 4, ChREBP loop variant and Mlx loop variant. Proteins were separated on an SDS–PAGE gel and immunoblotted with a ChREBP antibody to detect ChREBP and ChREBP loop variant (left side) or an anti-HA antibody to detect Mlx and Mlx loop variant (right side).
The loop region of Mlx is critical for stable binding to the ChoRE but not to the single E box
To test our computational results experimentally, we examined the role of the loop regions of ChREBP and Mlx in binding a ChoRE sequence. The loop region of each of these proteins was substituted with that of MondoA, another bHLH/LZ protein that heterodimerizes with Mlx, while the rest of the protein was left intact (Figure 4b). FLAG-tagged ChREBP or a ChREBP loop substitution variant was expressed with either HA-tagged Mlx or its loop substitution variant in 293 cells. Immunoblotting using either anti-HA or anti-ChREBP antibody was performed to confirm protein expression (Figure 4d). Each protein was stably expressed in 293 cells; however, the wild-type proteins were expressed at higher levels than the loop substitution variants. An EMSA performed using either the PK ChoRE (lanes 1–7) or single E box WBSCR14 oligonucleotide (lanes 8–14) demonstrated the importance of the Mlx loop region for binding to the ChoRE (Figure 4c). When ChREBP and Mlx were co-expressed in the 293 cells (lanes 4 and 11), a slower migrating complex appeared on the PK ChoRE oligonucleotide, whereas a more rapidly migrating complex was identified on the WBSCR14 oligonucleotide, above bands observed for mock-transfected 293 cells (lanes 1 and 8). However, extracts from 293 cells expressing the Mlx loop variant and ChREBP exhibited a greatly reduced level of the slower migrating band in the EMSA (lanes 5 and 7). Such an effect was not observed when the ChREBP loop substitution variant was co-expressed with Mlx (lane 6). In comparison, no differences in binding of the Mlx loop variant and ChREBP to the single E box oligonucleotide were observed (lanes 12 and 14). These results demonstrate clearly that the Mlx loop region is required for binding to the PK ChoRE, but not to the WBSCR14 oligonucleotide. In contrast, substitution of the ChREBP loop region did not have a significant effect on the binding to either PK ChoRE or WBSCR14. These data support the model presented in Figure 3a in which binding of two ChREBP/Mlx heterodimers is mediated by intermolecular contacts formed between residues of the Mlx loop regions.
The loop region of Mlx is critical for its functional activity
To test the importance of ChREBP/Mlx interactions in binding to the ChoRE, we performed a functional assay. In particular, we used a dominant negative form of Mlx that blocks the glucose response from an ACC ChoRE-containing promoter and rescued this response by over expressing wild-type Mlx (18). We used this system as a functional assay to test whether the Mlx loop substitution variant is competent for the ChREBP/Mlx-mediated glucose response. Hepatocytes were first transduced with an adenovirus expressing dominant negative Mlx for 2 h and then transfected with increasing amounts of plasmids expressing Mlx or Mlx loop substitution variant together with a reporter plasmid that has multiple copies of the ACC ChoRE in its promoter region. Cells were kept in low (5.5 mM) glucose medium for the first 24 h, after which the level was either maintained (control) or increased (27.5 mM) to induce the glucose response. As expected, increasing amounts of wild-type Mlx progressively rescued the glucose response inhibited by over expressing the dominant negative Mlx in hepatocytes (Figure 5a). In contrast, expression of the Mlx loop variant failed to rescue the glucose response at all doses administrated. Together with the binding data, these results indicate the importance of the Mlx loop region and of the tandem binding of two ChREPB/Mlx heterodimers to the ChoRE in the glucose response.
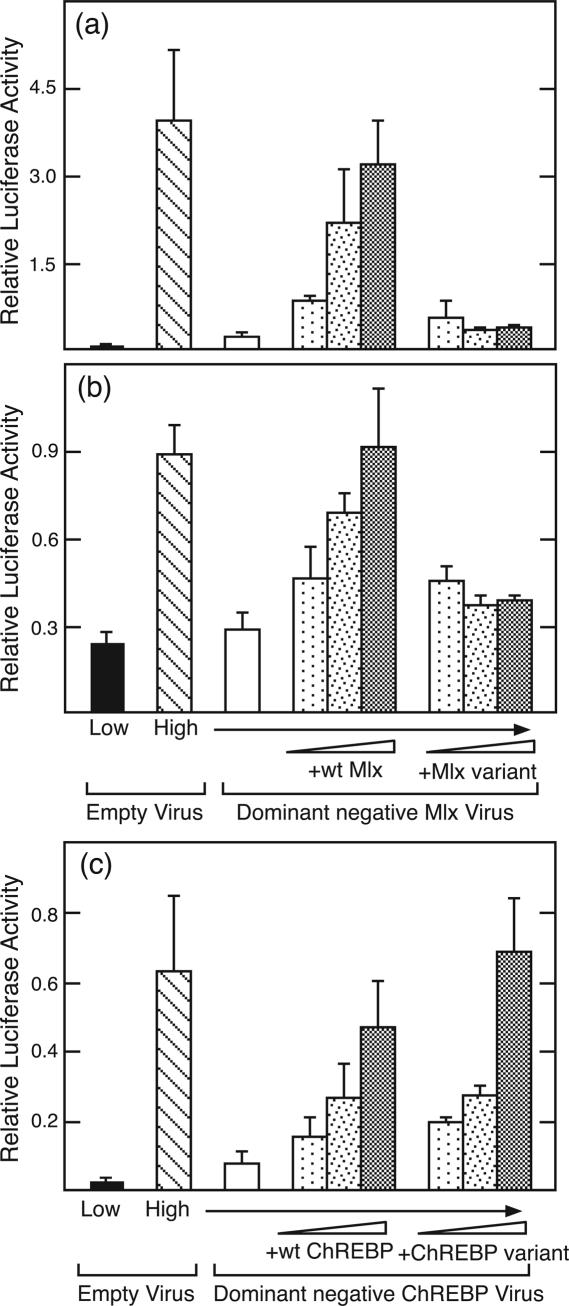
The loop region of Mlx is critical for functional activity. (a) Mlx function was measured by its ability to rescue a glucose-response in cells treated with a dominant negative Mlx adenovirus. Hepatocytes were treated with the dominant negative Mlx virus or control virus for 2 h. Plasmids expressing Mlx or the Mlx loop variant (50, 100 and 500 ng) were cotransfected into hepatocytes together with a reporter containing multiple copies of the ACC ChoRE linked to a basal promoter. Cells were incubated in low glucose for 24 h and then changed to either low or high glucose for an additional 24 h to induce the glucose response. Values represent the mean of triplicate samples (±SD). (b) Same as (a) except the reporter contained the PK promoter fragment from −183 to +12. (c) ChREBP function was measured in an identical manner to that described for Mlx in (a) except a dominant negative ChREBP adenovirus was used in place of the dominant negative Mlx virus.
To test the generality of this observation, we examined whether the Mlx loop variant could support a glucose response from the PK promoter fragment from −183 to +12. This region contains the functional ChoRE of the PK gene, as well as binding sites for additional transcription factors, HNF-1, HNF-4 and NF-1, involved in promoter function. The PK promoter supported an ~4-fold induction in response to treatment with high glucose, and this response was largely repressed by expression of the dominant negative Mlx (Figure 5b). When an expression vector for wild-type Mlx was transfected with the reporter plasmid, the response to glucose was restored in a dose-dependent manner. However, the Mlx loop variant was again unable to restore a functional glucose response. Hence, the Mlx loop region that was necessary for formation of the ChoRE complex on EMSA was also required to support the glucose response in hepatocytes.
We performed a similar experiment with the ChREBP loop variant. For this purpose, we used a dominant negative ChREBP in which two conserved basic residues in its basic region were mutated, analogous to the Mlx dominant negative form. Hepatocytes transduced with adenovirus expressing this dominant negative ChREBP were blocked in their ability to respond to glucose (Figure 5c). Introduction of increasing amounts of wild-type ChREBP into these hepatocytes progressively rescued the glucose response. Notably, the ChREBP loop variant was able to rescue glucose response as effectively as the wild-type protein. These results, together with the binding data, suggest that the loop region of Mlx, but not that of ChREBP, is critical for binding to the ChoRE and for glucose regulation.
F164, I166 and K170 in the Mlx loop region are critical for ChoRE binding and functional activity
Based on the ChREBP/Mlx structural model, we targeted several residues in the Mlx loop region as playing potentially important roles in promoting heterodimer interaction. F164 and I166 are two bulky hydrophobic residues in the loop region likely to form favorable intermolecular hydrophobic interactions. In addition, K170 is a basic residue located at the C-terminal end of loop and a likely candidate for forming favorable electrostatic interactions with the DNA backbone. We mutated these three residues separately or together to alanine residues. No effect on Mlx binding or function was observed with any single mutation (data not shown); however, an Mlx mutant in which all three residues were mutated lost the ability to bind the ChoRE. In particular, an EMSA performed with the ACC ChoRE showed greatly reduced binding when the triple mutant Mlx was used in place of the wild-type Mlx (Figure 6a, lanes 2 and 3). In contrast, when binding to the single E box WBSCR14 oligonucleotide was tested, the binding supported by triple mutant and wild-type Mlx were comparable (lanes 5 and 6). Immunoblotting confirmed that the triple mutant Mlx protein was expressed at levels comparable to wild-type Mlx (Figure 6b), and therefore these results indicate that the combined contribution of F164, I166 and K170 is essential for the coordinate binding of two ChREBP/Mlx heterodimers to the ChoRE, but not for binding of a single heterodimer to a perfect E box. Next, we tested the functional impact of mutating F164, I166 and K170 to alanine. As before, we performed a rescue experiment (Figure 6c) and confirmed that wild-type Mlx could rescue the glucose response. In contrast, the triple mutant Mlx was unable to support a glucose response. Therefore, these three residues play an important role in DNA binding to the ChoRE and supporting a functional response to glucose in hepatocytes.
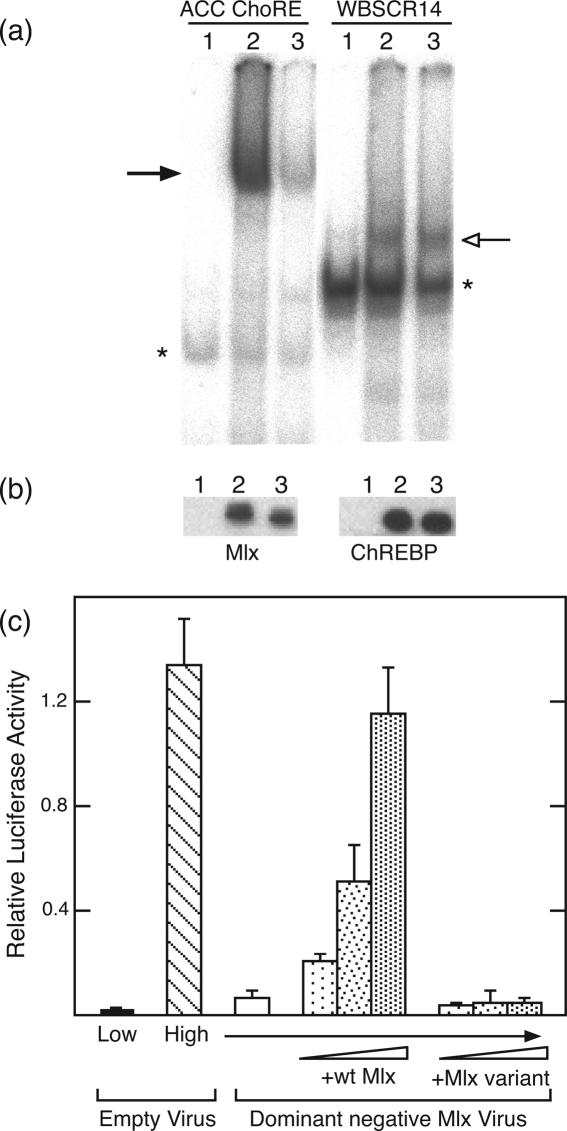
Three residues in the loop region are critical for Mlx binding and function. A mutant form of Mlx in which three loop residues—F164, I166 and K170—were changed to alanine was constructed. (a) EMSA was performed with an oligonucleotide containing the ACC ChoRE (5) or the WBSCR14 probe. Lanes 1 and 4 are the negative controls of mock-transfected 293 whole cell lysate. Lanes 2 and 5 are 293 cell whole cell extracts expressing both ChREBP and Mlx. Lanes 3 and 6 are ChREBP and Mlx loop mutant co-expressed in 293 cells. The solid arrow indicates the slower migrating and the open arrow the faster migrating ChREBP/Mlx complexes. Asterisks indicate background binding. (b) Protein levels of wild-type and mutant Mlx (left) and ChREBP (right). The loading corresponds to the loading used in the EMSA in panel (a). Proteins were separated on a SDS–PAGE gel and immunoblotted with an anti-HA antibody to detect wild-type and loop mutant Mlx, while an anti-FLAG antibody was used to detect ChREBP. (c) Hepatocytes were treated with dominant negative Mlx virus or control virus for 2 h. Increasing amounts of plasmids (50, 100 and 500 ng) expressing wild-type Mlx or Mlx variant (F, I, K/A) were cotransfected with the ACC ChoRE-containing reporter into hepatocytes. Cells were incubated in low glucose for 24 h and then changed to either low or high glucose for an additional 24 h to induce the glucose response. Values represent the mean of triplicate samples (±SD).
DISCUSSION
In this study, we demonstrate that the loop region of the bHLH/LZ domain of Mlx is required for binding to the ChoRE as well as for ChREBP/Mlx function in the glucose response. Our data suggest that two ChREBP/Mlx heterodimers bind coordinately to the tandem E box-like sites in the ChoRE, stabilized by interactions between Mlx loop regions. The interaction between ChREBP/Mlx heterodimers occurs when the two E boxes of the ChoRE are separated by five intervening bp. Binding of ChREBP/Mlx to the ChoRE likely occurs sequentially, with one heterodimer binding weakly and then stabilizing the binding of the second heterodimer in a cooperative manner. Alternatively, ChREBP and Mlx could exist as a tetramer that binds in a concerted manner to the ChoRE. In either case, we conclude that in contrast with a perfect E box motif, the affinity of ChREBP/Mlx for a single E box-like motif as present in the ChoRE is not sufficient for stable binding. This conclusion is supported by the observation that mutations introduced into either of the E box-like motifs of the PK ChoRE results in a complete loss of ChREBP/Mlx binding. It is worth noting that our experiments do not preclude the possibility that another protein assists the binding of ChREBP/Mlx to the ChoRE via interactions involving the Mlx loop region. In this scenario, ChREBP/Mlx dimer would occupy one E box as the other is occupied by another unknown dimer bHLH/LZ protein(s). Our data would similarly implicate the loop region of Mlx as essential for such a complex. However, we do not favor this possibility as sequence requirements for the two E box motifs in the ChoRE are similar to each other, suggesting they interact with a common component (22).
All of the ChoREs identified thus far with the exception of the S14 ChoRE contain two imperfect CACGTG motifs (22). For the S14 ChoRE, one perfect and one imperfect E box are found. We expect that the coordinate binding of multiple ChREBP/Mlx heterodimers plays an important role in supporting the recognition of such naturally occurring E box-like sequences in ChoREs. The bHLH/LZ family contains many members that bind with relative promiscuity to the perfect CACGTG motif. The presence of variant E boxes that only allow binding through tandem interactions of heterodimers in naturally occurring ChoREs may provide specificity for the actions of ChREBP/Mlx.
In addition to providing specificity in binding, the interaction of ChREBP/Mlx heterodimers on the ChoRE likely plays an important role in supporting a functional response to glucose. As shown by the EMSA, a single perfect E box site is sufficient to support binding of a ChREBP/Mlx heterodimer to DNA. However, reporter constructs that contain perfect E box sites do not respond to glucose unless two such sites are present and separated by 5 bp. The inability to respond is not merely due to the simple number of ChREBP/Mlx heterodimers recruited to the DNA. Constructs that contain two single perfect E box sites separated by 7–15 bp are not active in supporting a glucose response (9). These observations infer that there is a functional significance to the stereo-specific arrangement of ChREBP/Mlx heterodimers in the ChoRE complex. One possible explanation is that this arrangement may facilitate the recruitment of other proteins such as transcriptional co-activators or chromatin remodeling complexes.
The binding of two ChREBP/Mlx heterodimers to the ChoRE is likely supported by favorable protein–protein interactions that are promoted by the DNA, which performs a scaffolding role. Our structural models suggest that the loop regions of Mlx and ChREBP could be candidates for mediating interactions between the two heterodimers. Indeed, we observed that the Mlx loop variant not only displayed greatly reduced binding to the ChoRE, but also lost functional activity. In contrast, binding to the ChoRE and functional activity was not affected by replacing the ChREBP loop region. That the Mlx loop variant retained normal binding to a single E box sequence indicates that the mutation did not alter its protein structure. We further examined the contribution of specific residues in the Mlx loop region and found three that appear to stabilize heterodimer interactions. Hydrophobic residues F164 and I166 are likely to form hydrophobic protein–protein interactions, whereas the basic residue K170 is positioned favorably to interact with the DNA backbone and thereby stabilize DNA binding. Altogether our data favor a model in which Mlx loop regions are situated in the middle of the complex and interact homotypically. This is the first published evidence for a specific function for the loop region of any bHLH proteins in promoting protein–protein interactions.
Our model provides an explanation for the stringent spacing requirements of the E boxes in the ChoRE. In particular, the five intervening bp between the E box motifs likely enables the appropriate distance over which favorable interactions can occur between the Mlx loop regions. The loop regions of most bHLH proteins range from 6 to 10 residues in length (11). This minimal size is required for physically linking the two overlapping helices of the HLH domain with proper orientation. However, a few family members have notably longer loop regions. These include Mlx (15 residues), Hairy (14 residues) and ASCT5 (21 residues). For Mlx, we postulate that the longer loop enables favorable interactions between two closely positioned heterodimers bound to the ChoRE. Whether other bHLH proteins with longer loop regions utilize them to stabilize interactions between tandemly arranged DNA binding sites will be interesting to explore.
Certain members of the bHLH/LZ family have been shown to interact to form tetramers, but through a different mechanism than that proposed for ChREBP/Mlx heterodimer interaction. Both Myc-Max and USF are capable of forming tetramers that are stabilized through interactions of the leucine zipper domains in a head-to-tail assembly (12,26). This arrangement results in the formation of an antiparallel four-helix bundle that does not interfere with the DNA binding basic region. In this configuration, Myc-Max and USF tetramers can simultaneously bind to two E box motifs. As opposed to what we propose for ChREBP/Mlx, these interactions are not required for DNA binding and are not dependent on E box spacing. Instead, they allow interactions with E boxes that are widely and variably spaced through a DNA looping mechanism.
STAT family proteins bind to consensus STAT binding sites as dimers, but more commonly form complexes of interacting dimers when bound to tandemly linked imperfect STAT binding sites in natural promoters (27,28). As seen for ChREBP/Mlx, heterodimer interactions allow STAT proteins to extend the range of target sites to non-consensus binding sequences. The ability of STAT heterodimers to interact is mediated through an N-terminal region that is not part of the DNA binding domain (29,30). This interaction of STAT dimers is essential for transcriptional regulation of their target genes. The similar binding of STAT and ChREBP/Mlx dimers to tandemly arranged binding motifs underscores the importance of transcription factor interactions in activating gene transcription.
In conclusion, we have demonstrated that ChREBP/Mlx heterodimers bind to the ChoRE in a manner that requires residues of the Mlx, but not the ChREBP, loop region. The ability of ChREBP/Mlx heterodimers to interact likely enables these proteins to achieve stable binding to weakly conserved E boxes. The arrangement of ChREBP/Mlx heterodimers in this complex may facilitate interactions with other proteins that allow for the activation of genes containing ChoRE sequences in response to glucose.
Acknowledgments
This work was supported by grants DK26919 and P30 DK50456 (Minnesota Obesity Center) from the National Institutes of Health. Molecular modeling and visualization was performed in the Minnesota Supercomputing Institute Basic Sciences Computing Lab. Funding to pay the Open Access publication charges for this article was provided by NIH DK 26919.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Citations & impact
Impact metrics
Citations of article over time
Article citations
Suppression of hepatic ChREBP⍺-CYP2C50 axis-driven fatty acid oxidation sensitizes mice to diet-induced MASLD/MASH.
Mol Metab, 85:101957, 11 May 2024
Cited by: 0 articles | PMID: 38740087 | PMCID: PMC11145360
Normal and Neoplastic Growth Suppression by the Extended Myc Network.
Cells, 11(4):747, 21 Feb 2022
Cited by: 12 articles | PMID: 35203395 | PMCID: PMC8870482
Review Free full text in Europe PMC
Mechanisms of Binding Specificity among bHLH Transcription Factors.
Int J Mol Sci, 22(17):9150, 24 Aug 2021
Cited by: 38 articles | PMID: 34502060 | PMCID: PMC8431614
Review Free full text in Europe PMC
Modification of a Constitutive to Glucose-Responsive Liver-Specific Promoter Resulted in Increased Efficacy of Adeno-Associated Virus Serotype 8-Insulin Gene Therapy of Diabetic Mice.
Cells, 9(11):E2474, 13 Nov 2020
Cited by: 2 articles | PMID: 33202992 | PMCID: PMC7696068
LXRα Regulates ChREBPα Transactivity in a Target Gene-Specific Manner through an Agonist-Modulated LBD-LID Interaction.
Cells, 9(5):E1214, 13 May 2020
Cited by: 2 articles | PMID: 32414201 | PMCID: PMC7290792
Go to all (44) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes.
J Biol Chem, 279(15):15662-15669, 23 Jan 2004
Cited by: 140 articles | PMID: 14742444
Direct role of ChREBP.Mlx in regulating hepatic glucose-responsive genes.
J Biol Chem, 280(12):12019-12027, 20 Jan 2005
Cited by: 121 articles | PMID: 15664996
Flightless I homolog negatively regulates ChREBP activity in cancer cells.
Int J Biochem Cell Biol, 45(11):2688-2697, 17 Sep 2013
Cited by: 14 articles | PMID: 24055811
The regulation and role of carbohydrate response element-binding protein in metabolic homeostasis and disease.
J Neuroendocrinol, 29(10), 01 Oct 2017
Cited by: 6 articles | PMID: 28370553
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (4)
Grant ID: R01 DK026919
Grant ID: P30 DK050456
Grant ID: P30 DK50456
Grant ID: DK26919



