Abstract
Free full text

Intraflagellar Transport and Functional Analysis of Genes Required for Flagellum Formation in Trypanosomes
Abstract
Intraflagellar transport (IFT) is the bidirectional movement of protein complexes required for cilia and flagella formation. We investigated IFT by analyzing nine conventional IFT genes and five novel putative IFT genes (PIFT) in Trypanosoma brucei that maintain its existing flagellum while assembling a new flagellum. Immunostaining against IFT172 or expression of tagged IFT20 or green fluorescent protein GFP::IFT52 revealed the presence of IFT proteins along the axoneme and at the basal body and probasal body regions of both old and new flagella. IFT particles were detected by electron microscopy and exhibited a strict localization to axonemal microtubules 3–4 and 7–8, suggesting the existence of specific IFT tracks. Rapid (>3 μm/s) bidirectional intraflagellar movement of GFP::IFT52 was observed in old and new flagella. RNA interference silencing demonstrated that all individual IFT and PIFT genes are essential for new flagellum construction but the old flagellum remained present. Inhibition of IFTB proteins completely blocked axoneme construction. Absence of IFTA proteins (IFT122 and IFT140) led to formation of short flagella filled with IFT172, indicative of defects in retrograde transport. Two PIFT proteins turned out to be required for retrograde transport and three for anterograde transport. Finally, flagellum membrane elongation continues despite the absence of axonemal microtubules in all IFT/PIFT mutant.
INTRODUCTION
Intraflagellar transport (IFT) is the bidirectional movement of protein particles in the matrix of cilia and flagella, independent of flagellum beating (Kozminski et al., 1993). IFT is driven by two microtubule-associated motor complexes: the heterotrimeric kinesin II that moves particles toward the tip (Kozminski et al., 1995; Cole et al., 1998; Snow et al., 2004) and a dynein complex that displaces them from the tip to the base of the organelle (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999). IFT particles have been purified by differential centrifugation from the matrix of isolated flagella from the green algae Chlamydomonas. They are composed of at least 17 proteins that can be separated into a complex A, made of six proteins, and a complex B that contains 11 proteins (Piperno and Mead, 1997; Cole et al., 1998). These IFT proteins are rich in protein–protein interaction domains of the tryptophan-aspartic acid (WD)-40, tetratricopeptide repeat protein (TPR), or coil-coiled families (Cole, 2003). Several of these proteins have been localized along the length of growing and mature cilia or flagella, but a significant proportion is also found around the basal body area (Cole et al., 1998; Deane et al., 2001; Pedersen et al., 2005).
IFT is required for the transport of flagellar precursors to the distal tip of the flagellum, the site of construction of the organelle (Qin et al., 2004). Consequently, inhibition of IFT prevents flagellum assembly in most organisms studied so far (Kozminski et al., 1995; Nonaka et al., 1998; Brown et al., 2003; Kohl et al., 2003). The only exceptions are encountered in cell types that assemble their flagella in the cytoplasm, such as male gametes of Drosophila (Han et al., 2003; Sarpal et al., 2003) and of Plasmodium (Avidor-Reiss et al., 2004; Briggs et al., 2004a).
Analysis of Chlamydomonas and Caenorhabditis elegans IFT mutants indicates that complex B is associated to anterograde transport, whereas complex A would be linked to retrograde transport (Cole, 2003). Genetic, biochemical, and morphological analysis from such mutants contributed to the understanding of the mode of action of IFT (Pedersen et al., 2006; Ou et al., 2007). Particles are assembled at the base of the flagellum, and they are moved toward the tip by kinesin II directly associated to complex B proteins (anterograde movement). Proteins from complex A, the inactive dynein motor complex, and axonemal proteins are cotransported and unloaded at the distal end. Dynein becomes the active motor and associates to proteins of the A complex and travels back to the base of the flagellum where the cycle can reinitiate. Relatively few data are available about IFT regulation and the specific function of individual IFT proteins (Hou et al., 2007). IFT is involved in both construction and maintenance of cilia and flagella, but the differences between these two situations in the life of an organelle are poorly understood (Kozminski et al., 1995).
Trypanosoma brucei is a flagellated protist responsible for sleeping sickness in central Africa. It is a fascinating organism to study because it exhibits a number of original features, including at the flagellar level (Kohl and Bastin, 2005). It possesses a single motile flagellum whose axoneme is assembled on a basal body flanked by an immature probasal body (Sherwin and Gull, 1989). The axoneme contains a central pair and peripheral doublets carry outer and inner dynein arms and radial spokes. Molecular components of these structures are conserved and functional analysis revealed their role in flagellum beating (Branche et al., 2006; Broadhead et al., 2006; Ralston et al., 2006; Baron et al., 2007). An extra-axonemal structure called the paraflagellar rod (PFR) is present alongside the axoneme from its point of emergence from the flagellar pocket until its distal tip (Bastin et al., 1996). It is tightly linked to the axoneme via physical connections to microtubule doublets 4–7. This means that trypanosomes need to assemble separate structures during flagellum construction, a feature also encountered in spermatozoa (Escalier, 2003). PFR proteins are effectively transported toward the distal tip of the flagellum as demonstrated in mutants that fail to assemble a full PFR structure (Bastin et al., 1998, 1999b), although the actual motors are unknown. Like most protozoa, trypanosomes assemble a new flagellum at each cell cycle while conserving the old flagellum (Sherwin and Gull, 1989), thereby providing the unique opportunity to compare mature and constructing flagella in the same cell. The presence of two flagella of different age and of different length raises the question as to how trypanosomes control IFT. We previously demonstrated that IFT88 (an IFT complex B protein also called Polaris or OSM-5) and the heavy chain dynein motor DHC1b were both required for flagellum formation in T. brucei, suggesting that IFT is functional in trypanosomes (Kohl et al., 2003). Induction of RNA interference (RNAi) silencing any of these two genes first leads to the production of shorter new flagella without affecting the existing old flagellum. Analysis of cells inheriting the shorter new flagellum revealed a surprising link with cell size suggesting that trypanosomes could control IFT to regulate flagellar length and subsequently cell size. Longer induction times completely blocked flagellum formation, producing short cells that rapidly lose polarity, exhibit defects in basal body positioning and fail to undergo cytokinesis (Kohl et al., 2003; Absalon et al., 2007). However, the existing old flagellum remains in place and seems intact. Some of these results have recently been reproduced upon silencing of the IFT80 (CHE-2) gene that encodes another complex B protein (Davidge et al., 2006).
Inducible expression of tagged flagellar proteins showed a strong bias toward the distal tip of the new flagellum with little incorporation in the mature flagellum (Bastin et al., 1999a). To determine the role of IFT in this discrimination, we investigated IFT proteins, IFT particles and IFT movement in the old and the new flagella of T. brucei. We performed an extensive functional analysis of nine genes known to be involved in IFT in other species and of five novel genes identified by comparative genomics as putatively involved in IFT (putative intraflagellar transport [PIFT] genes).
MATERIALS AND METHODS
Trypanosome Cell Lines and Cultures
All cell lines used for this work were derivatives of strain 427 of T. brucei and cultured in SDM79 medium supplemented with hemin and 10% fetal calf serum. Cell lines DHC1bRNAi, IFT88RNAi (Kohl et al., 2003), IFT20RNAi (Absalon et al., 2007), and PF16RNAi and PF20RNAi (Branche et al., 2006) have been described previously. They all express complementary single-stranded RNA corresponding to a fragment of the gene of interest (300–900 base pairs) from two tetracycline-inducible T7 promoters facing each other in the pZJM vector (Wang et al., 2000) transformed in 29-13 cells that express the T7 RNA polymerase and the tetracycline-repressor (Wirtz et al., 1999). Addition of tetracycline to the medium induces expression of sense and antisense RNA strands that can anneal to form double-stranded RNA and trigger RNAi.
The T. brucei genome has been fully sequenced (Berriman et al., 2005), and the GeneDB data base was searched for homologues of Chlamydomonas or C. elegans IFT genes. The list of genes studied here, their reference number in GeneDB and the name of homologues in the C. elegans and D. melanogaster genome data bases is given at Figure 1.
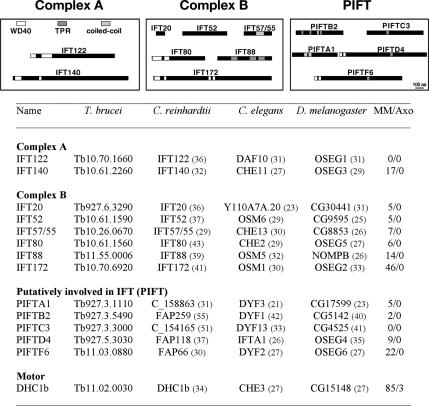
IFT and PIFT proteins investigated in this study. Top, diagram (to scale) illustrating the various IFT proteins from complex A and B and PIFT with their specific domains. Bottom, list of IFT proteins and name of their orthologues in C. reinhardtii, in C. elegans, and in D. melanogaster. The numbers in parentheses show the percentage of identity between the trypanosome and the indicated orthologous proteins. Most IFT proteins are found in the membrane + matrix fraction of Chlamydomonas proteome. MM/Axo, the number of peptides corresponding to each protein found in the membrane + matrix (MM) or the axoneme (Axo) fractions. Data are from the proteomic analysis of the C. reinhardtii flagellum (Pazour et al., 2005).
Plasmid Construction and Transformation in Trypanosomes
For generation of cell lines expressing double-stranded RNA (dsRNA) for RNAi knockdown, sequences were selected according to their lack of significant identity with other genes to avoid cross-RNAi (Durand-Dubief et al., 2003) by using the RNAit algorithm (Redmond et al., 2003). Primers are available on request from the authors. Gene fragments were amplified by polymerase chain reaction (PCR) on T. brucei genomic DNA, purified on QIAquik columns (QIAGEN, Hilden, Germany), digested with HindIII and XhoI, and ligated in the corresponding sites of the pZJM vector.
For expression of IFT20 fused to a tap-tag, the whole IFT20 sequence was amplified by PCR with the proof-reading enzyme PfuI by using GCGTGCCAAGCTTATGGATGATGATAAACTTGTG as forward primer (HindIII site underlined) and GCATGATATCCTCACGCGAGGCGTGACTCAG (HpaI site underlined) as reverse primer. The amplified product contains the complete coding sequence of IFT20 but the stop codon was not included. It was ligated in the HindIII and EcoRV sites of the pHD918 (Estevez et al., 2001) vector in such a way that the insertion of the IFT20 coding sequence was in-frame with that coding for protein A, generating plasmid pIFT20TAPTAG918. For inducible expression of GFP::IFT52, the full coding sequence of IFT52 was amplified by PCR by using Phusion (Finnzyme, Espoo, Finland), with GCATCATCTAGAATGACGGATGCCCCAAGG (XbaI site underlined) as forward primer and GCATCGGGATCCTCAAAGCTCCTCAATTTC as reverse primer (BamHI site underlined), cloned in the pPCEGFPN2PFR vector (Absalon, Buisson, and Bastin, unpublished data), and the fusion gene encoding GFP::IFT52 was transferred to the pHD430 inducible expression vector (Wirtz and Clayton, 1995). All inserted sequences and flanking regions were sequenced to confirm correct integration and fusion (Genome Express, Meylan, France).
For transfection, pZJM plasmids were linearized with NotI and individually transformed in 29-13 cells. pIFT20TAPTAG918 and pGFPIFT52430 were transformed in the rDNA locus of the PTP or PTH cell lines that express the tet-repressor from the pHD449 or the pHD360 vectors, respectively (Wirtz and Clayton, 1995; Bastin et al., 1999a). In all cases, transfected cells were immediately cloned, and all antibiotic-resistant cell lines were characterized by immunofluorescence with the anti-PFR2 antibody L8C4 or with the anti-protein A antibody (Sigma, St. Louis, MO) or by direct fluorescence observation (GFP). Clones with the clearest distinction between noninduced and induced samples were selected for subcloning by limiting dilution. RNAi was induced by addition of 1 μg of tetracycline per ml of medium, and fresh tetracycline was added at each cell dilution.
Protein Expression and Antibody Production
Given the large size of IFT172, only a fragment was cloned and expressed in Escherichia coli. The forward primer GATGAACGGATCCGCTATTCAAGCATACAAAAAGG was used (BamHI site underlined) and CCGTGCGAATTCGCTACACGAACAGCATCCTCC served as reverse primer (EcoRI site underlined), representing nucleotides 2500–3270 of the IFT172 coding sequence. The PCR product was amplified using a proof-reading enzyme, digested with BamHI and EcoRI, and ligated in compatible sites of the pGEXB vector for protein expression. The plasmids were sequenced to confirm identity and correct fusion with GST. Plasmids were transformed in the competent BL21 strain of E. coli, and protein expression was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie staining. Glutathione transferase (GST)-coupled proteins were purified as described previously (Smith and Johnson, 1988), and 20 μg was injected to BALB/C mice for immunization. After bleeding, sera were absorbed against GST. Sera from mice immunized with GST alone were used as negative controls.
Immunofluorescence and Light Microscopy Analysis
For immunofluorescence with the anti-IFT172 antiserum, intact cells or detergent-extracted cytoskeletons (see below) were washed, settled on poly-l-lysine–coated slides, and fixed either in 4% paraformaldehyde (PFA) for 10 min or in methanol for a maximum of 5 min at −20°C. For PFA fixation, cells were permeabilized with 0.1% Nonidet P-40 for 10 min, and samples were rinsed to remove the excess of detergent. Blocking was performed by an incubation of 45–60 min in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin, and washed slides were incubated with 1:200 dilution of antiserum for 45–60 min. Slides were washed and incubated with anti-mouse secondary antibodies coupled to Alexa 488 (Invitrogen, Paisley, United Kingdom). For detergent treatment, cells were settled on poly-l-lysine–coated slides and exposed to 1% Nonidet P-40 in PEM buffer (100 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.9, 1 mM MgCl2, and 1 mM EGTA). For cells expressing the IFT20::TAP tag, both PFA and methanol fixations were used, with a dilution of 1:10,000 of the anti-protein A antibody (Sigma). For cytoskeletons of GFP::IFT52-expressing cells, samples were settled on slides and briefly exposed to 1% Nonidet P-40 in 4 M glycerol, 10 mM PIPES, pH 6.5, 10 mM MgCl2, and 5 mM EGTA, and then they were washed and processed as described above. The monoclonal antibody (mAb) L8C4 (IgG1) that specifically recognizes PFR2 was used as marker of flagellum assembly (Kohl et al., 1999). The MAb22 is an IgM that detects an as yet unidentified antigen found at the proximal zone of both the mature and the probasal body (Bonhivers and Robinson, unpublished data; Pradel et al., 2006). MAb25 is an IgG2a that recognizes a protein found all along the axoneme (M. Bonhivers and D. Robinson, unpublished data)(Pradel et al., 2006). The MAb 20H5 recognizes centrins and was used at a 1:400 dilution (Sanders and Salisbury, 1994). A rabbit antiserum raised against the Leishmania donovani centrin 1 was used at a 1:1000 dilution (Selvapandiyan et al., 2007). GFP was observed directly or upon immunofluorescence using an anti-GFP antibody (Invitrogen). Subclass-specific secondary antibodies coupled to fluorescein isothiocyanate (Sigma), Alexa 488 or Alexa 594 (Invitrogen), and cyanine (Cy) 3 or Cy5 (Jackson ImmunoResearch Laboratories, West Grove, PA) were used for double labeling. Slides were stained with 4,6-diamidino-2-phenylindole (DAPI) for visualization of kinetoplast and nuclear DNA content. For visualization of GFP on live cells, trypanosomes cultures were mixed with a solution of 3% low melting point agarose (Bio-Rad, Hercules, CA) to reduce cell movement. Samples were observed with a DMR Leica microscope, and images were captured with a CoolSNAP HQ camera (Roper Scientific, Trenton, NJ). Alternatively, slides were also viewed on a DMI4000 microscope (Leica, Wetzlar, Germany), and images were acquired with a Retiga-SRV camera (QImaging, Surrey, BC, Canada). Images were analyzed and cell parameters were measured using the IPLab Spectrum 3.9 software (Scanalytics, Fairfax, VA and BD Biosciences, San Jose, CA) or ImageJ (http://rsb.info.nih.gov/ij/). For movies, cells were filmed using a Cohu camera 460LI, and images were recorded using a DVD recorder.
Electron Microscopy
Cell fixation, embedding, and sectioning for transmission electron microscopy of whole cells or of detergent-treated samples was carried out as described previously (Branche et al., 2006). For scanning electron microscopy, cells were washed in PBS, fixed with 2.5% glutaraldehyde, and treated as reported previously (Absalon et al., 2007).
Reverse Transcription (RT)-PCR
Total RNA was extracted from cells grown with or without tetracycline for the indicated periods of time and purified using TRIzol Reagent (Invitrogen). DNA was eliminated by DNase treatment, and RNA purity was confirmed by conventional PCR. After primer calibration and determination of optimal conditions, semiquantitative RT-PCR was performed as described previously (Durand-Dubief et al., 2003). At least one primer was selected to be outside the region selected for dsRNA expression to avoid amplification of RNA deriving from the dsRNA trigger.
Western Blot
Cells were washed in PBS and then homogenized and boiled in gel sample buffer before SDS-PAGE. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and incubated with the anti-protein A antibody (1:1000) or with the anti-GFP antibody (1:800). Membranes were probed with the anti-PFR antibody L13D6 (dilution 1:50) or with a rabbit anti-aldolase (dilution 1:1000) antibody as loading control and revealed with ECL+ (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
RESULTS
Identification of IFT-related Genes in Trypanosomes
To identify trypanosome genes participating in flagellum formation, we first searched the T. brucei genome for genes encoding proteins known to be components of purified IFT particles in Chlamydomonas (Cole et al., 1998). At the time this study was initiated, the sequences of eight genes encoding IFT proteins were available: IFT122, IFT140 (complex A) and IFT20, IFT52, IFT57/55, IFT80, IFT88, IFT172 (complex B) (C. elegans and Drosophila melanogaster gene nomenclature is given in Figure 1). The corresponding trypanosome genes are all present in the T. brucei genome database (Kohl et al., 2003; Briggs et al., 2004a; Berriman et al., 2005; Absalon et al., 2007) and exhibit 21–55% overall identity with orthologues from various species (Figure 1), a value in the same range as what was found for axonemal proteins (Branche et al., 2006; Ralston et al., 2006; Baron et al., 2007). In addition, the gene encoding the IFT dynein heavy chain previously identified and functionally characterized in our laboratory was also included in this study, hence providing a total of nine IFT-related genes.
Next, we searched to identify novel genes encoding proteins possibly involved in the IFT process but that were not purified with IFT particles. Because the only flagella of Plasmodium are found in the male gametes and they are assembled in the cytoplasm, IFT genes are absent from the genome of this group of species. By contrast, genes encoding proteins of the dynein arms or central pair components of the axoneme are conserved (Avidor-Reiss et al., 2004; Kohl and Bastin, 2005). This provides the opportunity to compare the genomes of Plasmodium spp. with those of other ciliated species and to discriminate genes encoding proteins involved in IFT from genes encoding structural components of the flagellum. A similar approach was successfully used to identify genes encoding flagellar proteins involved in beating by comparison of species with motile or nonmotile flagella (Baron et al., 2007). Comparison of genomes from species assembling their flagella with or without IFT indicated that at least 27 genes are conserved in species assembling their flagella by IFT (Avidor-Reiss et al., 2004). We selected five of these genes that are conserved in T. brucei for functional analysis and termed them PIFT genes (Figure 1). The PIFTA1 protein presents coiled-coil domains, whereas the PIFTB2 and PIFTC3 protein contain TPR domains, a feature shared with IFT88 and IFT139 (Cole, 2003). PIFTD4 and PIFTF6 both contain two WD-40 domains at their amino terminal region and a TPR motif toward the center of their sequence. PIFTE5 exhibits a different structure and will be reported elsewhere. The five trypanosome PIFT genes are the homologues of the recently identified DYF-3, DYF-1, DYF-13, IFTA-1, and DYF-2, respectively, known to be linked to the formation of sensory cilia in C. elegans (Starich et al., 1995; Blacque et al., 2005, 2006; Murayama et al., 2005; Bell et al., 2006; Efimenko et al., 2006). PIFTD4 and PIFTF6 are the homologues of the OSEG4 and OSEG6 proteins that localize to the Outer SEGment of sensory cilia in Drosophila (Avidor-Reiss et al., 2004). Although these proteins were not detected in purified IFT particles, they are present in the membrane + matrix fraction of Chlamydomonas flagella (Pazour et al., 2005). The only exceptions are the IFT122 and the PIFTC3 proteins that were not detected in that analysis (Figure 1).
Localization of Endogenous and Tagged IFT Proteins in Mature and Elongating Flagella
To determine the location of IFT proteins in trypanosomes, mouse polyclonal antibodies were raised against a classic IFT marker, the IFT172 protein. It has been localized to the flagellum matrix and to the basal bodies of Chlamydomonas where it is required for flagellum assembly (Pedersen et al., 2005) and in C. elegans (Bell et al., 2006). It is also involved in hedgehog signaling in mouse (Huangfu et al., 2003). A fragment of the trypanosome IFT172 gene was expressed in E. coli to produce a fusion protein with GST for injection in mice. In a second approach, the genes encoding IFT20 or IFT52 were selected for fusion to a protein A tag (Estevez et al., 2001) or to GFP, respectively. IFT20 and IFT52 are proteins belonging to the B complex that have been well characterized in Chlamydomonas (Deane et al., 2001), C. elegans (Collet et al., 1998), or in mammalian cells (Follit et al., 2006). The expression of the fusion proteins is controlled by a tetracycline-inducible promoter, and it is activated by addition of tetracycline to the culture medium. Western blot analysis with an anti-protein A antibody or with an anti-GFP antibody confirmed that the IFT20-tagged protein and the GFP::IFT52 protein are expressed upon addition of tetracycline and that they show an electrophoretic motility corresponding to the expected size of the fusion protein (Supplemental Figure S1, A and B).
Indirect immunofluorescence assay (IFA) with anti-IFT172 antibodies produced a defined pattern on trypanosomes fixed in methanol: a strong signal is observed at the basal body region, and a succession of closely spaced spots is found all along the length of the flagellum until its distal tip (Figure 2A). The same pattern was observed in cells possessing two flagella, and no visible difference in signal could be detected between the new (no matter its length) and the old flagellum (see, for example, the cell at the bottom left of Figure 2A). The same antiserum used on PFA-fixed trypanosomes stained the flagellar compartment but not the basal body (data not shown), possibly due to a more difficult access as noted previously for IFT20 in mammalian cells (Follit et al., 2006). IFT20 fused to a protein A tag also localized to the flagellum and to the basal body, with a weak signal on the cell body (Figure 2B). Double staining with markers of various organelles (endoplasmic reticulum, lysosomes, mitochondria) failed to reveal a specific association, indicating that the protein is present in the cytosol, possibly as a consequence of overexpression (Supplemental Figure S1, C and D; data not shown). Finally, observation of live cells expressing GFP::IFT52 also demonstrated localization to the basal body and the probasal body and to the flagellar compartment of both old and new flagellum, with a more pronounced localization at the proximal part of the flagellum (data not shown). This pattern was preserved after methanol fixation, although it became less intense (Figure 2C). Staining with an anti-GFP antibody confirmed this localization (Supplemental Figure S1, F and G).
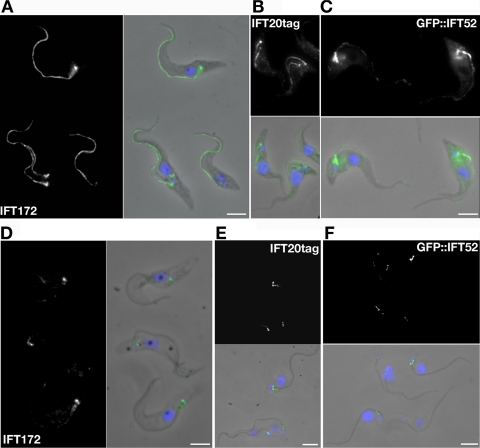
Localization of IFT proteins in trypanosome flagella. (A) Wild-type cells have been fixed in methanol for 5 min and stained with a 1:200 dilution of the anti-IFT172 antiserum (green). (B) Cells expressing IFT20 fused to a protein A tag stained with the anti-protein A antibody (green) after methanol fixation. (C) Direct observation of GFP (green) after methanol fixation of GFP::IFT52- expressing cells. (D–F) Cells from the same populations as shown in A–C but that were detergent-extracted to remove the membrane before PFA (D and E) or methanol (F) fixation. Only the basal body signal and sometimes the proximal part of the flagellum remain positive. All samples were counterstained with DAPI (blue) to visualize nuclear and kinetoplast DNA. Bar, 5 μm.
To further investigate IFT proteins, trypanosomes were treated with 0.1% Nonidet-P40 before fixation. Detergent addition removes the cell body and flagellum membranes but the subpellicular corset of microtubules, the axoneme, the PFR and the basal body complex remain intact (Sherwin and Gull, 1989). Detergent treatment abolishes most of the flagellum staining for IFT172, tagged IFT20, and GFP::IFT52, but a clear signal remains present at the basal body region in both methanol- and PFA-fixed cells (Figure 2, D and F). This indicates that a pool of IFT172 with possibly different biochemical characteristics is present around the basal bodies, as recently suggested for Bardet–Biedl syndrome (BBS) proteins (Nachury et al., 2007).
To further define the positioning of IFT proteins, GFP::IFT52-expressing cells were stained with several markers of the basal body (Figure 3, A–D). Double labeling with MAb22, a mAb marker of the proximal region of both mature and probasal bodies (Pradel et al., 2006; Absalon et al., 2007), showed that GFP::IFT52 was found in a more apical position (Figure 3A), a feature that was reproduced for tagged IFT20 (Supplemental Figure S1E). The antibody against trypanosome basal body component (TBBC) (Dilbeck et al., 1999), a protein found exclusively on the mature basal body that hence produces a single spot in IFA (Absalon and Bastin, unpublished data) showed that GFP::IFT52 was present in an even more apical position (Figure 3B). Costaining was also performed with two antibodies against centrins that localize to mature and probasal bodies, and to a “bilobed” structure found close to the Golgi apparatus (He et al., 2005; Selvapandiyan et al., 2007). The mAb 20H5 recognizes numerous centrin proteins from different organisms, and in detergent-extracted trypanosomes it was found to light up the flagellum, the mature and the probasal body, and a structure close to the flagellar pocket and the Golgi apparatus (Figure 3C). A similar pattern was obtained with the antiserum raised against centrin 1 from the related organism Leishmania donovani, except that the flagellum and basal body signals were weaker (Figure 3D). In both cases, GFP::IFT52 was found to merge with the centrin signal at the mature basal body. The relative position of IFT proteins themselves was investigated upon staining of GFP::IFT52-expressing cells with the anti-IFT172 antiserum, revealing a close but separate location for the two proteins. GFP::IFT52 was found to be in a more apical location compared with IFT172 (Supplemental Figure S2). Double staining of tagged IFT20-expressing cells with the anti-protein A antibody and the anti-IFT172 antiserum showed that these two proteins colocalized at the basal body region (Supplemental Figure S2).

GFP::IFT52 is found at the flagellum and the basal body and displays IFT in old and new flagella. (A–D) GFP::IFT52-expressing cells were fixed in methanol and stained with different antibody markers of the basal body: MAb22 (A), anti-TBBC (B), 20H5 (anti-centrins, C), and an antibody raised against the L. donovani centrin 1 (D). Direct GFP fluorescence is shown in green, the other antibodies are shown in red and DAPI in blue. Insets show 2.5-fold magnification of the indicated basal body regions. (E) Still images of Supplemental Video S1. The first panel shows a phase contrast image of the cell and the others show a succession of fluorescent images with the elapsed time indicated in seconds. The arrow points at a moving IFT particle. Bar, 5 μm.
In conclusion, three different IFT proteins are present along the length of the axoneme where they are sensitive to detergent, and at the mature basal body and frequently at the probasal body where they are resistant to detergent. Importantly, they localize to both old and new flagella, indicating that IFT could operate in mature and elongating flagella.
Movement of GFP::IFT52 Protein in Trypanosome Flagella
Presence of various IFT proteins in both old and new flagella does not necessarily prove that these proteins are trafficking in this compartment. We therefore examined the behavior of the GFP::IFT52 fusion protein in live trypanosomes. Agarose was added to the medium to restrict cell movement, and direct GFP fluorescent signals were monitored by videomicroscopy. This showed unambiguous movement of fluorescent particles in flagella, providing the first actual evidence for intraflagellar transport in trypanosomes (Supplemental Videos 1 and 2; still images of Movie 1 are presented at Figure 3E). Intraflagellar movement was detected in both old and new flagella, but it was more difficult to quantify on the new flagellum because this flagellum is regularly found on top of the cell body, which tends to obliterate the fluorescent signal. IFT was best visualized on the distal end of the mature flagellum (Supplemental Movies S1 and S2), but it was also detected in the new flagellum when it lied on the side of the cell (Supplemental Movie S1). Anterograde events were more frequent and easier to observe (Supplemental Movies S1 and S2), but a few retrograde events could also be detected (Supplemental Movie S2). Rate of anterograde IFT was measured at ~3 μm/s. Retrograde transport was faster, but it could not be estimated accurately due to its low frequency and to the weak GFP signal. These data demonstrate that IFT is indeed active in trypanosomes and operates in both mature and assembling flagella.
IFT-like Particles in the Trypanosome Flagellum
In Chlamydomonas, IFT particles can be recognized on flagellum sections as electron-dense granules found between the membrane and the axoneme (Kozminski et al., 1993). Occasional viewing of such particles has been previously reported in T. brucei, but detailed analysis is lacking (Bastin et al., 2000b). We examined a large number of cross sections of flagella from wild-type and several RNAi motility mutants (Branche et al., 2006) and noticed the frequent presence of granule-like structures with a diameter of 20–30 nm (Figure 4, A–F, and Supplemental Figure S3). Usually, only one particle is visible per section (Figure 4, A and C) but in rare instances (<5%), several particles can be seen on the same section (Figure 4B). Remarkably, particles show a very specific location relative to the axoneme doublets (Figure 4D): they are either associated to doublets 3–4 (Figure 4A) or 7–8 (Figure 4C), but they are never found next to doublets 5–6, doublet 9, and rarely close to doublets 1 and 2 (Figure 4D). Longitudinal sections showed that the particles look like flat rafts with one side that is very close to the microtubules, and the other side that seems almost in contact with the flagellar membrane (Figure 4E). Presence of the IFT-like particles seems to position the flagellar membrane more closely toward the axonemal microtubules (Figure 4E).

IFT particles in flagella of trypanosomes. (A–C) Cross sections through the flagella of wild-type (WT) trypanosomes (A and B) or PF16RNAi (C) cells induced for 3 d. The presence of the PFR attached to the axoneme allows for unambiguous identification of individual doublets, shown by white numbers. IFT-like particles are indicated by black arrows. (D) Position of IFT-like particles relative to axoneme doublets on cross sections of WT, PF16RNAi, or PF20RNAi induced cells for 3 d (n = 42, 81, and 21, respectively). (E) Longitudinal section through an IFT-like particle. Note the closer positioning of the flagellar membrane at the particle position. (F) Cross section through both the old (bottom) and the new (top) flagella from a wild-type trypanosome revealing the presence of IFT-like particles in the old flagellum. (G) Cross section through the transition zone showing the 9 + 0 arrangement but without IFT-like particles. (H and I) Cross sections of flagella of WT trypanosomes close to the top of the flagellar pocket. Central pair and dynein arms are visible, but the PFR is not yet present. Abundant particular material can be recognized. (J and K) Cross section through flagella of wild-type or PF20RNAi cells induced for 3 d after detergent removal of the membrane. IFT particles are less abundant in detergent-treated samples but can sometimes be recognized (arrow in K). (L) Abundance of IFT particles in cross sections of flagella from whole cells or detergent-extracted cytoskeletons of wild-type (n = 42 and 41, respectively), PF16RNAi (n = 82 and 73, respectively) or PF20RNAi (n = 21 and 74, respectively) cells induced for 3 d.
We also looked at flagellum sections of RNAi mutants where expression of two proteins of the central pair (PF16 or PF20) had been separately silenced, leading to inhibition of motility but without interfering with flagellum construction (Branche et al., 2006; Ralston et al., 2006). Similar particles are visible in these flagella (Figure 4C) although at a slightly lower frequency: 31% of sections for PF16RNAi cells and 24% for PF20RNAi cells instead of 57% for wild-type (Figure 4L). Nevertheless, they all show the preferential association to the same doublets as was found in wild-type cells (Figure 4D).
Because the new flagellum is always found in the same position relative to the old flagellum (Briggs et al., 2004b), it can be identified in the few sections where both flagella are clearly visible. This revealed that both flagella contain these particles (a clear example for the old flagellum is shown at Figure 4F). IFT-like particles are never recognized in the transition zone of the basal body, although some amorphous, relatively electron-dense, material is visible between the microtubule doublets and the membrane (Figure 4G). This was clearly different from the inside of the axoneme shaft that seems much more translucent (Figure 4G). The central pair and the dynein arms are already visible in flagellum sections close to the top of the flagellar pocket (as confirmed by the short diameter of the flagellar pocket lumen). The PFR is not yet present but a large amount of material is found around the microtubule doublets, sometimes resembling the granules identified above (Figure 4, H–I).
Detergent-treatment suppresses most of the IFT172, tagged IFT20 or GFP::IFT52 signal at the flagellum. To relate this result with granules seen by electron microscopy, wild-type, PF16RNAi, or PF20RNAi cells were treated with 1% Nonidet P-40. This procedure removes the cell membrane, but it ensures an excellent conservation of the whole trypanosome cytoskeleton for electron microscopy analysis, including the axoneme and the PFR (Sherwin and Gull, 1989). In these conditions, the frequency of IFT-like particles drops, with <10% of samples showing a possible remnant at the expected position on doublets 3–4 or 7–8 (Figure 4, J–L). This result shows that these granules display the same characteristics as the three IFT proteins studied above.
IFT Genes Are Necessary for Flagellum Construction
Expression of the trypanosome genes IFT122, IFT140 (complex A) and IFT20, IFT52, IFT55, IFT80, IFT88, IFT172 (complex B) was individually silenced by inducible RNAi in trypanosomes, and knockdown efficiency was controlled at the RNA level by semiquantitative RT-PCR (Supplemental Figure S4). In all cases, RNAi silencing led to the inhibition of new flagellum formation and to the emergence of apparently nonflagellated trypanosomes (Figure 5, A and B). However, the old flagellum is not depolymerized and remains present. Analysis of cells fixed at different stages of RNAi silencing followed by IFA staining using a mAb recognizing a major PFR protein (Figure 5, A and B) showed a mixture of different cell types: cells retaining the old flagellum, cells with a shorter flagellum and cells without PFR signal, that could correspond to cells with very short flagella missing the PFR or to nonflagellated cells (see below). Cells with shorter flagella exhibited a reduced cell size, with those without a visible flagellum being the shortest (Figure 5, A and B), in agreement with the central role of the flagellum in the definition of cell size (Kohl et al., 2003). Trypanosomes are difficult to synchronize reliably, meaning that RNAi induction is likely to have different effects according to the position of a cell in its cycle. Therefore, it is important to examine carefully cells throughout the induction. IFA with the anti-PFR was first used to score proportions of positive and negative cells during the course of RNAi silencing of IFT and genes (Figure 5, D and E). Significant differences were noted in the rate of emergence of nonflagellated cells. In complex A proteins, knockdown of IFT140 leads to faster emergence of nonflagellated cells compared with IFT122 (Figure 5D). Differences are also observed in the knockdown of RNA-encoding complex B proteins. For example, it takes only 30 h to obtain 50% of nonflagellated cells during silencing of IFT172, whereas it takes an extra 40 h to generate a similar result for IFT52 knockdown (Figure 5E). Hence, these differences in kinetics are not linked to genes encoding members of a specific complex. Inhibition of any of the IFT genes invariably leads to a growth arrest whose timing is correlated with the kinetics of emergence of nonflagellated cells (Figure 5, D and G and E and H, for proteins of complex A and B, respectively). For example, in complex A members, silencing of IFT140 rapidly produces a large number of nonflagellated cells and leads to a more premature growth arrest compared with IFT122RNAi grown and induced in the same conditions (Figure 5G).
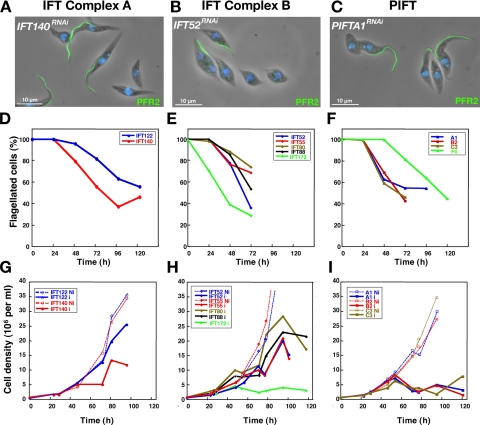
RNAi silencing of IFT and PIFT genes inhibits new flagellum formation and results in growth arrest. Cell lines were grouped as mutants of genes encoding proteins of complex A (A, D, and G), B (B, E, and H) or PIFT (C, F, and I). (A–C) Fields of cells from the indicated cell lines that had been induced for 3 d were stained with the anti-PFR2 L8C4 antibody (green) and with DAPI (blue). A mixture of nonflagellated and flagellated cells was present in all situations. (D–F) Proportion of flagellated cells left in the culture of the indicated RNAi mutants during the course of induction of RNAi silencing. (G–I) Growth curves of induced (plain lines) and noninduced (discontinuous lines) cells from the indicated RNAi mutants.
These differences in kinetics could reflect variable efficiency of RNA destruction or different rates of protein turnover (or both). RT-PCR analysis was performed on total RNA extracted from cell lines noninduced or induced for up to 5 d. Abundance of each target RNA was normalized with a control unrelated mRNA (Supplemental Figure S4). Kinetics of RNA silencing is very reproducible for all RNAi cell lines investigated: a potent silencing is observed 2 d after induction and a slight increase in the amount of the target RNA is noted the following days. This reproduces kinetic patterns observed for other RNAi cell lines developed in separate experiments (Durand-Dubief, Absalon, and Bastin, unpublished data). In summary, the differences in kinetics of emergence of nonflagellated cells during the course of RNAi are not due to a variable efficiency of RNA degradation, but they are likely to reflect differences at the protein level.
Distinct Phenotype upon Silencing of Genes Required for Anterograde or Retrograde Transport
In Chlamydomonas and C. elegans, inhibition of proteins associated to anterograde transport (IFT B and kinesin II complexes) leads to complete or severe inhibition of flagella or cilia formation. By contrast, blocking of retrograde transport (IFTA and dynein complexes) produces short cilia or flagella, filled with IFT material that can penetrate the flagellar compartment but cannot be recycled to the base (Cole, 2003). We analyzed IFT88RNAi (mutant of a complex B protein) and IFT140RNAi (mutant of a complex A protein) induced cells by double staining with the anti-IFT172 antiserum as a marker of the IFT particles and with the MAb25 antibody as a marker of the axoneme (Figure 6, A and C, and E). In IFT88RNAi, the majority of short cells did not exhibit a flagellum (no MAb25 signal) and only showed a weak signal for IFT172, indicating that absence of IFT88 severely blocks flagellum formation and leads to a reduction or a dispersion of IFT172 (Figure 6A, stars, and C). This result was supported by scanning electron microscopy that revealed that most short cells did not exhibit a flagellum. In the rare cases where a flagellum could be detected, it often seemed short (<1 μm) and thin (Figure 6D). Cells that retained the old flagellum were still positive for both the MAb25 axoneme marker and the anti-IFT172, no matter the length of the flagellum (Figure 6A, arrows for long flagellum and arrowheads for flagella of intermediate lengths). A very different result was obtained for the IFT140RNAi cells stained with the same antibodies (Figure 6, B and E). Numerous short cells were seen with a short line as MAb25 signal, indicating formation of a short axoneme, and with a very intense signal for IFT172 (Figure 6, B and E). The presence of short flagella filled with IFT172 proteins, producing very bright spots of fluorescence, increased rapidly during the course of RNAi silencing, whereas it was virtually never detected in the case of IFT88RNAi (Figure 6G). Scanning electron microscopy demonstrated the presence of cells with short flagella (1–3 μm) with large dilations (Figure 6F), that very likely results from the accumulation of IFT proteins. However, cells that retained an old flagellum of normal length still exhibited the usual MAb25 and anti-IFT172 signals (Figure 6B, arrows). By contrast, cells that possessed a flagellum of intermediate length (~5 μm) showed normal MAb25 signal but much increased abundance of IFT172 protein (Figure 6B, arrowhead).
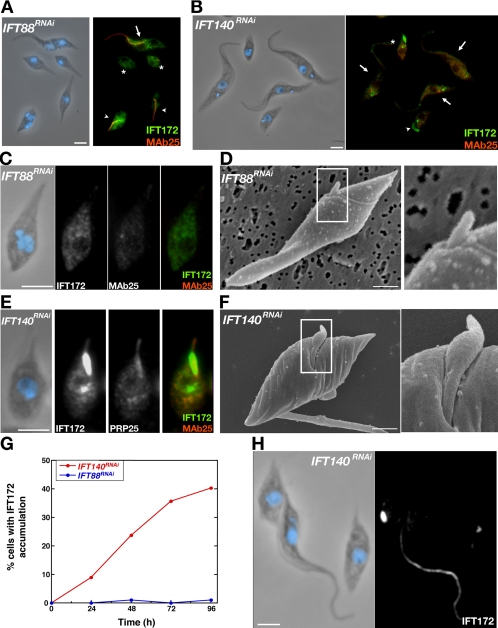
IFT proteins from complex A and B are required for flagellum formation but differ in function. (A and B) Fields of IFT88RNAi (A) or IFT140RNAi (B) cells induced for 3 d stained with the anti-IFT172 antiserum (green) and with the MAb25 axoneme marker (red). DNA was stained with DAPI (blue). Cells with a normal-length flagellum (arrows), with a short flagellum (arrowhead), or without visible flagellum (stars) are indicated. (C and D) IFT88RNAi induced cells have a tiny or no flagellum. (E and F) IFT140RNAi-induced cells exhibit short and dilated flagella accumulating a large amount of IFT172 protein. (C and E) Cells induced for 3 d and stained with the anti-IFT172 antiserum (green) and with MAb25 (red). DNA was stained with DAPI (blue). (D and F) Scanning electron micrographs. Bar, 1 μm. Inset shows a twofold magnification of the indicated area. (G) Quantification of the number of cells exhibiting accumulation of IFT172 as bright spots during the course of RNAi silencing in the indicated cell lines. (H). IFT140RNAi cell induced for 3 d with a short new flagellum and an old flagellum. Only the new flagellum is too short, dilated and accumulates IFT172 protein. Cells were stained with the anti-IFT172 antiserum (green) and with DAPI (blue). Bar, 5 μm.
Although construction of the new flagellum is inhibited or strongly reduced, the old flagellum remains present and is still motile. We examined IFT140RNAi cultures at early stages of RNAi, with special emphasis on cells that were binucleated, so at late stages of their cell cycle (Figure 6H). The mature flagellum was normal and exhibited the expected signal for the anti-IFT172 antiserum whereas the new flagellum was very short and filled with IFT172 protein. This shows that the old and the new flagellum are independent from each other and despite the arrest of new IFT140 production, IFT140 protein present in the old flagellum is still functional. The ultrastructure of the old flagellum of IFT88RNAi and IFT172RNAi (anterograde transport mutants) and DHC1bRNAi (retrograde transport mutant) was examined by transmission electron microscopy. Induction times were selected in such a way that cells were at a stage when they had stopped constructing a new flagellum, so that almost all flagellar sections should correspond to the old flagellum. This was carried out on whole cells and detergent-extracted cytoskeletons (Supplemental Figure S3). Sections through flagella were less frequent on induced cells compared with wild-type or noninduced controls, as expected from the drop in the proportion of flagellated cells upon RNAi induction. However, the ultrastructure of the axoneme seemed intact: the nine doublets microtubules, the central pair, the dynein arms and radial spokes were all present (Supplemental Figure S3). Their flagellar pocket also seemed structurally normal, as well as the flagellum adhesion zone (Supplemental Figure S3). Importantly, IFT particles are still visible on the old flagellum (Supplemental Figure S3). We observed IFT-like particles on six of 19 such profiles, i.e., a proportion similar to that observed for populations of cells that build their flagellum normally, indicating that IFT particles remain present in the old flagellum. This is confirmed by IFA with the anti-IFT172 that still stains the old flagellum of IFT140RNAi (Figure 6H), IFT88RNAi and DHC1bRNAi (data not shown) cells induced for 2 or 3 d, indicating a slow turnover of IFT proteins in mature flagella.
Transmission electron microscopy was used to examine the new flagellum in control wild-type cells or IFT88RNAi and DHC1bRNAi cells (Figure 7). The basal body is rooted within the cell body, whereas the transition zone is covered by the flagellum membrane and localized in the lumen of the flagellar pocket of wild-type cells (Figure 7A). The flagellar pocket exhibits a typical asymmetric structure with the larger lumen side found at the anterior end. When a new flagellum is assembled, it is made in the same flagellar pocket (Grassé, 1961) found on the large, anterior, side of the lumen (Figure 7B, arrow). The transition zone is clearly recognizable and a mass of amorphous material is found at the distal growing end (Figure 7, B and C), wrapped by the flagellum membrane that sometimes seems distorted (Figure 7B). The basal bodies of such flagella are “bald”, i.e., axoneme microtubules have not yet elongated. These basal bodies seem shorter, suggesting that they have not fully matured yet. This was encountered in five of the 72 analyzed sections of the flagellar pocket (7%) (Table 1). The new flagellum grows and axoneme microtubules are now visible in continuity with those of the transition zone. Nevertheless, this new flagellum remains in the same flagellar pocket as the old flagellum (Figure 7D). In nonsynchronous cultures of wild-trypanosomes, 16% of flagellar pockets contain two flagella (n = 72). Finally, two individual flagellar pockets are visible, each containing a single flagellum (Figure 7E).
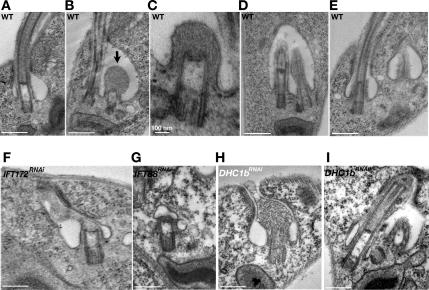
Formation of the new flagellum in wild type cells and its inhibition in IFTRNAi cell lines. Sections through the flagellar pocket of WT (A–E) or IFT172RNAi cells induced for 48 h (F) or of IFT88RNAi (G) or DHC1bRNAi (H and I) induced for 72 h. Notice the large amount of material only in very short flagella of wild-type cells (B and C). The arrow in B indicates the new flagellum. The new flagellum grows in the same flagellar pocket as the old flagellum (B and D) and elongates until two separate flagellar pockets are recognized (E). Bald basal bodies (F and G) were frequent in sections of induced IFT172RNAi and IFT88RNAi mutants whereas short flagella and accumulation of IFT material was common in DHC1bRNAi induced cells (H and I). Bars, 500 nm except where indicated.
Table 1.
Effects of RNAi silencing of IFT genes on axonemal microtubule elongation
| Cell line | % BB with axoneme microtubules | % BB with interrupted microtubules | % Bald BB | n |
|---|---|---|---|---|
| Wild-type | 89 | 0 | 11 | 46 |
| IFT88RNAi | 14 | 7 | 79 | 14 |
| DHC1bRNAi | 28 | 15 | 56 | 32 |
Ultrastructure of the basal body and the flagellum was analyzed in IFT88RNAi, IFT172RNAi (mutants of anterograde transport), and DHC1bRNAi (mutant of retrograde transport) cells at induction times where assembly of the new flagellum is drastically reduced (Figure 7, F–I). Longitudinal sections through basal bodies were grouped in three categories: those where axonemal microtubules are visible in direct continuity of the transition zone and seem normal, those where axonemal microtubules are present but interrupted, and those where only a bald basal body is visible (Table 1). In all IFTRNAi mutants analyzed, the frequency of bald basal bodies rose significantly reaching close to 80% in the IFT88RNAi cell line (Table 1). Some material is frequently detected on top of the basal body and looks like flagellar membrane extension (see below), but it does not resemble to what was observed for wild-type flagella (compare Figure 7F with B and C). The proximal region of all basal bodies still exhibits triplet microtubules, and the transition zone looks apparently normal. Nevertheless, these basal bodies seem slightly shorter, like the basal bodies of short new flagella in wild-type cells (Figure 7, B and C). These data demonstrate that axoneme formation is strongly inhibited in the absence of a protein of the IFT B complex. In the case of DHC1bRNAi cells, about half of the sections revealed the presence of a bald basal body but other situations were encountered. Large accumulation of electron-dense material resembling IFT particles was observed (Figure 7H). Moreover, several sections showed the presence of short microtubules and some material trapped between this axoneme and the tip of the flagellar membrane (Figure 7I). In these situations, the basal body displayed a normal length. This accumulation fits with the dilation of short flagella of retrograde transport mutants observed by scanning electron microscopy and by IFA with the anti-IFT172 antiserum (Figure 6).
We conclude that in the absence of retrograde transport, members of the IFT complex B can still access the flagellum and accumulate there as they cannot be recycled due to the absence of retrograde activity. This would allow the formation of short and dilated flagella. Conversely, in the absence of anterograde transport, entry of IFT protein in the flagellum is restricted and flagellum formation is more strongly inhibited.
PIFT Genes Are Required for Flagellum Construction and Can Be Classified as Involved in Anterograde or Retrograde Transport
Expression of PIFT genes was knock downed by RNAi, it was monitored in parallel to the mutants of conventional IFT genes, revealing that all five selected PIFT genes were required for flagellum formation (Figure 5, C and F). Significant differences were also observed in the emergence of nonflagellated cells (Figure 5F), with fast phenotypes for silencing of PIFTA1, PIFTB2 or PIFTC3 (50% nonflagellated cells in 48–60 h), and the slower kinetics for PIFTF6 knockdown (50% of nonflagellated cells after 120 h of induction) and PIFTD4 (3–5% of nonflagellated cells after 120–240 h of induction; data not shown). These differences correlated with the timing of growth arrest (Figure 5I; see Supplemental Figure S5, A and B, for PIFTF6RNAi), as observed in the case of silencing conventional IFT genes (Figure 5, G and H).
To evaluate the participation of these novel genes in IFT, IFA staining with the anti-IFT172 was performed on the five PIFTRNAi cell lines (Figure 8 and Supplemental Figure S6). Flagellum presence and IFT172 signal were drastically reduced in PIFTA1RNAi, PIFTB2RNAi, and PIFTC3RNAi. In the latter case, an increase of the cytoplasmic signal was consistently observed, indicating a possible redistribution of IFT172 in the cytoplasm (Figure 8B). In contrast, cells with short flagella brightly stained by the anti-IFT172 antiserum were abundant in PIFTD4RNAi and PIFTF6RNAi induced cells. Quantification of the proportion of short cells with bright, weak or no IFT172 signal was performed on all five PIFTRNAi cell lines and revealed that PIFTA1RNAi, PIFTB2RNAi and PIFTC3RNAi behave similarly to IFT88RNAi (complex B), whereas PIFTD4RNAi and PIFTF6RNAi behave similarly to IFT140RNAi (complex A) (Figure 8D). Scanning electron microscopy demonstrated that when visible, short flagella of PIFTA1RNAi were smaller and thinner compared with those from PIFTD4RNAi or PIFTF6RNAi (Supplemental Figure S6). These results support the hypothesis that PIFTA1, PIFTB2, and PIFTC3 proteins are involved in anterograde transport whereas PIFTD4 and PIFTF6 are involved in retrograde transport.
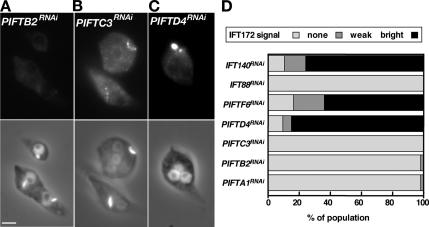
PIFT are involved in anterograde or retrograde transport. PIFTB2RNAi (A) and PIFTC3RNAi (B) induced for 3 d and PIFTD4RNAi (C) induced for 5 d were stained with the anti-IFT172 antiserum. (D) Proportion of short cells displaying no, weak or bright signal with the anti-IFT172 antiserum (n ≥ 100). Bar, 5 μm.
Flagellar Membrane Extension Can Take Place in the Absence of IFT
While analyzing IFTRNAi cells by scanning electron microscopy, we frequently noticed the presence of membrane extensions of various length and shape. To understand the signification of this phenotype, glutaraldehyde-fixed samples from IFT20RNAi, IFT172RNAi (complex B), IFT140RNAi (complex A), PIFTA1RNAi and PIFTF6RNAi cell lines at various stages of RNAi silencing were investigated (Figure 9). At least five categories of short cells could be defined and their proportion was quantified in the various RNAi cell lines analyzed (Figure 9, A and B): 1) “smooth” cells with no visible flagella nor other recognizable structures (however, these could be hidden at the other face of the sample; Figure 9Aa); 2) cells without flagella but with an unusual surface “depression” that might correspond to an “empty” flagellar pocket (identification is difficult without specific markers; Figure 9A, b and c); 3) cells with a “filament” that is often resolved as a close succession of small “balls” or sometimes as large vesicles that emerge from a depression without recognizable flagellar structure (yellow arrowheads on the Figure 9Ad); 4) short flagella (yellow arrow) alone (Figure 9Ae); and 5) short flagella with a thin extension (Figure 9Af). This last case could correspond to the “flagellar sleeve,” an amazing, long and thin extension of the flagellar membrane described for short flagella of the IFT80RNAi mutant (Davidge et al., 2006).
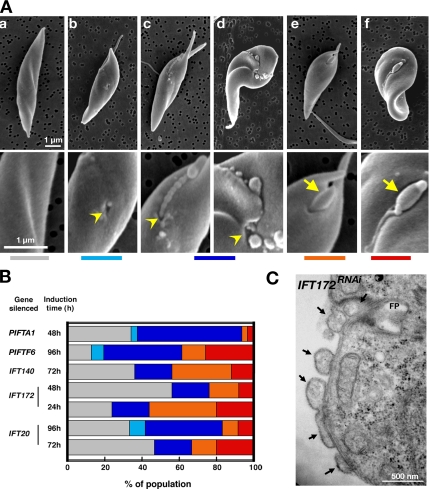
Inhibition of IFT does not prevent flagellum membrane elongation. (A) After RNA silencing of IFT and PIFT genes, five categories of cells were defined, illustrated here in PIFTF6RNAi cells induced for 4 d: cells with no recognizable structures (gray code) (a); cells without flagella but with a surface depression (light blue) (b); cells with tight (c) or disperse (d) rows of vesicles that emerge from a depression without recognizable flagellar structure (dark blue); short flagella alone (orange) (e); and short flagella with a flagellar sleeve (f) as described by Davidge et al. (2006) (red). Yellow arrowheads and arrows indicate cell depressions and short flagella, respectively. (B) Frequency of each cell types in the indicated cell lines (cells with flagella longer than 2 μm were not included). (C) Section through the flagellar sleeve of IFT172RNAi cells induced for 2 d.
As induction of RNAi silencing goes on, the proportion of cells with a short flagellum drops rapidly (Figure 9B). This drop is more severe in the case of knockdown of members of the IFT complex B (IFT20 and IFT172, <25% of flagellated cells) and of PIFTA1 compared with complex A (IFT140, close to 50% of flagellated cells) or PIFTF6 (Figure 9B). Cells with a flagellar sleeve were observed in all IFTRNAi and PIFTRNAi mutants, and in the DHC1bRNAi mutant, indicating that this is a consequence of inhibition of flagellum formation rather than a phenotype specific to IFT80 (Davidge et al., 2006) (Figure 9). Examination of cells that still possess an old flagellum demonstrated that when a new short flagellum is still visible, the flagellar sleeve is present starting from the distal tip of this flagellum and connecting to the old flagellum, presumably at the position of the flagellar connector, a structure that links the tip of the new flagellum to the side of the old flagellum (Moreira-Leite et al., 2001; Kohl et al., 2003; Davidge et al., 2006) (Figure 9C and Supplemental Figures S5C and S6). When no new flagellum was visible, a succession of vesicle-like structures was found at the expected location of the flagellar sleeve (Supplemental Figure S6C). Transmission electron microscopy confirmed that these vesicles emanate from the flagellar pocket and adhere to the cell surface (Figure 9C). These results indicate that inhibition of axoneme construction does not prevent flagellum membrane elongation. This was observed in anterograde transport mutants despite the total inhibition of axoneme assembly (Figure 7F where only a bald basal body is present). These results suggest that separate pathways are acting for formation of the cytoskeletal and the membrane compartment of the flagellum (Davidge et al., 2006).
DISCUSSION
IFT in Flagellum Construction and Maintenance
This work provides the first actual demonstration of intraflagellar transport in T. brucei, and it reveals the activity of this process in both old and new flagella. Three different IFT proteins were found in old and new flagella using different methodologies: IFA with antibodies raised against an endogenous protein (IFT172), expression of tagged protein followed by IFA (IFT20) and direct visualization of an expressed GFP fusion protein (IFT52). IFT particles were identified by transmission electron microscopy between the axoneme doublets and the flagellum membrane. These particles and the IFT proteins found in the flagella are lost upon detergent treatment that removes the membrane of the flagellum, in agreement with protein complexes that are displaced and are not tightly associated to microtubules. This is further supported by the absence of all IFT and PIFT proteins studied here from the proteomic analysis of T. brucei flagella purified with detergent and high salt concentrations (Broadhead et al., 2006).
The restricted location of IFT particles to two sets of specific doublet microtubules (3–4 and 7–8) could be partially explained by physical constraints resulting from the presence of the PFR. This large extra-axonemal structure runs along the axoneme from its point of emergence from the flagellar pocket until its distal tip, and it is tightly linked to the axoneme via physical connections to microtubule doublets 4–7 (Bastin et al., 1996). This could interfere with movement of molecular motors and their cargo along microtubules. However, it does not explain why particles are so infrequent close to doublets 1, 2, and 9. As IFT particles travel in both directions in the flagella, we propose that some doublets serve as specific tracks for anterograde or retrograde transport exclusively, hence reducing the risk of collision and offering the opportunity for precise regulation of each set of motor.
Movement of GFP::IFT52 particles has been visualized in both directions, anterograde tracks being much more abundant. In Chlamydomonas, particles trafficking from tip to base are smaller than those traveling in the opposite direction (Kozminski et al., 1993). If the same was true in trypanosome, retrograde IFT particles could be at the limit of detection. IFT rates are closer to what was reported for Chlamydomonas (Kozminski et al., 1993) compared with C. elegans (Orozco et al., 1999) or to primary cilia of mammalian cells (Follit et al., 2006). Faster IFT could be linked to 1) longer flagella (10 μm for Chlamydomonas and 20 μm for T. brucei, compared with 3–6 μm for the analyzed mammalian or C. elegans cilia); and/or to 2) motile axoneme, where several structures made of protein complexes such as dynein arms and radial spokes are transported for assembly (Qin et al., 2004). Functional studies will be required to understand the reasons for these differences. In trypanosomatids, two distinct genes encode IFT dynein heavy chains (Adhiambo et al., 2005), and functional analysis indicates that both are required for retrograde IFT (our unpublished data).
In addition to the flagellum compartment, IFT proteins are also abundant at the base of the flagellum where they localize to the apical region of the basal body. By contrast to IFT proteins in the flagellum, these proteins are resistant to detergent extraction of the cytoskeleton. This suggests different biochemical properties and could be explained if proteins were sequestered in a complex at the base of the flagellum, as proposed for BBS proteins (Nachury et al., 2007). A slightly different localization was noted for GFP::IFT52 that occurs in a more apical position compared with IFT20 or IFT172. In green algae, IFT52 localizes to fibers that delimitates the flagellum and the cell body cytoplasmic compartments (Deane et al., 2001). Different localizations for IFT proteins in the basal body region have also been reported in Chlamydomonas (Hou et al., 2007).
Importantly, both old and new flagella possess equal concentration of IFT proteins showing the presence of IFT in an organelle that is either in construction (the new flagellum) or in maintenance (the old flagellum) in the same cell. When the new flagellum is assembled, incorporation of new subunits take place at its distal tip, whereas only a small amount of material is turned over in the old flagellum (Bastin et al., 1999a). The presence of IFT material in both flagella indicates that this differential targeting is not due to more prominent IFT activity in the new flagellum.
Knockdown of each individual IFT or PIFT gene leads to a potent inhibition of elongation of the new flagellum but without a visible effect on the existing flagellum, as initially reported for IFT88 and DHC1b (Kohl et al., 2003) and recently for IFT80 (Davidge et al., 2006). Duplication of the basal body is not affected as confirmed by the presence of a short new basal body in all IFTRNAi cell lines analyzed. However, elongation seems more limited. In the case of mutants of retrograde transport (complex A proteins IFT122 and IFT140, DHC1b dynein motor), short flagella can still be assembled in many cells but axoneme microtubules failed to elongate extensively and IFT material accumulates in the flagellar compartment. A similar situation has been encountered in the DHC1b mutants of Chlamydomonas (Pazour et al., 1999; Porter et al., 1999) and of C. elegans (Signor et al., 1999) where retrograde transport is also blocked. No mutants of complex A has been reported to date in the green algae whereas cilia of worms lacking functional proteins of this complex also accumulate IFT proteins (Blacque et al., 2006). However, they are only slightly shorter (~4 μm) compared with wild-type cilia (~6 μm). Inhibition of proteins of the B complex leads to strong inhibition of flagellum or cilium formation in trypanosomes as in Chlamydomonas, C. elegans (Rosenbaum and Witman, 2002), or Tetrahymena (Brown et al., 2003), showing the functional conservation of this process.
In Chlamydomonas, transfer of thermosensitive mutants at the restrictive temperature leads to inactivation of kinesin II within existing flagella, revealing the role of IFT in both flagellum construction and maintenance (Kozminski et al., 1995). In RNAi silencing, only RNA is targeted and existing proteins are lost according to their turnover rate (Bastin et al., 2000a). A constant feature in RNAi silencing of IFT genes in trypanosomes is the persistence of the old flagellum despite the total block in new flagellum formation. Two explanations can be proposed: 1) IFT is not required for flagellum maintenance and 2) IFT proteins engaged in old flagella maintenance are turned over very slowly. Old flagella in wild-type contain IFT particles and IFT proteins, supporting the second hypothesis. Videomicroscopy revealed similar trafficking of GFP::IFT52 in old and new flagella, showing that IFT activity is not slowed down in the old flagellum. In the case of induced IFTRNAi cells, the persisting old flagella still beat and conserve an apparently normal ultrastructure with the 9 + 2 arrangement, dynein arms, and radial spokes. A possible clue to explain this situation could lie in the formation of new flagella in normal cells: they incorporate a large amount of IFT-like material in the flagellar compartment, even before the beginning of axonemal microtubule polymerization, a feature previously reported in Trypanosoma equiperdum (Grassé, 1961). This has been observed in Chlamydomonas where short flagella are filled with IFT material that is progressively spread out along the length of the flagellum (Marshall and Rosenbaum, 2001; Dentler, 2005; Marshall et al., 2005). A flagellum could be assigned a defined amount of IFT protein before assembly and these proteins would circulate within this flagellum compartment with possible exchange with IFT proteins at its basal body but not with the rest of the cytoplasm (and hence not with the new basal body complex). This could explain why RNAi silencing does not reduce significantly the amount of IFT material in the old flagellum and thereby the remarkable long-life of this flagellum.
PIFT Proteins Are Required for Flagellum Formation
The five PIFT genes identified by comparative genomics all turned out to be essential for flagellum formation, indicating the usefulness of this approach, also applied to identify genes involved in flagellum beating (Baron et al., 2007). The PIFT proteins are likely linked to anterograde or retrograde transport, as reported recently for the corresponding genes in C. elegans (Blacque et al., 2005, 2006; Murayama et al., 2005; Efimenko et al., 2006). These genes are conserved in Chlamydomonas, but their encoded proteins are not purified with IFT particles. This could be explained by a lower stoichiometry in the IFT particle or a weaker association that could be disrupted during purification. Alternatively, PIFT proteins could function in assembly or remodeling of the IFT particle without being a structural component per se.
Membrane Trafficking to the Trypanosome Flagellum
In addition to axoneme and PFR components, a significant amount of membrane is required to construct a flagellum. It has been proposed that vesicles are targeted to the base of the flagellar compartment to deliver both membranes and membrane proteins (Rosenbaum and Witman, 2002). In trypanosomes, all trafficking takes place in the flagellar pocket, the only site for endocytosis and exocytosis. Our results show that in the absence of a new flagellum, a flagellar pocket structure remains associated to the bald basal body. This was first noticed by Davidge et al. (2006) who recently reported that a flagellar sleeve was extending from the basal body region of IFT80RNAi-induced cells, passing through the neck of the pocket, and whose tip would be maintained on the existing flagellum by the flagellar connector, a structure that holds the distal end of the new flagellum to the side of the old flagellum (Moreira-Leite et al., 2001). The flagellar sleeve was visible in the seven IFTRNAi or PIFTRNAi mutants examined here, but intermediate structures with succession of vesicles were as frequent. This shows that despite the absence of IFT activity and of microtubule elongation, a flagellar compartment is still defined.
ACKNOWLEDGMENTS
We thank M. Bonhivers and D. R. Robinson (University of Bordeaux, France) for providing antibodies before publication and for numerous discussions; A. Estevez (Instituto de Parasitologia y Biomedicina Lopez-Neyra, Spain) for advise on the tap-tag system; J. Beisson (Centre de Génétique Moleculaire, France), G. Cross (Rockefeller University, New York, NY), P. Englund (Johns Hopkins School of Medicine, Baltimore, MD), K. Gull (University of Oxford, Oxford, United Kingdom), P. Michels (Christian de Duve Institute of Cellular Pathology, Belgium), and H. Nakhasi (Food and Drug Administration, Bethesda, MD) for providing trypanosome cell line, plasmids, or various antibodies. We acknowledge the electron microscopy departments of the Muséum National d'Histoire Naturelle and of the Pasteur Institute for providing access to equipment, and the advise from M. C. Prévost, N. Cayet, and S. Guadagnini. This work was funded by the Centre National de la Recherche Scientifique, by a Programme Protéomique et Génie des Protéines (to D. Robinson and P.B.), an ACI grant in Development Biology, by a Groupement d'Intérêt Scientifique grant on rare genetic diseases (to P.B.) and by sanofi-aventis. S.A. was funded by a Ministère de la Recherche et de l'Enseignement, Fondation pour la Recherche Médicale, and Pasteur-Weizmann fellowships. D.J. is funded by Agence Nationale de la Recherche grant “Maladies Rares” and by a Roux fellowship.
Abbreviations used:
| IFA | immunofluorescence assay |
| IFT | intraflagellar transport |
| MAb | monoclonal antibody |
| PFA | paraformaldehyde |
| PFR | paraflagellar rod |
| PIFT | putatively involved in intraflagellar transport. |
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0749) on December 19, 2007.
REFERENCES
- Absalon S., Kohl L., Branche C., Blisnick T., Toutirais G., Rusconi F., Cosson J., Bonhivers M., Robinson D., Bastin P. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS ONE. 2007;2:e437. [Europe PMC free article] [Abstract] [Google Scholar]
- Adhiambo C., Forney J. D., Asai D. J., LeBowitz J. H. The two cytoplasmic dynein-2 isoforms in Leishmania mexicana perform separate functions. Mol. Biochem. Parasitol. 2005;143:216–225. [Abstract] [Google Scholar]
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. [Abstract] [Google Scholar]
- Baron D. M., Ralston K. S., Kabututu Z. P., Hill K. L. Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 2007;120:478–491. [Abstract] [Google Scholar]
- Bastin P., Ellis K., Kohl L., Gull K. Flagellum ontogeny in trypanosomes studied via an inherited and regulated RNA interference system. J. Cell Sci. 2000a;113:3321–3328. [Abstract] [Google Scholar]
- Bastin P., MacRae T. H., Francis S. B., Matthews K. R., Gull K. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 1999a;19:8191–8200. [Europe PMC free article] [Abstract] [Google Scholar]
- Bastin P., Matthews K. R., Gull K. The paraflagellar rod of kinetoplastida: solved and unsolved questions. Parasitol. Today. 1996;12:302–307. [Abstract] [Google Scholar]
- Bastin P., Pullen T. J., Moreira-Leite F. F., Gull K. Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes Infect. 2000b;2:1865–1874. [Abstract] [Google Scholar]
- Bastin P., Pullen T. J., Sherwin T., Gull K. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 1999b;112:3769–3777. [Abstract] [Google Scholar]
- Bastin P., Sherwin T., Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. [Abstract] [Google Scholar]
- Bell L. R., Stone S., Yochem J., Shaw J. E., Herman R. K. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. [Europe PMC free article] [Abstract] [Google Scholar]
- Berriman M., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. [Abstract] [Google Scholar]
- Blacque O. E., Li C., Inglis P. N., Esmail M. A., Ou G., Mah A. K., Baillie D. L., Scholey J. M., Leroux M. R. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell. 2006;17:5053–5062. [Europe PMC free article] [Abstract] [Google Scholar]
- Blacque O. E., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. [Abstract] [Google Scholar]
- Branche C., Kohl L., Toutirais G., Buisson J., Cosson J., Bastin P. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 2006;119:3443–3455. [Abstract] [Google Scholar]
- Briggs L. J., Davidge J. A., Wickstead B., Ginger M. L., Gull K. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 2004a;14:R611–R612. [Abstract] [Google Scholar]
- Briggs L. J., McKean P. G., Baines A., Moreira-Leite F., Davidge J., Vaughan S., Gull K. The flagella connector of Trypanosoma brucei: an unusual mobile transmembrane junction. J. Cell Sci. 2004b;117:1641–1651. [Abstract] [Google Scholar]
- Broadhead R., et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. [Abstract] [Google Scholar]
- Brown J. M., Fine N. A., Pandiyan G., Thazhath R., Gaertig J. Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Mol. Biol. Cell. 2003;14:3192–3207. [Europe PMC free article] [Abstract] [Google Scholar]
- Cole D. G. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4:435–442. [Abstract] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. [Europe PMC free article] [Abstract] [Google Scholar]
- Collet J., Spike C. A., Lundquist E. A., Shaw J. E., Herman R. K. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. [Europe PMC free article] [Abstract] [Google Scholar]
- Davidge J. A., Chambers E., Dickinson H. A., Towers K., Ginger M. L., McKean P. G., Gull K. Trypanosome IFT mutants provide insight into the motor location for mobility of the flagella connector and flagellar membrane formation. J. Cell Sci. 2006;119:3935–3943. [Abstract] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. [Abstract] [Google Scholar]
- Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol. 2005;170:649–659. [Europe PMC free article] [Abstract] [Google Scholar]
- Dilbeck V., Berberof M., Van Cauwenberge A., Alexandre H., Pays E. Characterization of a coiled coil protein present in the basal body of Trypanosoma brucei. J. Cell Sci. 1999;112:4687–4694. [Abstract] [Google Scholar]
- Durand-Dubief M., Kohl L., Bastin P. Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;129:11–21. [Abstract] [Google Scholar]
- Efimenko E., Blacque O. E., Ou G., Haycraft C. J., Yoder B. K., Scholey J. M., Leroux M. R., Swoboda P. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol. Biol. Cell. 2006;17:4801–4811. [Europe PMC free article] [Abstract] [Google Scholar]
- Escalier D. New insights into the assembly of the periaxonemal structures in mammalian spermatozoa. Biol. Reprod. 2003;69:373–378. [Abstract] [Google Scholar]
- Estevez A. M., Kempf T., Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. [Europe PMC free article] [Abstract] [Google Scholar]
- Follit J. A., Tuft R. A., Fogarty K. E., Pazour G. J. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. [Europe PMC free article] [Abstract] [Google Scholar]
- Grassé P. P. La reproduction par induction du blepharoplaste et du flagelle de Trypanosoma equiperdum. C R Acad. Sci. 1961;252:3917–3921. [Google Scholar]
- Han Y. G., Kwok B. H., Kernan M. J. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr. Biol. 2003;13:1679–1686. [Abstract] [Google Scholar]
- He C. Y., Pypaert M., Warren G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science. 2005;310:1196–1198. [Abstract] [Google Scholar]
- Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., Witman G. B. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 2007;176:653–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. [Abstract] [Google Scholar]
- Kohl L., Bastin P. The flagellum of trypanosomes. Int. Rev. Cytol. 2005;244:227–285. [Abstract] [Google Scholar]
- Kohl L., Robinson D., Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. [Europe PMC free article] [Abstract] [Google Scholar]
- Kohl L., Sherwin T., Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 1999;46:105–109. [Abstract] [Google Scholar]
- Kozminski K. G., Beech P. L., Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. [Europe PMC free article] [Abstract] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. [Europe PMC free article] [Abstract] [Google Scholar]
- Marshall W. F., Qin H., Rodrigo Brenni M., Rosenbaum J. L. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol. Biol. Cell. 2005;16:270–278. [Europe PMC free article] [Abstract] [Google Scholar]
- Marshall W. F., Rosenbaum J. L. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 2001;155:405–414. [Europe PMC free article] [Abstract] [Google Scholar]
- Moreira-Leite F. F., Sherwin T., Kohl L., Gull K. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science. 2001;294:610–612. [Abstract] [Google Scholar]
- Murayama T., Toh Y., Ohshima Y., Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 2005;346:677–687. [Abstract] [Google Scholar]
- Nachury M. V., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. [Abstract] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. [Abstract] [Google Scholar]
- Orozco J. T., Wedaman K. P., Signor D., Brown H., Rose L., Scholey J. M. Movement of motor and cargo along cilia. Nature. 1999;398:674. [Abstract] [Google Scholar]
- Ou G., Koga M., Blacque O. E., Murayama T., Ohshima Y., Schafer J. C., Li C., Yoder B. K., Leroux M. R., Scholey J. M. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell. 2007;18:1564–1569. [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. [Europe PMC free article] [Abstract] [Google Scholar]
- Pazour G. J., Dickert B. L., Witman G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 1999;144:473–481. [Europe PMC free article] [Abstract] [Google Scholar]
- Pedersen L. B., Geimer S., Rosenbaum J. L. Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr. Biol. 2006;16:450–459. [Abstract] [Google Scholar]
- Pedersen L. B., Miller M. S., Geimer S., Leitch J. M., Rosenbaum J. L., Cole D. G. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 2005;15:262–266. [Abstract] [Google Scholar]
- Piperno G., Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 1997;94:4457–4462. [Europe PMC free article] [Abstract] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. [Europe PMC free article] [Abstract] [Google Scholar]
- Pradel L. C., Bonhivers M., Landrein N., Robinson D. R. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J. Cell Sci. 2006;119:1852–1863. [Abstract] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. [Europe PMC free article] [Abstract] [Google Scholar]
- Ralston K. S., Lerner A. G., Diener D. R., Hill K. L. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell. 2006;5:696–711. [Europe PMC free article] [Abstract] [Google Scholar]
- Redmond S., Vadivelu J., Field M. C. RNAit: an automated Web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;128:115–118. [Abstract] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. [Abstract] [Google Scholar]
- Sanders M. A., Salisbury J. L. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1994;124:795–805. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarpal R., Todi S. V., Sivan-Loukianova E., Shirolikar S., Subramanian N., Raff E. C., Erickson J. W., Ray K., Eberl D. F. Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 2003;13:1687–1696. [Abstract] [Google Scholar]
- Selvapandiyan A., Kumar P., Morris J. C., Salisbury J. L., Wang C. C., Nakhasi H. L. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol. Biol. Cell. 2007;18:3290–3301. [Europe PMC free article] [Abstract] [Google Scholar]
- Sherwin T., Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond B Biol. Sci. 1989;323:573–588. [Abstract] [Google Scholar]
- Signor D., Wedaman K. P., Orozco J. T., Dwyer N. D., Bargmann C. I., Rose L. S., Scholey J. M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 1999;147:519–530. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. [Abstract] [Google Scholar]
- Snow J. J., Ou G., Gunnarson A. L., Walker M. R., Zhou H. M., Brust-Mascher I., Scholey J. M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. [Abstract] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., Schuyler M. W., Collet J., Thomas J. H., Riddle D. L. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E., Englund P. T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. [Abstract] [Google Scholar]
- Wirtz E., Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1183. [Abstract] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.e07-08-0749
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2262991?pdf=render
HAL Open Archive
http://hal-pasteur.archives-ouvertes.fr/pasteur-00217549
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Whole cell reconstructions of Leishmania mexicana through the cell cycle.
PLoS Pathog, 20(2):e1012054, 28 Feb 2024
Cited by: 1 article | PMID: 38416776 | PMCID: PMC10927142
Monoallelic pathogenic IFT140 variants are a common cause of autosomal dominant polycystic kidney disease-spectrum phenotype.
Clin Kidney J, 17(2):sfae026, 15 Feb 2024
Cited by: 1 article | PMID: 38404363 | PMCID: PMC10894029
Genome-wide investigation to assess copy number variants in the Italian local chicken population.
J Anim Sci Biotechnol, 15(1):2, 03 Jan 2024
Cited by: 1 article | PMID: 38167097 | PMCID: PMC10763469
Implications of Flagellar Attachment Zone Proteins TcGP72 and TcFLA-1BP in Morphology, Proliferation, and Intracellular Dynamics in Trypanosoma cruzi.
Pathogens, 12(11):1367, 18 Nov 2023
Cited by: 1 article | PMID: 38003831 | PMCID: PMC10675206
Matching variants for functional characterization of genetic variants.
G3 (Bethesda), 13(12):jkad227, 01 Dec 2023
Cited by: 0 articles | PMID: 37933433 | PMCID: PMC10700107
Go to all (114) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Bidirectional intraflagellar transport is restricted to two sets of microtubule doublets in the trypanosome flagellum.
J Cell Biol, 217(12):4284-4297, 01 Oct 2018
Cited by: 19 articles | PMID: 30275108 | PMCID: PMC6279389
Intraflagellar transport is required for the maintenance of the trypanosome flagellum composition but not its length.
J Cell Sci, 129(15):3026-3041, 24 Jun 2016
Cited by: 24 articles | PMID: 27343245
The GTPase IFT27 is involved in both anterograde and retrograde intraflagellar transport.
Elife, 3:e02419, 24 Apr 2014
Cited by: 38 articles | PMID: 24843028 | PMCID: PMC4003483
[The flagellum: from cell motility to morphogenesis].
J Soc Biol, 197(4):379-387, 01 Jan 2003
Cited by: 1 article | PMID: 15005520
Review
 *†
*†




