Abstract
Free full text

Functionally distinct monomers and trimers produced by a viral oncoprotein
Abstract
While the process of homo-oligomer formation and disassembly into subunits represents a common strategy to regulate protein activity, reports of proteins in which the subunit and homo-oligomer perform independent functions are scarce. Tumorigenesis induced by the adenovirus E4-ORF1 oncoprotein depends on its binding to a select group of cellular PDZ proteins, including MUPP1, MAGI-1, ZO-2 and Dlg1. We report here that in cells E4-ORF1 exists as both a monomer and trimer and that monomers specifically bind and sequester MUPP1, MAGI-1 and ZO-2 within insoluble complexes whereas trimers specifically bind Dlg1 and promote its translocation to the plasma membrane. This work exposes a novel strategy wherein the oligomerization state of a protein not only determines the capacity to bind separate related targets but also couples the interactions to different functional consequences.
Introduction
Among human adenoviruses, adenovirus type 9 uniquely elicits mammary tumors in experimentally infected animals. The primary oncogenic determinant of this virus is the E4 region open-reading-frame 1 (E4-ORF1) gene, which codes for a 125-residue polypeptide possessing two separate protein-interaction elements, the C-terminal PDZ domain-binding motif (PBM) and centrally located Domain 2 (Lee et al., 1997; Chung et al., 2007). These two critical elements determine the capacity of E4-ORF1 to associate with the plasma membrane and/or membrane vesicles, to activate cellular phosphatidylinositol 3-kinase (PI3K), and to promote cellular transformation in vitro and mammary tumorigenesis in vivo (Frese et al., 2006; Chung et al., 2007).
While the function of Domain 2 remains poorly understood because the 70-kDa cellular phosphoprotein targeted by this element has not yet been identified, the PBM has been shown to mediate interactions with the five cellular multi-PDZ domain proteins MUPP1, PATJ, MAGI-1, ZO-2 and Dlg1. Significantly, cellular transformation induced by the E6 oncoprotein of high-risk human papillomavirus (HPV) or by the Tax oncoprotein of human T-cell leukemia virus type 1 (HTLV-1) likewise depends on a C-terminal PBM that mediates binding to cellular PDZ proteins (Kiyono et al., 1997; Hirata et al., 2004), including one or more targeted by E4-ORF1 (Lee et al., 1997, 2000; Glaunsinger et al., 2000; Latorre et al., 2005). These cellular factors act as membrane-associated scaffolds to assemble protein complexes that control fundamental processes such as protein trafficking, cell signaling, cell polarity, cell migration and tumor suppression (Humbert et al., 2006). Consequently, investigating interactions of E4-ORF1 with PDZ proteins promises to reveal important new mechanisms involved in the development of human cancers.
E4-ORF1 has been shown to alter the subcellular localization of its PDZ protein targets in one of two strikingly different ways. In a Domain 2-independent manner, E4-ORF1 sequesters MUPP1, PATJ, MAGI-1 and ZO-2 in insoluble complexes associated with cytoplasmic membrane vesicles or triggers Dlg1 to translocate to the plasma membrane (Glaunsinger et al., 2000; Lee et al., 2000; Latorre et al., 2005; Frese et al., 2006; Chung et al., 2007). These two effects on localization also yield distinct functional consequences. Evidence suggests that sequestered PDZ proteins are functionally inactivated because, in epithelial cells, E4-ORF1-induced sequestration of the PATJ polarity protein and ZO-2, which are critical for tight junction (TJ) biogenesis (Shin et al., 2005; Umeda et al., 2006), disrupts the TJ barrier function and causes a loss of apicobasal polarity (Latorre et al., 2005). In fibroblasts, sequestration of ZO-2 also blocks its tumor suppressor activity (Glaunsinger et al., 2001). Conversely, results indicate that E4-ORF1-induced translocation of Dlg1 to the plasma membrane activates a Dlg1 oncogenic function required for E4-ORF1 to promote Ras-dependent PI3K stimulation (Frese et al., 2006).
Adenovirus E4-ORF1 evolved from an ancestral cellular dUTP pyrophosphatase (dUTPase) (Weisset al., 1997). Polypeptides encoded by dUTPase genes are comparable in length to E4-ORF1 and form homotrimeric enzymes that hydrolyse dUTP and, in so doing, prevent detrimental incorporation of this nucleotide into replicating cellular DNA (Mol et al., 1996). Nonetheless, E4-ORF1 and dUTPase have functionally diverged (Weiss et al., 1997). In this paper, we present evidence indicating that, despite their functional divergence, these two related proteins share a conserved protein fold, as well as a capacity to form homo-trimers, though E4-ORF1 differs from dUTPase by additionally producing monomers in cells. The existence of two different forms of E4-ORF1 led to the discovery that each has distinct functions, with monomers specifically binding and sequestering MUPP1, MAGI-1 and ZO-2 within insoluble complexes and trimers specifically binding Dlg1 and promoting its translocation to the plasma membrane. Therefore, E4-ORF1 employs a novel strategy to increase protein complexity by producing monomers and trimers that bind separate related targets and promote different functional outcomes.
Results
Predicted structural similarity with trimeric dUTPases leads to discovery that E4-ORF1 exists as a trimer and monomer in cells
Comparative modeling analyses predicted that E4-ORF1 adopts a protein fold like trimeric dUTPase enzymes and that the only major difference between the two proteins is located at the C terminus where a short 4-residue E4-ORF1 PBM element replaces a longer 20-residue dUTPase peptide containing the nucleotide-binding P-loop motif (Supplementary Figure S1). We therefore tested whether E4-ORF1 likewise forms a trimer. Extracts of 293 T cells expressing either 14-kDa wild-type (wt) E4-ORF1 or 16-kDa His-tagged human dUTPase (His-dUTPase) were prepared in NETN buffer containing NP-40 detergent and analysed by size-exclusion chromatography (SEC) with elution buffer lacking detergent. Under these mild conditions, His-dUTPase eluted at 45-kDa, similar to the predicted 48-kDa mass of a trimer, whereas E4-ORF1 eluted as two peaks, one at 38-kDa and a more prominent one at 13-kDa (Figure 1a), comparable to the predicted 42- and 14-kDa masses of a trimer and monomer, respectively. This finding suggests that E4-ORF1 exists as a trimer and monomer in cells.
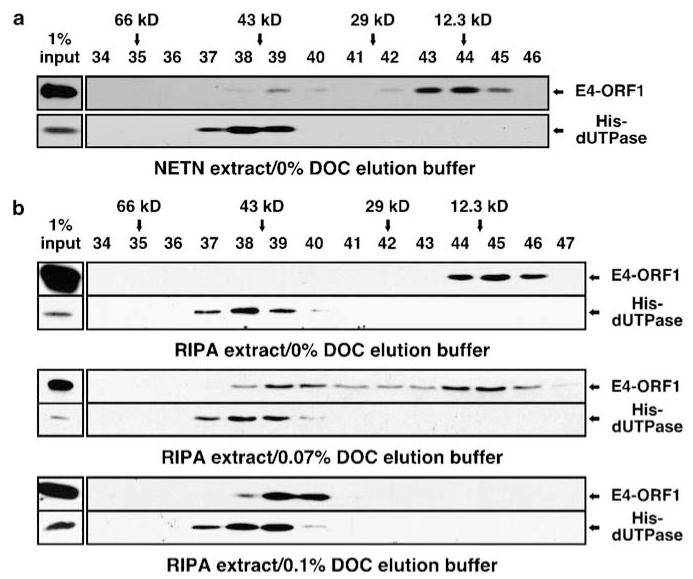
E4-ORF1 exists as a monomer and trimer. (a) E4-ORF1 elutes as a monomer and trimer by SEC. 293 T cells were transfected with pGW1 encoding wt E4-ORF1 or His-dUTPase. Extracts in NETN buffer were fractionated by SEC with elution buffer lacking detergent and blotted with E4-ORF1 or His antibody. Fractions lacking E4-ORF1 or dUTPase signal are not shown. Arrows mark elution peaks of size markers. (b) Stringent detergents disrupt E4-ORF1 trimers and DOC reverses this effect. Extracts in RIPA buffer were fractionated by SEC with elution buffer lacking or containing DOC and analyzed as described in (a).
In SEC analyses otherwise identical to those described above, we discovered that preparation of cell extracts in stringent RIPA buffer containing detergents SDS, deoxycholate (DOC), and NP-40 rather than in mild NETN buffer eliminates the larger putative E4-ORF1 trimer peak but not the smaller monomer peak (Figure 1b, top panel). Conversely, the smaller but not larger peak was eliminated either completely by using RIPA buffer as the SEC elution buffer or in a dose-dependent manner by supplementing the SEC elution buffer with DOC but not SDS, NP-40, or Triton X-100 (Figure 1b, middle and bottom panels) (unpublished data). The fact that purified E4-ORF1 produced only the larger peak with DOC present in the SEC elution buffer (Supplementary Figure S2) verifies formation of bona fide homo-trimers and supports the hypothesis that E4-ORF1 and dUTPase have related protein structures. By contrast, His-dUTPase eluted as a trimer regardless of extract buffer stringency or DOC inclusion in the SEC elution buffer (Figure 1). These observations suggest that cell extract preparation under stringent conditions displaces a small, unknown cellular factor required to stabilize E4-ORF1 trimers and that DOC complements this loss when maintained at low concentrations during SEC.
A common C-terminal element stabilizes trimer formation by dUTPase and E4-ORF1
The crystal structure of human dUTPase predicts that NH2-terminal β-strand 1 (β1) from one subunit and C-terminal β-strand 8 (β8) from an adjacent subunit mediate intermolecular interactions that stabilize trimer formation (Mol et al., 1996). Supporting this prediction and the hypothesis that E4-ORF1 and dUTPase adopt a related protein fold, results from SEC analyses with deletion mutants of dUTPase (ΔC16 and ΔC20) or E4-ORF1 (ΔC5 and ΔC7) demonstrated a requirement for β8-residues in trimer formation by both proteins (Figure 2a, Supplementary Figures S3 and S4).
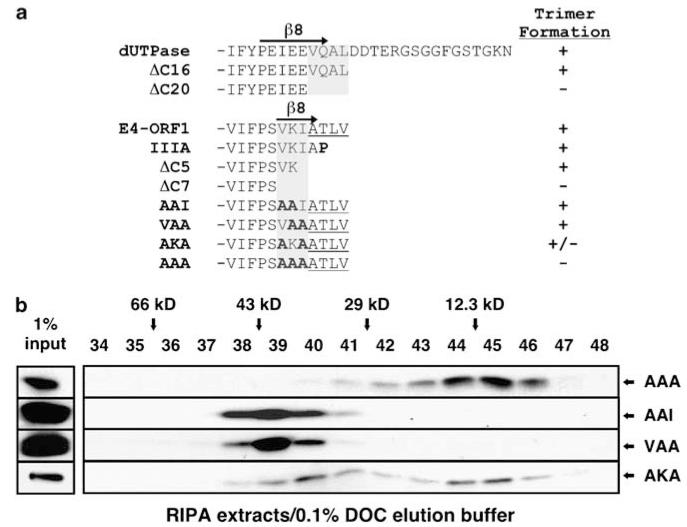
A C-terminal element determines dUTPase and E4-ORF1 trimerization. (a) C-terminal mutants of human dUTPase and E4-ORF1. Gray shaded regions indicate residues needed for trimer formation. Substitution mutations are indicated in bold. E4-ORF1 PBM residues are underlined. (b) β8 mutations abolish E4-ORF1 trimer formation. 293 T cells were transfected with pGW1 encoding the indicated mutant E4-ORF1. Extracts in RIPA buffer were fractionated by SEC with elution buffer containing 0.1% DOC and blotted with E4-ORF1 antibody.
Identical experiments were conducted with four E4-ORF1 mutants having double (AAI, VAA and AKA) or triple (AAA) alanine substitutions in β8-residues VKI (Figure 2a). In SEC analyses, AAI and VAA eluted as trimers, whereas AKA eluted as a mixture of trimers and monomers, reflecting a partial defect in trimer formation (Figure 2b). By contrast, AAA eluted primarily as a monomer and, unlike VAA, failed to self-associate in a co-IP assay (Figure 2b, Supplementary Figure S4b). These results define β8-residues VKI as a crucial E4-ORF1 trimerization (TRI) element.
E4-ORF1 binds cooperatively to PDZ domains in Dlg1 but not MAGI-1 and MUPP1
The existence of E4-ORF1 monomers and trimers in cells might indicate that each form has distinct functions. In this regard, oligomeric ligands that target multi-PDZ domain proteins often bind cooperatively to two closely situated PDZ domains, termed a PDZ tandem (Grootjans et al., 2000; Long et al., 2003). As the E4-ORF1 trimer possesses oligomeric PBM ligands, this feature might uniquely permit a cooperative interaction with the Dlg1 PDZ1 + 2 tandem, which mediates binding to E4-ORF1 (Frese et al., 2006). Supporting this idea, E4-ORF1 co-IP’d wt Dlg1 but not Dlg1 mutants having PDZ1 and PDZ2 dually or individually inactivated (Supplementary Figure S5a).
Unlike the interaction with Dlg1, E4-ORF1 does not target a PDZ tandem in other PDZ proteins but rather binds either one PDZ domain in ZO-2 (PDZ1) and PATJ (PDZ8) or two non-tandem PDZ domains in MUPP1 (PDZ7 and PDZ10) and MAGI-1 (PDZ1 and PDZ3) (Glaunsinger et al., 2000, 2001; Lee et al., 2000; Latorre et al., 2005). While E4-ORF1 would not be expected to bind cooperatively to one PDZ domain, such an interaction is feasible with two non-tandem PDZ domains. Nonetheless, in co-IP assays, wt E4-ORF1 bound HA-tagged wt MAGI-1 but not MAGI-1 having non-tandem PDZ1 and PDZ3 dually deleted (PDZΔ1Δ3), yet retained binding to MAGI-1 having PDZ1 or PDZ3 individually deleted (PDZΔ1 and PDZΔ3), albeit at reduced levels compared to wt MAGI-1 (Supplementary Figure S5b) (Glaunsinger et al., 2000). The amount of E4-ORF1 binding to wt MAGI-1 equaled the sum of binding to PDZΔ1 and PDZΔ3 (Supplementary Figure S5b), revealing non-cooperative binding to these non-tandem PDZ domains. A similar conclusion was drawn for E4-ORF1 binding to non-tandem PDZ domains in MUPP1 (Lee et al., 2000).
These observations led to the hypothesis that E4-ORF1 monomers mediate non-cooperative interactions with individual PDZ domains in MUPP1, MAGI-1, ZO-2 and PATJ whereas E4-ORF1 trimers mediate cooperative interactions with the PDZ tandem in Dlg1.
The E4-ORF1 monomer binds and sequesters MUPP1, MAGI-1 and ZO-2
Our hypothesis predicted that monomeric AAA, as well as partially TRI-defective AKA and TRI-competent AAI and VAA, all of which retain an intact PBM element (see Figure 2a), should display wt binding to MUPP1, PATJ, MAGI-1 and ZO-2. In co-IP assays with epitope-tagged PDZ proteins, these predictions were affirmed for MUPP1 and MAGI-1 (Figure 3a, Supplementary Figure S6), as well as PATJ (unpublished data). All four mutants also exhibited wt proficiency for sequestering endogenous MUPP1 and MAGI-1 within insoluble complexes in CREF cells (Figure 3b), which do not express PATJ (Latorre et al., 2005). By contrast, neither wt nor mutant E4-ORF1 proteins sequestered endogenous Dlg1 (Figure 3b), consistent with previous findings (Frese et al., 2006). Based on results with AAA, we conclude that the E4-ORF1 monomer is sufficient to bind and sequester MUPP1 and MAGI-1 in cells. As will be discussed in a subsequent section, a similar conclusion was drawn for ZO-2, despite its reduced binding to TRI element mutants.
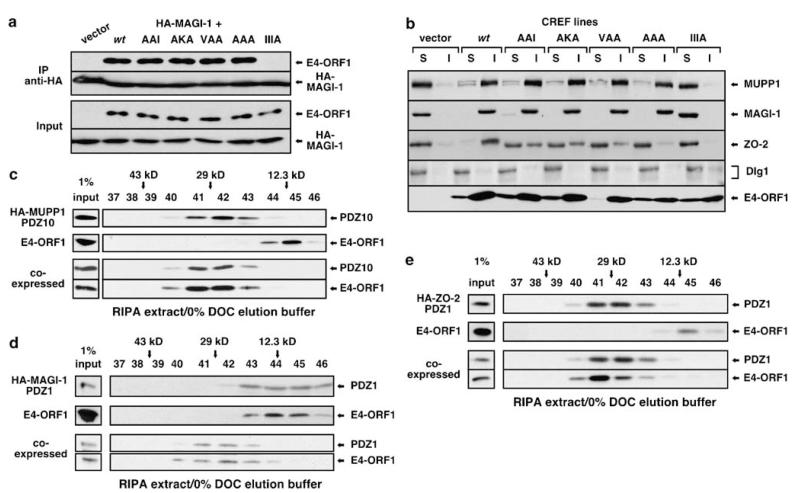
E4-ORF1 monomers bind and sequester MUPP1, MAGI-1 and ZO-2. (a) Monomeric AAA binds MAGI-1. 293 T cells were co-transfected with pGW1 encoding HA-MAGI-1 and either pGW1 (vector) or pGW1 encoding wt or the indicated mutant E4-ORF1. Extracts in RIPA buffer were IP’d with HA antibody. Recovered proteins and total cell extracts (Input) were blotted with HA or E4-ORF1 antibody. E4-ORF1 PBM mutant IIIA (see Figure 2a) is a negative control. (b) Monomeric AAA sequesters endogenous MUPP1 and MAGI-1. CREF cells were transduced with pBABE (vector) or pBABE encoding wt or the indicated mutant E4-ORF1. Extracts in RIPA buffer were separated into soluble supernatant (S) and insoluble pellet (I) fractions. Equivalent amounts of each fraction were blotted with MUPP1, MAGI-1, ZO-2, SAP97 (Dlg1) or E4-ORF1 antibody. (c–e) E4-ORF1 monomers bind PDZ domains of MUPP1, MAGI-1 and ZO-2. 293 T cells were transfected with pGW1 encoding wt E4-ORF1 and/or pGW1 encoding (c) HA-MUPP1 PDZ10, (d) HA-MAGI-1 PDZ1, or (e) HA-ZO-2 PDZ1. Extracts in RIPA buffer were fractionated by SEC with elution buffer lacking DOC and blotted with E4-ORF1 or HA antibody.
We also directly tested whether an E4-ORF1 monomer binds the PDZ domains of MUPP1, MAGI-1 and ZO-2. Extracts of 293 T cells expressing HA-tagged MUPP1 PDZ10 (12-kDa), MAGI-1 PDZ1 (14-kDa), or ZO-2 PDZ1 (13-kDa) alone or in combination with 14 kDa wt E4-ORF1 were prepared in RIPA buffer and analysed by SEC with elution buffer lacking DOC, conditions that convert E4-ORF1 into monomers. When expressed alone, E4-ORF1 and MAGI-1 PDZ1 eluted as 12-kDa monomers, whereas MUPP1 PDZ10 and ZO-2 PDZ1 eluted as 26-kDa dimers (Figures 3c–e). When co-expressed, E4-ORF1 and each PDZ domain co-eluted at approximately 27-kDa (Figures 3c–e), similar to the predicted 26–28-kDa masses for heterocomplexes composed of one E4-ORF1 molecule bound to one PDZ domain molecule. Elution sizes of MUPP1 PDZ10 and ZO-2 PDZ1 dimers seemed unchanged by heterocomplex formation as the incorporated E4-ORF1 monomer displaced a PDZ domain having a similar mass. By contrast, identical experiments conducted with SEC elution buffer containing 0.1% DOC, which converts E4-ORF1 into its trimeric form, showed that the E4-ORF1 trimer neither co-elutes with nor changes the elution size of the same PDZ domains (Supplementary Figure S7). Thus, the E4-ORF1 monomer specifically binds the PDZ domains of MUPP1, MAGI-1 and ZO-2.
Two residues in the E4-ORF1 TRI element also influence PBM-mediated binding to ZO-2 and Dlg1
As only the E4-ORF1 monomer binds ZO-2 PDZ1 (see Figure 3e), we predicted that all four TRI element mutants would bind proficiently to ZO-2. Compared to wt E4-ORF1, however, AAI and AKA or VAA and AAA displayed either modest or moderate binding defects, respectively, to HA-tagged ZO-2 (Figure 4a). Our hypothesis additionally predicted that TRI-competent AAI and VAA would exhibit wt binding to Dlg1 whereas partially TRI-defective AKA or monomeric AAA would show a reduced ability or fail to bind Dlg1, respectively. While AKA and AAA yielded anticipated results, AAI or VAA unexpectedly displayed a reduced capacity or failed to interact with HA-tagged Dlg1, respectively (Figure 4b). In CREF cells, TRI element mutants showed corresponding defects in sequestering endogenous ZO-2 within insoluble complexes (Figure 3b) and in binding endogenous Dlg1 (Supplementary Figure S8). In agreement with the fact that the oncogenic potential of E4-ORF1 depends on binding to ZO-2 and Dlg1 (Glaunsinger et al., 2001; Frese et al., 2006), AAI and AKA or VAA and AAA also showed a reduced ability or failed, respectively, to promote phosphorylation of the PI3K-effector protein kinase B (PKB) and focus formation (Supplementary Figure S9).
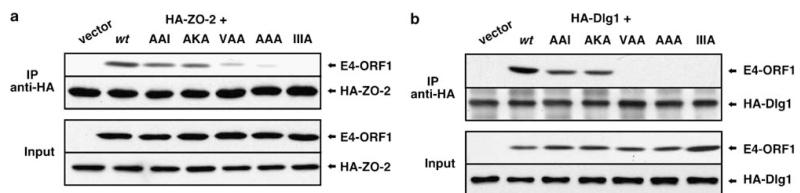
Residues KI in the TRI element additionally control E4-ORFl binding to ZO-2 and Dlg1. (a) E4-ORF1 TRI element mutants have trimerization (TRI)-independent defects in binding ZO-2. 293 cells were co-transfected with pGW1 encoding HA-ZO-2 and either pGW1 (vector) or pGW1 encoding wt or the indicated mutant E4-ORF1. Extracts in RIPA buffer were IP’d with HA antibody. Recovered proteins and total cell extracts (Input) were blotted with HA or E4-ORF1 antibody. (b) E4-ORF1 TRI element mutants have TRI-independent defects in binding Dlg1. Identical experiments as those described in (a) were conducted, except pGW1 encoding HA-Dlg1 was used.
We drew three conclusions from these unexpected results. (1) The observation that TRI-competent VAA exhibited reduced binding to ZO-2 and failed to bind Dlg1 indicates that the mutated KI residues function not only in E4-ORF1 trimer formation but also in enhancing or determining the ability of the adjacent PBM (residues ATLV) (see Figure 2a) to mediate binding to ZO-2 or Dlg1, respectively. Thus, the KI residues dually control both TRI and PBM element activity, redefining the E4-ORF1 PBM element as the expanded six-residue sequence KIATLV. (2) Because KI residues enhance E4-ORF1 binding to ZO-2, this interaction was reduced for TRI-competent VAA and monomeric AAA; however, the fact that these mutants showed comparable weak binding to ZO-2 supports the conclusion that an E4-ORF1 monomer is sufficient to bind this protein. (3) Because KI residues are required for E4-ORF1 to interact with Dlg1, this protein failed to bind both TRI-competent VAA and monomeric AAA, confounding our initial attempt to show that an E4-ORF1 trimer specifically mediates this interaction.
The E4-ORF1 trimer binds Dlg1 and promotes its translocation to the plasma membrane
To evaluate the hypothesized requirement for the E4-ORF1 trimer in Dlg1 binding, we needed to circumvent the problem of the KI residues dually controlling activities of the TRI and PBM elements. We reasoned that duplication of this pair of residues in wt E4-ORF1, creating insertion mutant +KI (Figure 5a), would separate the TRI and PBM elements by providing each with its own set of KI residues. Based on this premise, we compared TRI and Dlg1 binding by mutant +KI and related mutants additionally having substitution mutations in the TRI element (AAI + KI, VAA + KI, AAA + KI) or PBM element (+AA) (Figure 5a). As expected, all mutants except AAA+KI formed trimers (Figure 5a, Supplementary Figure S10) (unpublished data). In co-IP assays with CREF cells, TRI-competent +KI, AAI+KI and VAA+KI bound endogenous Dlg1 with wt proficiency, whereas monomeric AAA+KI, as well as TRI-competent PBM mutant +AA, instead behaved identically to negative-control PBM mutant IIIA (Figure 5b). Unlike Dlg1, ZO-2 failed to bind parental mutant +KI (unpublished data), hinting that E4-ORF1 residues upstream of the expanded PBM also control this interaction. More important, the Dlg1 binding defect of AAA+KI was specific and caused by a failure to oligomerize because AAA+KI sequestered endogenous MAGI-1 in CREF cells (unpublished data) and fusion of AAA+KI but not PBM mutant IIIA to dimeric GST restored Dlg1 binding to wt levels in a GST pulldown assay (Supplementary Figure S11). In 293 T cells, wt E4-ORF1 and TRI-competent +KI and VAA+KI also promoted GFP-Dlg1 accumulation at the plasma membrane, whereas monomeric AAA+KI lacked this activity, identical to negative-control empty plasmid and PBM mutant IIIA (Supplementary Figure S12). Besides confirming that each set of KI residues in mutant +KI contributes separately to TRI or PBM element activity, these findings importantly demonstrate that the E4-ORF1 trimer but not monomer binds Dlg1 and promotes its translocation to the plasma membrane.
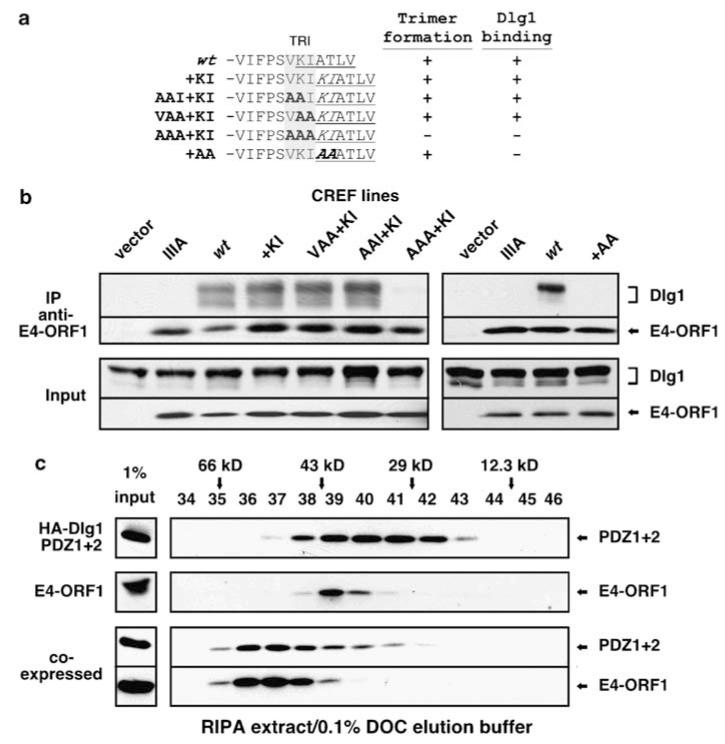
E4-ORF1 trimers bind Dlg1. (a) C-terminal sequence of E4-ORF1 mutants and their capacities to trimerize and bind Dlg1. Results summarized from Figure 5b, Supplementary Figure S10, or unpublished data. The TRI or expanded PBM element are gray shaded or underlined, respectively. Inserted residues are italicized. Alanine substitutions are indicated in bold. (b) Monomeric AAA + KI fails to bind endogenous Dlg1. CREF cells were transduced with pBABE (vector) or pBABE encoding wt or the indicated mutant E4-ORF1. Extracts in RIPA buffer were IP’d with E4-ORF1 antibody. Recovered proteins were blotted with Dlg1 or E4-ORF1 antibody. Left and right panels represent two independent experiments. (c) E4-ORF1 trimers bind the Dlg1 PDZ1 + 2 tandem. 293 T cells were transfected with pGW1 encoding wt E4-ORF1 or HA-Dlg1 PDZ1 + 2 alone or in combination. Extracts in RIPA buffer were fractionated by SEC with elution buffer containing 0.1% DOC and blotted with E4-ORF1 or HA antibody.
To obtain direct evidence that an E4-ORF1 trimer binds to the Dlg1 PDZ tandem, we expressed 22-kDa HA-tagged Dlg1 PDZ1+2 alone or in combination with 14-kDa wt E4-ORF1 in 293 T cells and analysed cell extracts prepared in RIPA buffer by SEC with 0.1% DOC elution buffer. When expressed alone, E4-ORF1 eluted as a 39-kDa trimer, whereas Dlg1 PDZ1+2 eluted as broad peak having a size range consistent with a mixture of 44-kDa dimers and 22-kDa monomers (Figure 5c). When co-expressed, both proteins co-eluted at 60-kDa, similar to the predicted 64-kDa mass for a heterocomplex composed of an E4-ORF1 trimer bound to one Dlg1 PDZ1+2 molecule (Figure 5c). Thus, the E4-ORF1 trimer uniquely binds to and disrupts dimers of the Dlg1 PDZ tandem. This finding, coupled with the inability of wt E4-ORF1 to bind Dlg1 having PDZ1 or PDZ2 inactivated (Supplementary Figure S5a) and conversely of monomeric AAA+KI to bind wt Dlg1 (Figure 5b), imply that two of three PBM in an E4-ORF1 trimer mediate a cooperative interaction with the two PDZ domains in a Dlg1 PDZ tandem. Results from identical SEC analyses conducted with elution buffer lacking DOC supported this conclusion (Supplementary Figure S13).
Discussion
The tumorigenic potential of E4-ORF1 depends on a PBM that mediates binding to five cellular PDZ proteins. Depending on the PDZ protein, these interactions result in two different functional consequences, yet the basis for this divergence has not been determined. Here, we presented evidence indicating that E4-ORF1 and trimeric dUTPase share a similar protein fold and that E4-ORF1 exists not only as a trimer but also a monomer in cells (Figures 1 and and2,2, Supplementary Figure S1). This information, along with data suggesting that E4-ORF1 binds non-cooperatively to MUPP1 and MAGI-1 yet cooperatively to Dlg1 (Supplementary Figure S5) (Lee et al., 2000), led to the discovery that monomers specifically bind and sequester MUPP1, MAGI-1 and ZO-2 (Figures 3 and and4a,4a, Supplementary Figure S6) whereas trimers specifically bind and promote Dlg1 to translocate to the plasma membrane (Figure 5, Supplementary Figures S8 and S12). Considering that E4-ORF1 interacts with Dlg1 to trigger PI3K activation (Frese et al., 2006) or with PATJ and ZO-2 to disrupt the TJ and cell polarity (Glaunsinger et al., 2001; Latorre et al., 2005), we conclude that E4-ORF1 monomers and trimers perform distinct functions in cells.
One question arising from this work is: How do E4-ORF1 monomers and trimers target different cellular PDZ proteins? While the mechanism for this selectivity is not known, the extended C-terminal arm carrying the PBM is free and more exposed in an E4-ORF1 monomer (Supplementary Figure S1) than in a trimer (Chung et al., 2007), where the arm engages in TRI element-dependent interactions with adjacent subunits. We speculate that monomers specifically interact with MUPP1, PATJ, MAGI-1 and ZO-2 because unidentified critical E4-ORF1 residues only accessible in the free C-terminal arm of a monomer selectively stabilize binding to individual PDZ domains in these proteins but not Dlg1. Conversely, perhaps trimers specifically interact with Dlg1 because oligomeric PBM elements synergistically stabilize otherwise weak monomer binding to individual PDZ domains of the Dlg1 PDZ tandem whereas an inaccessibility of postulated critical E4-ORF1 residues in the trimer precludes binding to MUPP1, PATJ, MAGI-1 and ZO-2.
We found that DOC specifically induces TRI of E4-ORF1 (Figure 1b) and possibly dimerization of the Dlg1 PDZ tandem (Figure 5c). DOC is a bile acid produced in the liver by oxidation of cholesterol, an important lipid of animal cells found at the plasma membrane and internal membranes, including membrane vesicles (Raffy and Teissie, 1999). Interestingly, cholesterol and DOC also induce homo-oligomerization of pore-forming bacterial toxins at host cell membranes (Bhakdi et al., 1981; Forti and Menestrina, 1989; Ramachandran et al., 2004). Perhaps under normal physiological conditions, cholesterol likewise promotes E4-ORF1 trimer formation and thereby interactions with Dlg1 at the plasma membrane (Frese et al., 2006).
E4-ORF1 represents one of four known proteins having subunits and oligomers that perform separate functions. For instance, β-actin polymers that form actin filaments are fundamental components of the cytoskeleton, whereas β-actin monomers instead augment the activity of the BAF mammalian SWI/SNF chromatin remodeling complex (Zhao et al., 1998) or inhibit the MAL coactivator of the SRF pathway (Miralles et al., 2003). Furthermore, monomers or dimers of the Pit-1 transcription factor regulate expression from either the prolactin gene or the growth hormone gene, respectively (Holloway et al., 1995), and dimers but not monomers of the bovine papillomavirus E5 interact with the platelet-derived growth factor β receptor (DiMaio and Mattoon, 2001). Many growth factor receptors (for example, epithelial growth factor receptor (EGFR)) and transcription factors (for example, nuclear factor-kappa B (NF-κB)) are not mentioned here, as such proteins function solely as homo-dimers and hetero-dimers formed with closely related proteins. Thus, E4-ORF1 employs a rare strategy to increase protein complexity.
In conclusion, we showed that the combined actions of the overlapping TRI and PBM elements uniquely permit E4-ORF1 to produce two oligomeric forms that not only bind separate targets containing the same conserved modular domain but also promote different functional consequences. This novel, economical strategy for increasing protein complexity makes perfect sense for viruses, which have limited coding capacities, and also may have broader implications by exposing a new additional mechanism, besides conventional aminoacid sequence determinants, to permit specific binding to and differential functional regulation of modular signaling domains. As more proteins are examined for this property, this strategy may prove more common than presently recognized.
Materials and methods
Plasmids
E4-ORF1 and human dUTPase plasmids were constructed as previously described (Chung et al., 2007). The HA-MUPP1 PDZ10 (aa 1606–1706), HA-MAGI-1 PDZ1 (aa 431–545), or HA-ZO-2 PDZ1 (aa 1–113) cDNA was inserted into pGW1. pGW1 encoding HA-ZO-2, HA-MAGI-1, HA-Dlg1 or HA-Dlg1 PDZ1 2 were described (Glaunsinger et al., 2000, 2001; Frese et al., 2006).
Antibodies
E4-ORF1, MUPP1, MAGI-1, ZO-2 and SAP97/Dlg1 antisera were described (Lee et al., 1997; Weiss et al., 1997; Glaunsinger et al., 2000, 2001; Lee et al., 2000). Antibodies to Dlg1 (BD Transduction Laboratories, San Jose, CA, USA), HA (Covance Research Products, Emeryville, CA, USA), and His (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were purchased, as were HRP-conjugated goat anti-rabbit IgG and goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, AL, USA).
Cells and extracts
Transfections were performed with TransIT LT1 (Mirus Bio Corporation, Madison, WI, USA). MLV vectors (Soneoka et al., 1995) were produced by co-transfecting 293T cells with pHIT60, pME-VSVG (gift from K Maruyama, DNAX Research Institute), and pBABE-puro encoding E4-ORF1 genes. Cell extracts were prepared in RIPA buffer (50 mm Tris–HCl, pH 8.0, 0.15 m NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) or NETN buffer (20 mm Tris–HCl, pH 8.0, 0.1 m NaCl, 1 mm EDTA, 0.5% NP-40) containing protease and phosphatase inhibitors. Separation of extracts into soluble supernatant and insoluble pellet fractions was done as described (Latorre et al., 2005).
Immunoprecipitations and immunoblots
These assays were conducted as described (Frese et al., 2006).
SEC chromatography
Cell extracts mixed with size markers (200-kDa β-amylase, 66-kDa bovine serum albumin, 43-kDa ovalbumin, 29-kDa carbonic anhydrase and 12.4-kDa cytochrome c (Sigma-Aldrich)) were loaded on a Superose 6 10/300 size-exclusion column (Amersham Biosciences, Piscataway, NJ, USA) pre-equilibrated with elution buffer (50 mm Tris–HCl, pH 8.0, 0.15m NaCl, 10% glycerol) lacking or containing DOC. Samples were run through the column with elution buffer using a Pharmacia LKB Biotechnology FPLC system. Collected fractions (0.3 ml) were resolved by SDS–PAGE. Size markers were visualized by staining with Coomassie brilliant blue.
Acknowledgements
We thank Andy Rice and Chris Herrmann for use of their FPLC system and Richard Sutton for retroviral vector plasmids. RSW and KKF were supported by predoctoral fellowships from the US. Army (Breast Cancer Training Grant DAMD17-94 J4204) and the National Cancer Institute (Viral Oncology Training Grant T32 CA09197). This research was funded by Public Health Service Grants CA58541 (RTJ) and AI36040 (BVVP).
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- Bhakdi S, Fussle R, Tranum-Jensen J. Staphylococcal alpha-toxin: oligomerization of hydrophilic monomers to form amphiphilic hexamers induced through contact with deoxycholate detergent micelles. Proc Natl Acad Sci USA. 1981;78:5475–5479. [Europe PMC free article] [Abstract] [Google Scholar]
- Chung SH, Frese KK, Weiss RS, Prasad BV, Javier RT. A new crucial protein-interaction element that targets the adenovirus E4-ORF1 oncoprotein to membrane vesicles. J Virol. 2007;81:4787–4797. [Europe PMC free article] [Abstract] [Google Scholar]
- DiMaio D, Mattoon D. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene. 2001;20:7866–7873. [Abstract] [Google Scholar]
- Forti S, Menestrina G. Staphylococcal alpha-toxin increases the permeability of lipid vesicles by cholesterol- and pH-dependent assembly of oligomeric channels. Eur J Biochem. 1989;181:767–773. [Abstract] [Google Scholar]
- Frese KK, Latorre IJ, Chung SH, Caruana G, Bernstein A, Jones SN, et al. Oncogenic function for the Dlg1 mammalian homolog of the Drosophila discs-large tumor suppressor. EMBO J. 2006;25:1406–1417. [Europe PMC free article] [Abstract] [Google Scholar]
- Glaunsinger BA, Lee SS, Thomas M, Banks L, Javier R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene. 2000;19:5270–5280. [Europe PMC free article] [Abstract] [Google Scholar]
- Glaunsinger BA, Weiss RS, Lee SS, Javier R. Link of the unique oncogenic properties of adenovirus type 9 E4-ORF1 to a select interaction with the candidate tumor suppressor protein ZO-2. EMBO J. 2001;20:5578–5586. [Europe PMC free article] [Abstract] [Google Scholar]
- Grootjans JJ, Reekmans G, Ceulemans H, David G. Syntenin–syndecan binding requires syndecan–synteny and the co-operation of both PDZ domains of syntenin. J Biol Chem. 2000;275:19933–19941. [Abstract] [Google Scholar]
- Hirata A, Higuchi M, Niinuma A, Ohashi M, Fukushi M, Oie M, et al. PDZ domain-binding motif of human T-cell leukemia virus type 1 Tax oncoprotein augments the transforming activity in a rat fibroblast cell line. Virology. 2004;318:327–336. [Abstract] [Google Scholar]
- Holloway JM, Szeto DP, Scully KM, Glass CK, Rosenfeld MG. Pit-1 binding to specific DNA sites as a monomer or dimer determines gene-specific use of a tyrosine-dependent synergy domain. Genes Dev. 1995;9:1992–2006. [Abstract] [Google Scholar]
- Humbert PO, Dow LE, Russell SM. The Scribble and Par complexes in polarity and migration: friends or foes? Trends Cell Biol. 2006;16:622–630. [Abstract] [Google Scholar]
- Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. [Europe PMC free article] [Abstract] [Google Scholar]
- Latorre IJ, Roh MH, Frese KK, Weiss RS, Margolis B, Javier RT. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J Cell Sci. 2005;118:4283–4293. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee SS, Glaunsinger B, Mantovani F, Banks L, Javier RT. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J Virol. 2000;74:9680–9693. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee SS, Weiss RS, Javier RT. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. [Europe PMC free article] [Abstract] [Google Scholar]
- Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, et al. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol. 2003;327:203–214. [Abstract] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. [Abstract] [Google Scholar]
- Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. [Abstract] [Google Scholar]
- Raffy S, Teissie J. Control of lipid membrane stability by cholesterol content. Biophys J. 1999;76:2072–2080. [Europe PMC free article] [Abstract] [Google Scholar]
- Ramachandran R, Tweten RK, Johnson AE. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit beta-strand alignment. Nat Struct Mol Biol. 2004;11:697–705. [Abstract] [Google Scholar]
- Shin K, Straight S, Margolis B. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J Cell Biol. 2005;168:705–711. [Europe PMC free article] [Abstract] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. [Europe PMC free article] [Abstract] [Google Scholar]
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, et al. ZO-1 and ZO-2 independently determine where Claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. [Abstract] [Google Scholar]
- Weiss RS, Lee SS, Prasad BV, Javier RT. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, et al. Rapid and phosphoinositol-dependent binding of the SWI/ SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1038/sj.onc.1210784
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3471668?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
E4orf1: The triple agent of adenovirus - Unraveling its roles in oncogenesis, infectious obesity and immune responses in virus replication and vector therapy.
Tumour Virus Res, 17:200277, 28 Feb 2024
Cited by: 0 articles | PMID: 38428735
Review
A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer.
Cancers (Basel), 13(17):4264, 24 Aug 2021
Cited by: 7 articles | PMID: 34503076 | PMCID: PMC8428372
Review Free full text in Europe PMC
Upsetting the Balance: When Viruses Manipulate Cell Polarity Control.
J Mol Biol, 430(19):3481-3503, 20 Apr 2018
Cited by: 12 articles | PMID: 29680664 | PMCID: PMC7094317
Review Free full text in Europe PMC
Adenovirus E4-ORF1 Dysregulates Epidermal Growth Factor and Insulin/Insulin-Like Growth Factor Receptors To Mediate Constitutive Myc Expression.
J Virol, 89(21):10774-10785, 12 Aug 2015
Cited by: 10 articles | PMID: 26269183 | PMCID: PMC4621122
Hijacking Dlg1 for oncogenic phosphatidylinositol 3-kinase activation in human epithelial cells is a conserved mechanism of human adenovirus E4-ORF1 proteins.
J Virol, 88(24):14268-14277, 24 Sep 2014
Cited by: 12 articles | PMID: 25253337 | PMCID: PMC4249130
Go to all (11) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A new crucial protein interaction element that targets the adenovirus E4-ORF1 oncoprotein to membrane vesicles.
J Virol, 81(9):4787-4797, 21 Feb 2007
Cited by: 28 articles | PMID: 17314165 | PMCID: PMC1900153
Link of the unique oncogenic properties of adenovirus type 9 E4-ORF1 to a select interaction with the candidate tumor suppressor protein ZO-2.
EMBO J, 20(20):5578-5586, 01 Oct 2001
Cited by: 57 articles | PMID: 11598001 | PMCID: PMC125668
Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes.
J Virol, 71(3):1857-1870, 01 Mar 1997
Cited by: 48 articles | PMID: 9032316 | PMCID: PMC191256
Cell polarity proteins: common targets for tumorigenic human viruses.
Oncogene, 27(55):7031-7046, 01 Nov 2008
Cited by: 69 articles | PMID: 19029943 | PMCID: PMC3501650
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: T32 CA009197
Grant ID: CA58541
Grant ID: T32 CA09197
Grant ID: R01 CA058541
NIAID NIH HHS (3)
Grant ID: AI36040
Grant ID: R01 AI036040
Grant ID: R37 AI036040




