Abstract
Background
Association studies have examined the significance of several candidate genes based on biological pathways relevant to prostate carcinogenesis, including both the androgen and insulin-like growth factor pathways. Clinical and epidemiologic evidence suggest that androgens, specifically testosterone and dihydrotestosterone (DHT) are important not only in normal prostate growth but in the pathogenesis of prostate cancer. Similarly, the insulin-like growth factor-1 (IGF-1) signaling pathway regulates both cellular proliferation and apoptosis. Therefore, genes involved in the biosynthesis, activation, metabolism and degradation of androgens and the stimulation of mitogenic and antiapoptotic activities of prostate epithelial cells represent important candidates for affecting the development and progression of prostate cancer.Methods
Using resources from the Flint Men's Health Study, a population-based case control study of African-American men aged 40-79, we evaluated the associations between selected single-nucleotide polymorphisms (SNPs) in the CYP17, CYP3A4, CYP19A1, SDR5A2, IGF1, and IGFBP3 genes and prostate cancer diagnosis in 473 men (131 prostate cancer cases and 342 disease-free controls).Results
We found a significant association between prostate cancer and selected CYP17 SNP genotypes, with the heterozygous genotype conferring decreased risk. Suggestive evidence for association between IGF1 SNPs and prostate cancer were also found. No significant associations were observed between SNPs in the other genes and prostate cancer.Conclusions
These findings suggest that variation in or around CYP17 and/or IGF1 may be associated with prostate cancer development in the African-American population. Additional studies are needed to determine whether these polymorphisms are indeed associated with prostate cancer risk in African Americans.Free full text

Genetic Polymorphisms in CYP17, CYP3A4, CYP19A1, SRD5A2, IGF-1, and IGFBP-3 and Prostate Cancer Risk in African-American Men: The Flint Men’s Health Study
Abstract
BACKGROUND
Association studies have examined the significance of several candidate genes based on biological pathways relevant to prostate carcinogenesis, including both the androgen and insulin-like growth factor pathways. Clinical and epidemiologic evidence suggest that androgens, specifically testosterone and dihydrotestosterone (DHT) are important not only in normal prostate growth but in the pathogenesis of prostate cancer. Similarly, the insulin-like growth factor-1 (IGF-1) signaling pathway regulates both cellular proliferation and apoptosis. Therefore, genes involved in the biosynthesis, activation, metabolism and degradation of androgens and the stimulation of mitogenic and antiapoptotic activities of prostate epithelial cells represent important candidates for affecting the development and progression of prostate cancer.
METHODS
Using resources from the Flint Men’s Health Study, a population-based case control study of African-American men aged 40–79, we evaluated the associations between selected single-nucleotide polymorphisms (SNPs) in the CYP17, CYP3A4, CYP19A1, SDR5A2, IGF1, and IGFBP3 genes and prostate cancer diagnosis in 473 men (131 prostate cancer cases and 342 disease-free controls).
RESULTS
We found a significant association between prostate cancer and selected CYP17 SNP genotypes, with the heterozygous genotype conferring decreased risk. Suggestive evidence for association between IGF1 SNPs and prostate cancer were also found. No significant associations were observed between SNPs in the other genes and prostate cancer.
CONCLUSIONS
These findings suggest that variation in or around CYP17 and/or IGF1 may be associated with prostate cancer development in the African-American population. Additional studies are needed to determine whether these polymorphisms are indeed associated with prostate cancer risk in African Americans.
INTRODUCTION
Prostate cancer is the most common cancer among men in the United States with an expected 218,890 new cases and 27,050 deaths in 2007 [1]. In addition to increasing age, the etiology of prostate cancer suggests that race is one of the most important recognized risk factors for the disease [2]. African-American men have an approximately 1.6-fold greater chance of being diagnosed with prostate cancer compared to Caucasian men and a 2.5-fold greater chance of dying from the disease [3]. Numerous studies have demonstrated that positive family history is a strong risk factor for prostate cancer [4,5]. Genetic association studies are designed to identify genetic variants that increase the risk of developing disease. Unfortunately, despite their increased risk, African Americans are typically under-represented in genetic association studies of prostate cancer.
Prostate cancer association studies have focused on candidate genes based on biological pathways relevant to prostate carcinogenesis, including both the androgen and insulin-like growth factor pathways. Clinical and epidemiologic evidence suggest that androgens, specifically testosterone and dihydrotestosterone (DHT) are important not only in normal prostate growth but in the pathogenesis of prostate cancer. Androgens exert their effect via binding to the androgen receptor (AR) which subsequently acts as a transcriptional modifier of a variety of genes by binding to an androgen response element [6]. Polymorphic variants have been described in genes involved in the biosynthesis, activation, metabolism and degradation of androgens [7]. The insulin-like growth factor-1 (IGF-1) signaling pathway regulates both cellular proliferation and apoptosis [8]. Bioavailability of IGF-1 is modulated by a family of IGF binding proteins with over 90% of circulating IGF-1 bound in a complex with IGF binding protein-3 (IGFBP-3) [8]. Epidemiologic evidence suggests that higher serum levels of IGF-1 compared to IGFBP-3 are associated with increased risk of prostate cancer [9]. Polymorphic alleles in the CYP17, CYP3A4, CYP19, 5α-reductase, IGF-1 and IGFBP-3 genes have been associated with prostate cancer in several studies [7].
SRD5A2, located on chromosome 2 (2p23), encodes the steroid 5α-reductase type 2 enzyme, a membrane-bound enzyme which catalyzes the irreversible conversion of testosterone into the main prostatic androgen, DHT [10]. Certain SRD5A2 polymorphisms may encode for 5α-reductase variants with different activities, likely attributable to altered mRNA stability [11]. Levels of 5α-reductase-2 expression are greater in prostate cancer cells than in benign prostatic cells [12]. It has been hypothesized that polymorphic variants of the SRD5A2 gene influence 5α-reductase-2 activity. Notably, a substitution of valine to leucine in codon 89 in exon 1 of the 5α-reductase enzyme gene has been associated with varying levels of 5α-reductase-2 activity and with prostate cancer risk [13,14].
Two genes which encode for the cytochrome p-450 enzymes, CYP17, located on chromosome 10 (10q24.32) and CYP3A4 located on chromosome 7 (7q21.1), influence the rate of androgen metabolism [15,16]. Specifically, the CYP17 enzyme is the rate-limiting step in biosynthesis from 17-α hydroxylase and 17,20-lyase [15]. The thymine to cytosine substitution in the 5′ untranslated promoter region of the CYP17 gene has been associated with a 1.2- to 2.8-fold increased risk of prostate cancer in several studies [17,18]. Early studies suggested that the T to C substitution results in an additional Sp1-binding site (CCACC box) in the promoter region of the gene thus influencing androgen concentrations [19]. The CYP3A4 gene involved in the oxidation of testosterone metabolizes the hormone to less active metabolites [16]. A germ-line variant in the 5′ regulatory region of the CYP3A4 gene that substitutes an alanine for a glycine at codon 293 has been associated with a 1.7- to 9.5-fold increase in risk for prostate cancer [16,20].
The CYP19 gene is located on chromosome 15 (15q21.1) and encodes the enzyme aromatase that catalyzes the irreversible conversion of androstenedione to estrone and testosterone to estradiol [21]. Aromatase is present in the gonads and in the extra-gonadal tissue, including the prostate and adipose tissue. Aromatase mRNA and protein have both been detected in benign prostatic hyperplasia (BPH) and prostate cancer tissue [21]. Few studies have investigated the role of CYP19 SNPs in the development of prostate cancer [22,23].
Polymorphisms in the IGF-1 and IGFBP-3 genes have been found to be associated with serum IGF-1 and IGFBP-3 concentrations [24]. As serum concentrations of these growth factors and binding proteins have been demonstrated to influence prostate cancer risk [9], it has been hypothesized that these polymorphisms would be associated with increased risk of disease. Previous studies that have investigated the role of genetic variation in IGF-1 and IGFBP-3 in relation to prostate cancer risk have focused solely on a (CA)n in the IGF-1 gene [7] and a single polymorphism in the promoter region of the IGFBP-3 gene [24].
Given the increased risk of prostate cancer in African-American men and the under-representation of African Americans in past genetic association studies, the goal of the current study was to investigate the association between prostate cancer and androgen and IGF-common and haplotype mapping selected single-nucleotide polymorphisms (SNPs) in the CYP17, CYP3A4, CYP19A1, SDR5A2, IGF-1, and IGFBP-3 genes using samples from the Flint Men’s Health Study. This study of genes involved in the androgen and IGF pathways should provide additional and important insight into the genetic basis of the development and progression of prostate cancer.
MATERIALS AND METHODS
The Flint Men’s Health Study
In 1996, a probability sample of 943 African-American men was selected from households located in Genesee County, Michigan to participate in a study of prostate cancer [25,26]. Of the 817 men who agreed to participate (87% response rate), 87 were determined to be ineligible due to a history of prostate cancer or a prior operation on the prostate gland and excluded from the study. Trained interviewers from the University of Michigan Institute for Social Research contacted the 730 eligible subjects, and performed a detailed in-home interview which covered information on potential risk factors for prostate cancer; general health and medical history; and socio-demographic information. At the conclusion of the interview, voluntary participation in a comprehensive urologic examination (digital rectal exam (DRE), transrectal ultrasound (TRUS) and a screening serum PSA measurement), was elicited. Prostate biopsy was recommended in individuals with an elevated PSA(>4.0 ng/ml) or suspicious DRE. 379 of the 730 men who completed the interview participated in the clinical examination component of the study. After exclusion of men who were determined to be biopsy positive for prostate cancer at baseline and/or who subsequently developed prostate cancer after baseline (included in the present analyses as cases), a sufficient DNA sample was available for genotyping on 342 of the remaining controls.
Prostate cancer case recruitment from the same community was initiated in 1999. Eligible men include those who were between the ages of 40–79 at time of prostate cancer diagnosis (between 1995 and 2002). Cases completed a detailed epidemiologic interview as described above for controls and provided a blood sample. Diagnosis of prostate cancer was confirmed by review of pathology reports or medical records, and age at diagnosis calculated from the date of the first biopsy positive for prostate cancer.
A total of 136 cases were ultimately recruited to participate in the study. A sufficient DNA sample was available for genotyping on 131 cases. Informed consent was obtained from all study participants and the research protocol was approved by the University of Michigan Institutional Review Board. For both cases and controls, genomic DNA was isolated from whole blood using the Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN).
SNP Selection and Genotyping Methods
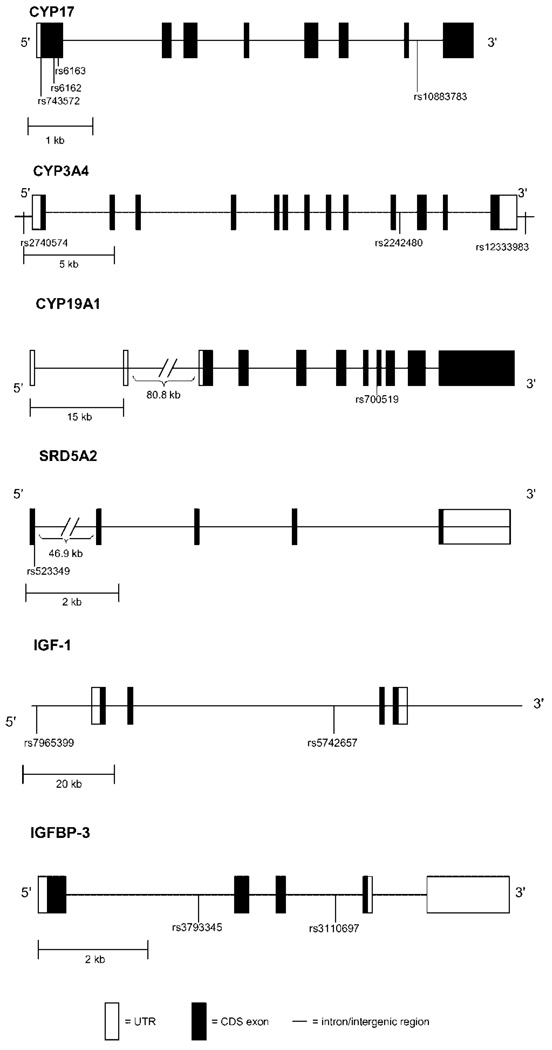
We selected 18 SNPs in the CYP3A4 (3 SNPs), CYP17 (4 SNPs), CYP19A1 (1 SNP), SDR5A2 (3 SNPs), IGF1 (5 SNPs), and IGFBP3 (2 SNPs) genes for genotyping in the FMHS sample. Eleven of these SNPs were previously reported to be associated with prostate cancer [7,21,27] and breast cancer [28] status. Two SNPs in the SDR5A2 gene (rs9282858, rs9332964) and 3 SNPs in the IGF-1 gene (rs1106381, rs12423791, rs5742723) were homozygous for the wild type allele in all subjects and therefore are not included leaving a total of 13 SNPs for the current analyses (Fig. 1).
Genotyping was performed by BioServe Biotechnologies Ltd. (Laurel, MD) using the MassARRAY iPLEX™ platform, a PCR process and mass spectrometry based system (Sequenom, CA). The genotyping assays were custom developed using the MassARRAY Assay Designer 3.0. All PCR and extension primers were synthesized at BioServe. Four positive controls for each assay and four no-template controls were included per 384-well plate. The quality control used for this high-throughput genotyping consists of repeated assays on ~10% of randomly selected samples from each experiment as well as the inclusion of blinded controls. The genotyping results of the DNA as a “sample” and as a “quality control duplicated sample” were compared to obtain an overall concordance level of 99.45%.
Statistical Analysis
For each SNP, the observed genotype distribution was tested for consistency with Hardy–Weinberg equilibrium expected proportions using Pearson’s chi-square test in SAS (SAS version 9.1, Cary NC). Lewontin’s D’ statistic [29] and the squared correlation statistic Δ2 [30] were used to estimate the degree of linkage disequilibrium and correlation between all possible pair-wise combinations of SNPs in the same gene using the computer software GOLD (www.sph.u-mich.edu/csg/abecasis/gold/index.html). Estimated proportion of African ancestry for each study participant was obtained using the statistical software Structure [31] as described previously [32]. To avoid potential bias due to the possibility of the younger sample of controls being diagnosed with prostate cancer at a later age, age was calculated based on the most recent date of follow-up for both cases and controls (July 29, 2002). For controls who died before this date, date of death was used to calculate age.
SNP association
For each SNP, unconditional multivariable logistic regression models were used to test whether SNP alleles/genotypes were associated with prostate cancer. Specifically, we conducted both allelicbased (using 1 degree of freedom likelihood ratio tests) and genotype-based (using 2 degree of freedom likelihood ratio tests) association tests. For the genotype-based association tests, no genetic mode-of-inheritance models were assumed. Two covariate-adjusted models were analyzed: the first adjusting for age and the second adjusting for age and estimated percent African ancestry.
Haplotype analysis
Tests of haplotype effects were performed using a score test developed by Schaid et al. [33] and implemented in Haplo.Stat [33]. This approach uses EM derived haplotype frequency estimates to assign probabilistic weights to every haplotype pairing (diplotype) that are consistent with the observed genotype data rather than assigning the “most likely” diplotype to an individual. Statistical differences in overall haplotype frequencies (excluding haplotypes with frequency less than 3%) were tested for association with prostate cancer. Covariate model adjustment was made for age, and for both age and estimated proportion of African descent. In addition to testing for an overall difference in haplotype frequencies, specific individual haplotype effects were also tested.
RESULTS
The sample consisted of 473 (131 prostate cancer cases, 342 disease-free controls) African-American subjects with both genotype and phenotype data. Mean age overall was 63.5 years (SD = 10.0) with cases being older than controls (cases mean age = 67.2 years, SD = 8.6; controls mean age = 62.1 years, SD = 10.1; P<0.0001). 21.4% of cases and 17.0% of controls reported a family history of prostate cancer in a first degree relative. There was no statistical difference in mean percent African descent between cases (70.5%) and controls (70.6%). Characteristics for the 131 prostate cancer cases are presented in Table I. The positions and corresponding observed genotype frequencies of the identified SNPs in all 473 FMHS case and control subjects are depicted in Table II.
TABLE I
Characteristics of Men With Prostate Cancer (n = 131)
| Characteristic | Mean (SD) or n (%) |
|---|---|
| Age at diagnosis (years) | 63.5 (8.7) |
| Serum prostate-specific antigen at diagnosis (ng/ml) | 24.3 (106.3) |
| Surgery (% yes) | 63 (48.1%) |
| Family history (% yes) | 28 (21.4%) |
| Stage | |
| Localized (T1) | 29 (23.0%) |
| Locally advanced (T2) | 78 (61.9%) |
| Regional (T3) | 19 (15.1%) |
| Gleason | |
| 2–6 | 37 (28.9%) |
| 7 | 77 (60.2%) |
| 8–10 | 14 (10.9%) |
| Clinically aggressive prostate cancer (% yes) | 95 (72.5%) |
TABLE II
Frequencies of Selected SNPs Among 131 Men With Prostate Cancer and 342 Controls
| SNP/genotype Chromosome | Chromosome | Location of SNP | Nucleotide change | Prostate cancer cases (n = 131) | Controls (n = 342) | Allelic-based age adjusted (1 df) P-value | Genotype-based age adjusted (2 df) P-value | Age adjusted OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| CYP17 | ||||||||
| rs10883783 | 10q24.3 | Intron 7 | A to T | |||||
| AA | 89 (67.94) | 231 (67.74) | — | |||||
| AT | 36 (27.48) | 97 (28.45) | 0.93 (0.58, 1.49) | |||||
| TT | 6 (4.58) | 13 (3.81) | 0.99 | 0.88 | 1.22 (0.44, 3.39) | |||
| rs6163 | 10q24.3 | Exon 1 (Codon 65) | G to T | |||||
| GG | 54 (41.22) | 114 (33.43) | — | |||||
| GT | 52 (39.69) | 192 (56.31) | 0.56 (0.35, 0.88) | |||||
| TT | 25 (19.08) | 35 (10.26) | 0.83 | 0.0014 | 1.59 (0.85, 2.98) | |||
| rs6162 | 10q24.3 | Exon 1 (Codon 46) | C to T | |||||
| CC | 53 (40.46) | 112 (33.23) | — | |||||
| CT | 53 (40.46) | 190 (56.38) | 0.57 (0.36, 0.90) | |||||
| TT | 25 (19.08) | 35 (10.39) | 0.80 | 0.0018 | 1.59 (0.85, 2.97) | |||
| rs743572 | 10q24.3 | 5′-UTR | T to C | |||||
| TT | 52 (41.27) | 106 (32.92) | — | |||||
| CT | 51 (40.48) | 183 (56.83) | 0.55 (0.35, 0.88) | |||||
| CC | 23 (18.25) | 33 (10.25) | 0.97 | 0.0028 | 1.51 (0.79, 2.88) | |||
| CYP3A4 | ||||||||
| rs12333983 | 7q21.1 | 3′-UTR | A to T | |||||
| AA | 55 (44.00) | 141 (43.52) | — | |||||
| AT | 57 (45.60) | 139 (42.90) | 1.11 (0.71, 1.74) | |||||
| TT | 13 (10.40) | 44 (13.58) | 0.75 | 0.62 | 0.79 (0.39, 1.60) | |||
| rs2242480 | 7q21.1 | Intron 10 | A to G | |||||
| AA | 67 (51.15) | 183 (53.51) | — | |||||
| AG | 53 (40.46) | 133 (38.89) | 1.07 (0.69, 1.66) | |||||
| GG | 11 (8.40) | 26 (7.60) | 0.55 | 0.82 | 1.28 (0.59, 2.77) | |||
| rs2740574 | 7q21.1 | 5′-UTR (−293) | G to A | |||||
| GG | 45 (35.16) | 150 (44.25) | — | |||||
| GA | 66 (51.56) | 149 (43.95) | 1.51 (0.96, 2.37) | |||||
| AA | 17 (13.28) | 40 (11.80) | 0.14 | 0.20 | 1.39 (0.71, 2.72) | |||
| CYP19 | ||||||||
| rs700519 | 15q21.1 | Exon 8 (Codon 264) | C to T | |||||
| CC | 91 (69.47) | 236 (69.21) | — | |||||
| CT | 37 (28.24) | 93 (27.27) | 0.97 (0.61, 1.53) | |||||
| TT | 3 (2.29) | 12 (3.52) | 0.67 | 0.85 | 0.68 (0.18, 2.61) | |||
| SRD5A2 | ||||||||
| rs523349 | 2p23 | Exon 1 (Codon 89) | G to C | |||||
| GG | 71 (54.20) | 177 (51.91) | — | |||||
| GC | 51 (38.93) | 136 (39.88) | 0.86 (0.56, 1.34) | |||||
| CC | 9 (6.87) | 28 (8.21) | 0.36 | 0.65 | 0.72 (0.32, 1.64) | |||
| IGF-1 | ||||||||
| rs5742657 | 12q22-q23 | Intron 2 | A to G | |||||
| AA | 97 (74.62) | 271 (82.12) | — | |||||
| AG | 30 (23.08) | 58 (17.58) | 1.57 (0.94, 2.63) | |||||
| GG | 3 (2.31) | 1 (0.30) | 0.020 | 0.058 | 7.69 (0.77, 77.10) | |||
| rs7965399 | 12q22-q23 | 5′ | T to C | |||||
| TT | 62 (47.69) | 181 (54.85) | — | |||||
| TC | 56 (43.08) | 128 (38.79) | 1.42 (0.91, 2.21) | |||||
| CC | 12 (9.23) | 21 (6.36) | 0.056 | 0.16 | 1.80 (0.82, 3.95) | |||
| IGFBP-3 | ||||||||
| rs3110697 | 7p13-p12 | Intron 3 | G to A | |||||
| GG | 53 (40.46) | 121 (35.59) | — | |||||
| GA | 61 (46.56) | 159 (46.77) | 0.92 (0.58, 1.44) | |||||
| AA | 17 (12.98) | 60 (17.65) | 0.28 | 0.53 | 0.69 (0.36, 1.31) | |||
| rs3793345 | 7p13-p12 | Intron 1 | T to C | |||||
| TT | 89 (67.94) | 201 (58.94) | — | |||||
| TC | 39 (29.77) | 125 (36.66) | 0.75 (0.48, 1.18) | |||||
| CC | 3 (2.29) | 15 (4.40) | 0.11 | 0.25 | 0.44 (0.12, 1.63) |
The observed distributions for genotype data for the CYP3A4, CYP19A1, SDR5A2, IGF1, and IGFBP3 SNPs were found to be consistent with expected Hardy–Weinberg equilibrium proportions in both the case and control samples (P>0.05, data not shown). Observed genotype frequencies for some CYP17 SNPs were, however, not consistent with Hardy–Weinberg equilibrium. Specifically, SNPs rs6163, rs6162, and rs743572 were all found to have genotypes with frequencies that were inconsistent with Hardy–Weinberg equilibrium (P = 0.001, for all three) in the larger control group. There was little evidence of historical recombination between these three CYP17 SNPs (estimated pair-wise D’ and Δ 2 estimates ranged from 0.99 to 1.0) in the control sample. CYP17 SNP rs10883783 genotype distribution was consistent with Hardy–Weinberg equilibrium (P = 0.57). There was no evidence for recombination, but the estimated degree of correlation was modest, between rs10883783 and the three aforementioned CYP17 SNPs (estimated D’ = 1.0 and Δ2 = 0.35 between rs10883783 and rs743572 in the control sample).
Statistical analyses showed that genotype (using 2 df test) was significantly associated with prostate cancer for CYP17 rs6163 (P = 0.0014), rs6162 (P = 0.0018), and rs743572 (P = 0.0028) SNPs, even after adjustment for multiple tests (conservative Bonferroni threshold value for family-wise error rate of alpha = 0.05 is P<0.0038 = 0.05/13; Table II). Specifically, heterozygotes were significantly at decreased risk of having prostate cancer compared to those with the homoyzygous wild type genotypes after adjustment for age. Further adjustment for percent African descent negligibly changed these results (data not shown). Because these three SNPS were in almost perfect linkage disequilibrium, the evidence for association between these SNPs and prostate cancer was nearly identical. No significant differences in genotype frequencies for CYP17 SNP rs10883783 or any SNPs in the CYP3A4, CYP19A1, SRD5A2 or IGFBP-3, genes were observed between prostate cancer cases and controls. A single IGF-1 SNP, rs5742657, demonstrated a trend (P = 0.058) towards an association between genotype and prostate cancer.
There were no statistically significant differences in allele frequencies, after adjustment for multiple tests, for any of the SNPs studied (Table II). In fact, the difference in allele frequencies between prostate cancer cases and controls for CYP17 SNPs rs6163, rs6162 and rs743572 were very modest (Table II) and thus the statistical significance observed for the genotype-based association tests for these SNPs was nearly entirely driven by the distribution of alleles into the different genotype categories within each group and not by differences in allele frequencies between the two groups. The marginal evidence for an association between IGF-1 SNP rs5742657 and prostate cancer increased modestly using the 1 df allelic association test (P = 0.02).
In order to examine whether specific phased combinations of allelic variants were associated with prostate cancer status, we performed haplotype-based association analyses. Consistent with the 1 df single SNP analyses, no evidence for haplotype associations with prostate cancer were detected with CYP17, CYP3A4, CYP19A1, SDR5A2, and IGFBP3 (data not shown). Modest evidence for association between IGF-1 haplotypes, defined by SNPs rs7965399 and rs5742657, and prostate cancer were observed after adjustment for age (P = 0.052) and age + percent African ancestry (P = 0.061; data not shown).
DISCUSSION
Using data from the FMHS, a population-based case control study among 473 African-American men aged 40–79, we sought to determine whether polymorphisms in the CYP17, CYP3A4, CYP19A1, SDR5A2, IGF1, and IGFBP3 genes were associated with prostate cancer. All six genes were examined because of their involvement in either (1) the synthesis and conversion of testosterone to dihydrotestosterone and estradiol (androgen pathway) or (2) the stimulation of mitogenic and antiapoptotic activities of prostate epithelial cells (insulin-like growth factor pathway) and subsequent potential influence on disease risk. Our most interesting results were for SNPs in CYP17. In this study, we observed significant associations between CYP17 SNPs rs6163, rs6162, and rs743572 genotypes and prostate cancer status. Specifically, we observed a strong decreased risk for prostate cancer in heterozygotes for these three CYP17 SNPs. However, we did not observe any difference in allele frequencies between case and controls for these three CYP17 SNPs. Previous evidence regarding associations between CYP17 SNPs and prostate cancer has been mixed [34], however this is the first observation to our knowledge that suggests a protective effect of being heterozygous. Although this finding has not been observed by others, it should be noted that there have been very limited studies on African-American prostate cancer cases for any of these genes, including CYP17 [35]. As noted previously, these three CYP17 SNPs had genotype distributions in the controls that were not consistent with Hardy–Weinberg equilibrium. We took considerable efforts to validate the genotype data for these three SNPs, including retyping one of the SNPs in all samples using a Taqman SNP Genotyping Assay (Applied Biosystems, Foster City, CA) as well as directly sequencing ~20% of the samples. We note that the three SNPs are known from HapMap data to be in strong LD and our data were consistent with this previous finding. We also note that the validity of our statistical significance estimates from our 2 df statistical tests do not depend on the SNPs being in HWE.
CYP3A4 encodes an enzyme that catalyses the 6β-hydroxylation of testosterone, suggesting that it may be involved in the oxidative metabolism of testosterone. Genetic variants that affect CYP3A4 activity could result in increased androgen levels. Previous studies have reported that the CYP3A4 variants are positively associated with prostate cancer [16]. Specifically, among white and African-American subjects with prostate cancer, higher clinical grade and stage were significantly associated with the frequency of this allele [20]. Furthermore, racial variations in the frequency of CYP3A4 alleles have also been reported [34]. However, consistent evidence regarding the contribution of alterations of CYP3A4 to prostate cancer risk is lacking and has led to the hypothesis that prior observations between prostate cancer and CYP3A4 may be attributed to LD with other genes, possibly CYP3A5 [36]. In this study, we found no evidence of an association between prostate cancer and selected CYP3A4 SNPs.
The SRD5A2 gene product, 5α-reductase type II, catalyzes the conversion of testosterone to dihydrotestosterone (DHT), its active metabolite. The possibility that different alleles of the SRD5A2 gene may be associated with different levels of 5α-reductase type II activity has been examined in numerous studies [13,37–39]. Furthermore, a higher SRD5A2 activity in African-American menhas been reported and supports observed ethnic differences in prostate cancer risk [40]. One missense substitution, a G to C transversion resulting in a valine to leucine substitution at codon 89 (V89L substitution, rs523349) has been hypothesized to reduce prostate cancer risk [37]. Although the V89L substitution is particularly common among Asians and purported to explain the low risk of prostate carcinoma in this population, several studies in white men have been unable to detect an association between the V89L variant and prostate cancer [41]. We observed no association between V89L genotypes and prostate cancer diagnosis in this population of African- American men.
The CYP19A1 gene encodes the enzyme aromatase which plays a key role in the conversion of androgen to estrogen [21]. As the prostate is influenced by estrogen from peripheral sources as well as through aromatase activity within its stroma, it has been suggested that genetic variations in the aromatase genes, that is, CYP19A1 alter an individual’s risk of prostate cancer [21]. CYP19A1 has been less well studied and although a few reports have suggested an association between the CYP19A1 R264C alteration (rs700519) and prostate cancer risk [23], other studies, including the current study were unable to replicate these findings.
The IGF-1 gene regulates cell proliferation, differentiation, apoptosis and transformation via the IGF-1 receptor with both paracrine and autocrine mechanisms and is required for the development of the prostate gland and reported to be involved with prostate cancer [8]. IGF-1 also activates the androgen receptor directly in the absence of androgens suggesting that IGF-1 may be responsible for prostate cancer cell proliferation in both an androgen-dependent and - independent manner [42]. Previous studies that have investigated the role of genetic variation in the IGF-1 gene in relation to prostate cancer risk have focused on a (CA)n repeat sequence located approximately 1 kilobase (kb) upstream from the IGF-1 transcription start site. Results of these studies have been inconsistent. Only one study to our knowledge (The Multiethnic Cohort Study) has since examined more systematically the genetic variation at the IGF-1 locus [43]. Our findings are consistent with Cheng et al. [43]in that we observed differences in allele frequencies between cases and controls for SNPs rs7965399 and rs5742657. The direction of the effects, with increased risk for individuals with at least one copy of the variant allele, were the same as those reported in the African-American subset of the Multiethnic Cohort Study. Cheng et al. [43] observed strong correlations between variants across the locus suggesting the existence of one signal that was detected at several sites. Interestingly, the strongest signal was located in block 1, a noncoding region which also contains the (CA)n repeat polymorphism previously identified to be associated with prostate cancer risk. Therefore it is possible that the (CA)n repeat allele is in linkage disequilibrium with the other SNPs examined.
IGFBP-3, which is a major circulating IGF binding protein, binds to IGF-1, forming a complex that limits the IGF-1 bioavailability for binding to the IGF-1 receptor [8]. IGFBP-3 suppresses mitogenic and anti-apoptopic action of IGF-1 and is therefore, associated with decreased risks of prostate cancer [8]. Further-more, plasma IGFBP-3 concentrations have been reported to be lower in African-American men which may partly explain the greater incidence of disease in this population [44]. Although a recent Physician’s Health Study report observed that the presence of A/C polymorphism at position −202 in the promoter region of the gene was correlated with circulating levels of IGFBP-3, a study of U.S. veterans found no association between the IGFBP-3 C allele and prostate cancer. We found no association between our IGFBP-3 selected SNPs and prostate cancer status. There are no other studies to our knowledge that have examined these SNPs in relation to prostate cancer to date.
As described above, the associations between variants in CYP17, CYP3A4, CYP19A1, SDR5A2, IGF-1, and IGFBP-3 and prostate cancer have been assessed in previous studies however, the findings are inconclusive. There are a number of possible explanations for the different results observed. Many of these studies have been limited by inclusion of primarily Caucasian men and have reported findings on specific individual variants in the candidate genes. Variation in study design might also account for some of the inconsistency between findings.
There are several limitations in our study which must be addressed. First, the relatively small sample size potentially limits our statistical power to detect small genotype effects on prostate cancer status. Second, we genotyped a relatively small number of SNPs within the selected genes and it is possible that other SNPs within these genes may be associated with prostate cancer status in African Americans. Our SNP selection process was based primarily on SNPs in the CYP17, CYP19A1, CYP3A4, SRD5A2, IGF-1, and IGFBP-3 genes that have been reported previously as being associated with prostate cancer.
In conclusion, our analysis of 473 African-American men with and without prostate cancer suggests that specific SNP genotypes in both the 5′-UTR and Exon 1 regions of the CYP17 gene and intron 2 region of the IGF-1 gene influence prostate cancer susceptibility. Given the strong LD between SNPs in the genotyped regions, it will be difficult to establish the underlying susceptibility polymorphism(s). Additional studies are needed to determine whether these polymorphisms are indeed associated with prostate cancer risk in African Americans.
Acknowledgments
Grant sponsor: Department of Defense, Prostate Cancer Research Program; Grant number: DAMD17-03-0270; Grant sponsor: National Institutes of Health; Grant numbers: P50 A69568, R01 CA79596.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1002/pros.20696
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2712831?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/pros.20696
Article citations
Predicting biochemical-recurrence-free survival using a three-metabolic-gene risk score model in prostate cancer patients.
BMC Cancer, 22(1):239, 04 Mar 2022
Cited by: 10 articles | PMID: 35246070 | PMCID: PMC8896158
The CYP19A1 (TTTA)n Repeat Polymorphism May Affect the Prostate Cancer Risk: Evidence from a Meta-Analysis.
Am J Mens Health, 15(3):15579883211017033, 01 May 2021
Cited by: 1 article | PMID: 34036824 | PMCID: PMC8161905
The Impact of African Ancestry on Prostate Cancer Disparities in the Era of Precision Medicine.
Genes (Basel), 11(12):E1471, 08 Dec 2020
Cited by: 17 articles | PMID: 33302594 | PMCID: PMC7762993
Review Free full text in Europe PMC
Polymorphism in the Androgen Biosynthesis Gene (CYP17), a Risk for Prostate Cancer: A Meta-Analysis.
Am J Mens Health, 14(5):1557988320959984, 01 Sep 2020
Cited by: 1 article | PMID: 32964792 | PMCID: PMC7518003
Review Free full text in Europe PMC
Association Between CYP17A1, CYB5A Polymorphisms and Efficacy of Abiraterone Acetate/Prednisone Treatment in Castration-Resistant Prostate Cancer Patients.
Pharmgenomics Pers Med, 13:181-188, 04 Jun 2020
Cited by: 6 articles | PMID: 32581567 | PMCID: PMC7280245
Go to all (41) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 18 of 18)
- (6 citations) dbSNP - rs743572
- (5 citations) dbSNP - rs6162
- (5 citations) dbSNP - rs6163
- (5 citations) dbSNP - rs5742657
- (4 citations) dbSNP - rs10883783
- (2 citations) dbSNP - rs7965399
- (1 citation) dbSNP - rs9332964
- (1 citation) dbSNP - rs2740574
- (1 citation) dbSNP - rs2242480
- (1 citation) dbSNP - rs12423791
- (1 citation) dbSNP - rs3110697
- (1 citation) dbSNP - rs9282858
- (1 citation) dbSNP - rs523349
- (1 citation) dbSNP - rs5742723
- (1 citation) dbSNP - rs3793345
- (1 citation) dbSNP - rs12333983
- (1 citation) dbSNP - rs1106381
- (1 citation) dbSNP - rs700519
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Prostate cancer risk and polymorphism in 17 hydroxylase (CYP17) and steroid reductase (SRD5A2).
Carcinogenesis, 20(9):1727-1731, 01 Sep 1999
Cited by: 111 articles | PMID: 10469617
Comprehensive evaluation of the association between prostate cancer and genotypes/haplotypes in CYP17A1, CYP3A4, and SRD5A2.
Eur J Hum Genet, 12(4):321-332, 01 Apr 2004
Cited by: 26 articles | PMID: 14560315
Eighteen insulin-like growth factor pathway genes, circulating levels of IGF-I and its binding protein, and risk of prostate and breast cancer.
Cancer Epidemiol Biomarkers Prev, 19(11):2877-2887, 01 Sep 2010
Cited by: 41 articles | PMID: 20810604 | PMCID: PMC2989404
GEN GEN: the genomic genetic analysis of androgen-metabolic genes and prostate cancer as a paradigm for the dissection of complex phenotypes.
Front Biosci, 4:D596-600, 15 Jul 1999
Cited by: 1 article | PMID: 10417059
Review
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: P50 CA069568
Grant ID: R01 CA79596
Grant ID: R01 CA079596-10
Grant ID: P50 CA069568-05S90006
Grant ID: R01 CA079596
PHS HHS (1)
Grant ID: P50 A69568