Abstract
Free full text

Common variants in the GDF5-BFZB region are associated with variation in human height
Associated Data
Abstract
Identifying genetic variants that influence human height will further our understanding of skeletal growth and development. A number of rare genetic variants have been convincingly and reproducibly associated with height in Mendelian syndromes, and common variants in HMGA2 were recently found to be associated with variation in height in the general population1. Here, we report genome-wide association analyses of 6,669 individuals from Finland and Sardinia, using genotyped and imputed markers, and follow-up in an additional 28,801 individuals. We show that common variants in the osteoarthritis-associated2 GDF5-BFZB locus are responsible for variation in height (estimated additive effect of 0.44 cm, overall p<10−15). Our results suggest a link between the genetic basis of height and osteoarthritis, potentially mediated through alterations in bone growth and development.
Height is a complex trait influenced by genes and a variety of environmental factors, including diet and the prenatal environment3. Heritability estimates suggest that ≥ 80% of variation in height may be genetically determined3,4. Rare mutations with large effects on height in Mendelian syndromes have been identified in FBN1, FGFR3, GH1, EVC1, and GPC3 (MIM 154700, 134934, 262400, 604831, and 312870). Despite the high heritability, numerous candidate gene and linkage studies to identify loci influencing height in individuals of “normal stature” have been inconclusive5. Overall, variation in human height is likely to be polygenic and heterogeneous. Recently, the first GWAS of height1 identified common variants in HMGA2 associated with normal variation in height, both in adults (p=4 × 10−16) and children (p=1 × 10−6); the variants account for a small fraction (~0.3%) of the overall variation in height.
To identify additional genetic variants associated with height, we analyzed genome-wide SNP data on 2,371 Finns from the FUSION study6 and 4,298 Sardinians from the SardiNIA study7 (Table 1). The two samples were originally genotyped with distinct sets of markers. We used genotype imputation methods6,8 to facilitate comparison of the two studies and evaluate association between height and ~2.28 million common genetic variants. After verifying the overall accuracy of imputed genotypes in a subset of markers, we conducted within-study analyses using a rapid variance components-based association test9 and then carried out a meta-analysis of the two studies (Supplementary Figure 1).
Table 1
Subject characteristics. Values are reported as means (standard deviation)
| Gender (M/F) | Study age in years | Standing height in cm | Sitting height in cm | Body mass index in kg/m2 | |
|---|---|---|---|---|---|
| FUSION T2D stage 1 | 617/467 | 62.7 (7.6) | 167.3 (9.0) | - | 30.2 (4.7) |
| FUSION NGT stage 1 | 640/647 | 60.9 (11.2) | 167.4 (9.3) | - | 27.0 (3.9) |
| SardiNIA | 1,883/2,415 | 43.6 (17.5) | 159.9 (9.0) | - | 25.3 (4.7) |
| FUSION T2D stage 2 | 718/490 | 59.4 (8.7) | 168.9 (9.6) | - | 30.9 (5.4) |
| FUSION NGT stage 2 | 768/490 | 58.4 (7.7) | 169.1 (9.3) | - | 26.9 (3.9) |
| DGI T2D | 772/745 | 58.8 (10.0) | 167.7 (9.1) | - | 28.5 (4.4) |
| DGI controls | 709/759 | 64.2 (10.0) | 168.9 (8.9) | - | 26.7 (3.7) |
| Old Order Amish | 1,253/1,458 | 49.6 (16.9) | 164.9 (8.8) | - | 27.2 (5.1) |
| ARIC European Americans | 5,374/5,996 | 54.4 (5.7) | 168.6 (9.4) | 89.4 (4.7) | 27.0 (4.9) |
| ARIC African Americans | 1,600/2,595 | 53.6 (5.8) | 168.1 (8.9) | 86.2 (4.4) | 29.6 (6.2) |
| Caerphilly | 1,389/0 | 56.7 (4.3) | 171.2 (6.4) | 91.1 (3.3) | 26.3 (3.5) |
| BWHHS | 0/3,685 | 68.9 (5.5) | 158.7 (6.1) | 83.0 (3.6) | 27.6 (5.0) |
|
| |||||
| Total samples | 15,723/19,747 | ||||
Our results provided confirmatory evidence of association with rs1042725 and rs7968682, the recently reported common variants in HMGA2 (p=.031 and .0093, respectively, both in the same direction as the original report: Supplementary Table 1)1. The five loci showing the most significant evidence of association in our study are shown in Supplementary Table 2. To our knowledge, common variants in these loci have not been associated with height previously.
The genes near our strongest signal (p<2 × 10−7), located on chromosome 20, have a plausible biological role in human height. Rare variants in growth differentiation factor 5 (GDF5) have been associated with disorders of skeletal development (see below), and BFZB (also known as C20orf44 and UQCC) encodes a ZIC-binding protein repressed by basic fibroblast growth factor10.
We pursued the chromosome 20 signal because it was the single best result in our initial scan, the surrounding region accounts for 40 of the 50 lowest p-values in our meta-analysis, and it overlaps an osteoarthritis susceptibility locus2. We focused our follow-up efforts for this locus on SNP rs6060369 (p = 9.7 × 10−7 in the initial meta-analysis, Table 2) because it exhibited the strongest evidence for association among all Affymetrix Mapping 500K SNPs, which had been genotyped in the majority of our GWAS samples; we favored genotyped over imputed SNPs for initial follow-up. The SNP most significantly associated with height in the meta-analysis of imputed HapMap SNPs was rs725908, which is in strong LD with rs6060369 (r2 = 0.96, Supplementary Table 2). SNP rs6060369 was initially imputed in the FUSION GWAS, but direct genotyping sustained strong evidence of association (meta-analysis p=1.5 × 10−6). In the GWAS samples, each copy of the C allele at rs6060369 was associated with an increase in height of 0.3 to 0.7 cm (accounting for 0.3–0.6% of the variance in height, after adjustment for age and sex).
Table 2
Association between rs6060369 and height.
| Standing height | N | C/C | C/T | T/T | Allele freq (C) | Effect (se) cm | Effect (se) standardizeda | p-value |
|---|---|---|---|---|---|---|---|---|
| FUSION T2D stage 1b | 1,084 | 167.4 (8.8) | 167.5 (8.9) | 167.0 (9.2) | .426 | .382 (.259) | .081 (.045) | .072 |
| FUSION NGT stage 1b | 1,287 | 167.5 (9.7) | 167.7 (9.2) | 166.7 (9.0) | .449 | .634 (.241) | .124 (.041) | .0024 |
| SardiNIA c | 4,298 | 158.8 (8.2) c | 158.5 (8.3) c | 158.0 (8.7) c | .384 | .700 (.186) | .083 (.021) | 4.35 × 10−4 |
|
| ||||||||
| 6,669 | Stage 1 meta-analysis | 9.73 × 10−7 | ||||||
|
| ||||||||
| FUSION T2D stage 2 | 1154 | 168.8 (9.3) | 169.4 (9.8) | 168.5 (9.5) | .458 | .477 (.262) | .071 (.041) | .084 |
| FUSION NGT stage 2 | 1,203 | 170.3 (9.4) | 168.6 (9.2) | 168.9 (9.3) | .451 | .456 (.244) | .103 (.040) | .0090 |
| DGI T2Db | 1,517 | 168.4 (9.3) | 167.6 (9.1) | 167.4 (9.1) | .447 | .449 (.234) | .062 (.038) | .044 |
| DGI controlsb | 1,468 | 169.0 (8.6) | 169.1 (9.0) | 168.3 (8.8) | .425 | .323 (.249) | 069 (.042) | .038 |
| Old Order Amish | 2,711 | 165.4 (9.1) | 165.3 (9.0) | 164.2 (8.5) | .383 | .424 (.178) | .044 (.020) | .028 |
| ARIC European Americans | 10,882 | 168.9 (9.3) | 168.7 (9.4) | 168.3 (9.5) | .398 | .252 (.085) | .029 (.009) | .0020 |
| ARIC African Americans | 3,860 | 168.4 (9.1) | 167.9 (8.7) | 167.3 (8.5) | .710 | .254 (.160) | .025 (.019) | .169 |
| Caerphilly | 1,097 | 171.6 (6.6) | 171.3 (6.5) | 170.8 (6.2) | .370 | .522 (.270) | .083 (.042) | .055 |
| BWHHS | 3,652 | 159.1 (6.0) | 159.0 (6.1) | 158.2 (6.2) | .362 | .560 (.147) | .093 (.240) | 9.71 × 10−5 |
|
| ||||||||
| 27,544 | Stage 2 meta-analysis | 1.05 × 10−11 | ||||||
| 34,213 | Standing height overall meta-analysis | 2.22 × 10−16 | ||||||
|
| ||||||||
| Sitting height | ||||||||
|
| ||||||||
| ARIC European Americans | 10,863 | 89.6 (4.6) | 89.4 (4.6) | 89.3 (4.6) | .398 | .137 (.046) | .029 (.009) | .0036 |
| ARIC African Americans | 3,857 | 86.3 (4.5) | 86.3 (4.2) | 86.1 (4.1) | .710 | −.043 (.085) | −.008 (.019) | .679 |
| Caerphilly | 1,092 | 91.3 (3.4) | 91.2 (3.4) | 90.9 (3.2) | .370 | .275 (.138) | .087 (.041) | .038 |
| BWHHS | 3,655 | 83.3 (3.6) | 83.2 (3.6) | 82.7 (3.4) | .362 | .345 (.083) | .100 (.023) | 1.73 × 10−5 |
|
| ||||||||
| 19,467 | Sitting height meta-analysis | 1.40 × 10−5 | ||||||
Association results are shown for an additive genetic model. Height means (SD) in cm are shown for each genotype class.
Motivated by previous reports of sex differences at osteoarthritis-associated loci11, we carried out an analysis stratified by sex. The stratified analyses show strong evidence of association in both males and females with no evidence of heterogeneity (Supplementary Table 3). We did not detect significant association of the SNP with other anthropometric measures (p>.05 for weight, body mass index, waist circumference, hip circumference, and waist-to-hip ratio).
We next tested the association of rs6060369 with height in nine follow-up samples, comprising 23,684 individuals of European ancestry and 3,860 African American individuals (Table 2). Six of the samples provided significant evidence of association (p<.05) and the other three showed a trend (p<.20) in the same direction (Table 2). The p-value for association in all 27,544 follow-up samples was 1.05 × 10−11, and in all 34,213 GWAS and follow-up samples combined was 2.22 × 10−16 (Figure 1). In the follow-up samples, each copy of the C allele at rs6060369 was associated with an increase in height of 0.2 – 0.6 cm (Table 2), and overall we estimate the SNP explains 0.3–0.5% of the variance in height, both in males and females (Supplementary Table 3). Further independent evidence for association between rs6060369 and adult height comes from the 1958 British Birth Cohort for which rs6060369 is associated with height measured at 44–45 years of age (p = .0046, explaining 0.5% of the variance) (http://www.b58cgene.sgul.ac.uk/, accessed October 2007).
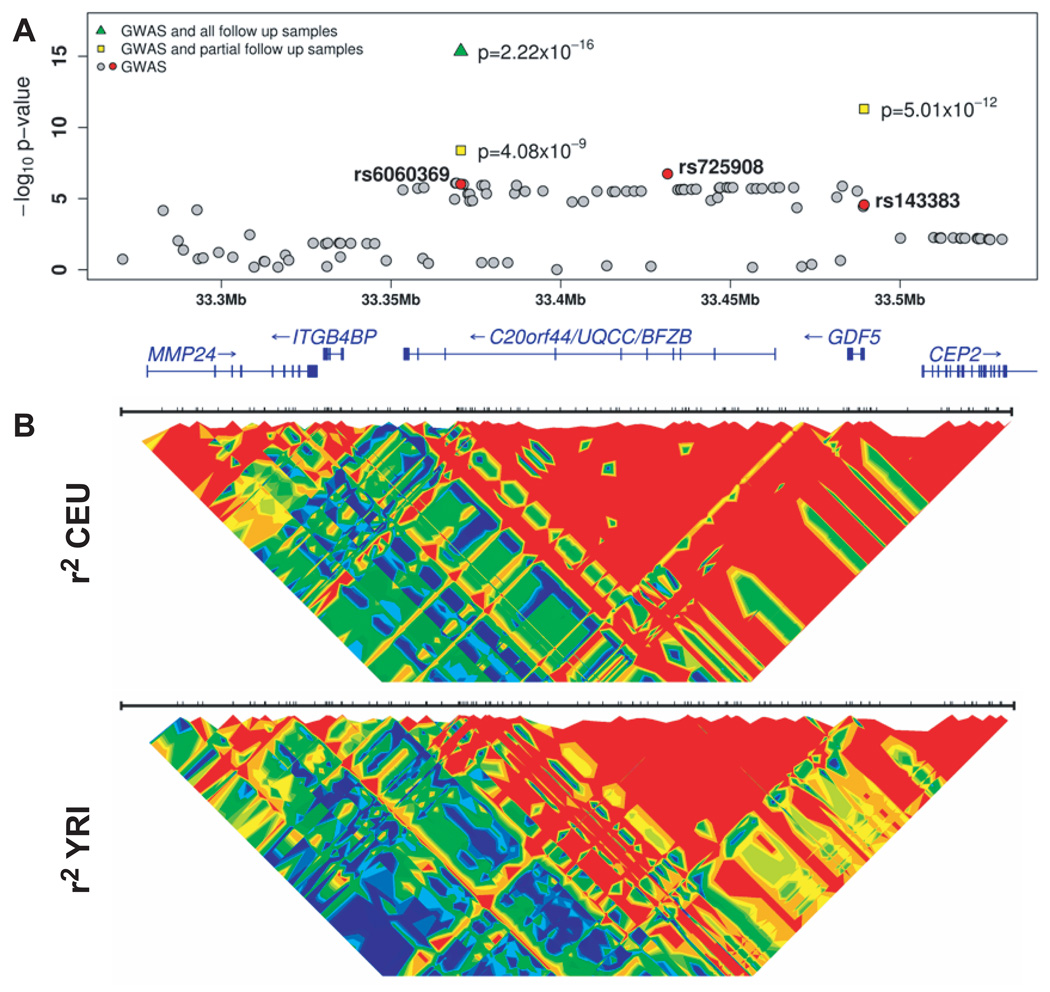
All genotyped or imputed SNPs in the SardiNIA and FUSION GWA studies are plotted with association p-values (additive test) compared to genomic position in NCBI Build 35 (gray circles, red circles for labeled SNPs). Yellow squares indicate p-values for SNPs analyzed in a portion of follow up samples (FUSION Stage 1 and 2, SardiNIA, DGI, and ARIC studies). The green triangle indicates rs6060369 analyzed in all GWAS and follow up samples. Patterns of linkage disequilibrium (r2) for two of the HapMap populations (CEU, Utah residents with European ancestry, and YRI, Yoruba)30 are plotted and colored with a 15 color red-green-blue scale, to represent values ranging from high (red), to intermediate (green), to low (blue).
Our association signal lies in a 136 kb stretch of linkage disequilibrium (LD) that contains two genes, GDF5 and BFZB (Figure 1, Supplementary Table 4). BFZB is expressed in differentiating chondrocytes12, starting at early stages of mesenchymal cell proliferation13. Studies in mouse embryonic stem cells have shown that BFZB is down-regulated on addition of FGF2 (bFGF)10, which acts in concert with bone morphogenic proteins and several Hox gene products to initiate and promote morphogenesis and growth of the skeleton. Thus, BFZB appears to be involved in a network of FGF2-regulated growth control. GDF5 is a member of the TGF-beta superfamily involved in bone growth and differentiation, both in adult and embryonic tissues14,15. GDF5 expression is typically restricted to the primordial cartilage of long bones, with little expression in the vertebrae and ribs14. Mutations in GDF5 are associated with several disorders of skeletal development (MIM 201250, 200700, 112600, 113100, 228900, 185800 and 186500). Recombinant human GDF5 has been shown to restore vertebral disc height in a rabbit disc degeneration model, perhaps through enhanced extracellular matrix production15. Other nearby genes do not appear to be involved in chondrocyte differentiation, bone growth, or development, but the locus that includes GDF5 and BFZB is also interesting because it was highlighted as showing very strong evidence for selection in a genome-wide search for regions that underwent recent selection16. The target of selection is presently unknown.
A SNP located in the 5’ untranslated region of GDF5, rs143383, is strongly associated with osteoarthritis2,17 and is estimated to be in very strong LD with rs6060369 in the HapMap, FUSION, and SardiNIA samples (r2=.83–.90). The SNP appears to influence GDF5 expression2,17 and, we reasoned, might be a causal variant. Thus, we analyzed this SNP in our screening samples and a subset of our follow-up samples. The rs143383 A allele previously associated with increased risk of osteoarthritis was associated with decreased height in our studies (p=5.01 × 10−12 versus p=4.08 × 10−9 for rs6060369 in the same subset of samples; Figure 1, Table 3). Analysis stratified by gender shows strong association in both males and females (Supplementary Table 5).
Table 3
Association between rs143383 and height.
| Standing height | n | G/G | G/A | A/A | Allele freq (G) | Effect (se) Cm | Effect (se) standardizeda | p-value |
|---|---|---|---|---|---|---|---|---|
| FUSION T2D stage 1b | 1,084 | 167.3 (8.8) | 167.8 (9.0) | 166.7 (9.0) | .406 | .461 (.281) | .088 (.049) | .071 |
| FUSION NGT stage 1b | 1,287 | 167.8 (9.8) | 167.7 (9.3) | 166.7 (9.0) | .425 | .697 (.263) | .129 (.044) | .0037 |
| SardiNIAc | 4,298 | 158.9 (8.2) | 158.5 (8.4) | 157.9 (8.6) | .403 | .546 (.189) | .065 (.021) | 6.73 × 10−3 |
|
| ||||||||
| 6,669 | Stage 1 meta-analysis | 2.70 × 10−5 | ||||||
|
| ||||||||
| FUSION T2D stage 2 | 1,167 | 168.9 (9.2) | 169.1 (9.8) | 168.5 (9.6) | .436 | .417 (.262) | .057 (.041) | .166 |
| FUSION NGT stage 2 | 1,216 | 170.2 (9.4) | 168.8 (9.4) | 168.8 (9.2) | .432 | .460 (.247) | .098 (.040) | .014 |
| DGI T2Dc | 1,517 | 168.4 (9.3) | 167.7 (9.1) | 167.3 (9.1) | .422 | .550 (.237) | .080 (.039) | .018 |
| DGI controlsc | 1,468 | 169.1 (8.5) | 169.1 (8.9) | 168.3 (8.9) | .392 | .359 (.255) | .070 (.043) | .036 |
| Old Order Amish | - | - | - | - | - | - | - | - |
| ARIC European Americans | 10,857 | 168.9 (9.3) | 168.8 (9.5) | 168.3 (9.5) | .387 | .257 (.086) | .029 (.009) | .0019 |
| ARIC African Americans | 3,881 | 168.2 (9.0) | 167.4 (8.5) | 167.3 (8.4) | .879 | .608 (.222) | .065 (.025) | .011 |
| Caerphilly | - | - | - | - | - | - | - | - |
| BWHHS | - | - | - | - | - | - | - | - |
|
| ||||||||
| 20,106 | Stage 2 meta-analysis | 8.48 × 10−8 | ||||||
| 26,775 | Standing height overall meta-analysis | 5.01 × 10−12 | ||||||
|
| ||||||||
| Sitting height | ||||||||
|
| ||||||||
| ARIC European Americans | 10,838 | 98.6 (4.5) | 89.5 (4.7) | 89.3 (4.6) | .387 | .122 (.046) | .026 (.010) | .0098 |
| ARIC African Americans | 3,878 | 86.3 (4.3) | 86.3 (4.1) | 86.5 (4.5) | .879 | −.100 (.119) | −.027 (.027) | .318 |
| Caerphilly | - | - | - | - | - | - | - | - |
| BWHHS | - | - | - | - | - | - | - | - |
|
| ||||||||
| 14,716 | Sitting height meta-analysis | 0.088 | ||||||
Association results are shown for an additive genetic model. Height means (SD) in cm are shown for each genotype class.
Interestingly, the ARIC African American samples that showed only a trend toward association with rs6060369 (p=.17) showed significant evidence of association with rs143383 (p=.011), illustrating the utility of studying different ancestral groups in the fine-mapping of complex disease genes18,19. In the ARIC African American samples, even when rs6060369 was included in a regression model, rs143383 remained marginally associated with height (p=.034, estimated additive effect = 0.579 cm), while the association of rs6060369 disappeared (p=.92, estimated additive effect = −0.019 cm) when conditioning on rs143383. These results suggest that GDF5 is more likely to influence height, although other nonsynonymous SNPs present in GDF5 and BFZB may affect function instead or as well.
Miyamoto and colleagues demonstrated that the A allele of rs143383 is associated with decreased GDF5 transcriptional activity in chondrogenic cells2. Decreased expression of GDF5 would logically lead to decreased limb bone growth, consistent with decreased height, as we observed. Decreased transcription of GDF5 may influence the amount of cartilage of the vertebrae, limb proportions, or joint angles, resulting both in a modest decrease in stature and susceptibility to osteoarthritis.
To evaluate the impact of osteoarthritis as a confounding factor, we repeated the association analysis restricted to younger individuals (age < 40). In SardiNIA, we analyzed 1,964 individuals and confirmed the association (p=.0018 for rs6060369, and p=.015 for rs143383) with effect size similar to estimates as for the combined sample (0.70 cm per copy of the C allele for rs6060369). In the Old Order Amish, the younger subgroup of 891 individuals showed a trend toward association of allele C with increased height (0.60 cm per copy of the C allele at rs6060369), but no significant association (p = .86), likely due to low statistical power.
To compare the evidence of association with length of long bones compared to vertebrae and skull, we tested rs6060369 and rs143383 for evidence of association with sitting height, which was measured in the ARIC and BWHHS studies. In ARIC European Americans and the BWHHS British sample, similar evidence of association was observed for both standing and sitting height. In ARIC African Americans, only rs143383 was significantly associated (p<.05) with height, and it was associated only with standing height, not with sitting height (Table 3), perhaps suggesting a stronger effect on long bones than on vertebrae.
Multiple regression analysis of our data suggests that a single common variant in the region may underlie the evidence of association. Specifically, multiple regression analysis in GWAS samples showed that after including rs6060369, rs143383, or rs725908 as a covariate, other association signals in the region become non-significant. One of these common variants or another nearby unmeasured variant in LD may influence height through expression of GDF5 2,17. However, either or both GDF5 and BFZB could be affected. Thoroughly evaluating the contribution of this locus to human height will require re-sequencing to catalog all genetic variants in the region and genotyping to evaluate their effects.
Combined, the variants identified here and previously reported in HMGA2 account for <1% of the variance in height, so that most of the 80% of variation in height estimated to be genetic remains unexplained. Our GWAS provides suggestive evidence for several other loci influencing height. For example, after excluding SNPs within 250 kb from GDF5, we observed a slight excess of SNPs with p-value <10−5 (38 observed versus 23 expected, Supplementary Figure 2). Still, it appears likely that many of the common variants influencing height will have only small effects. Follow-up of additional SNPs in larger meta-analyses will be necessary to define these variants20, which may also be relevant not only to normal variation in height but also to musculoskeletal or other diseases.
Methods
Study Subjects
Informed consent was obtained from all study participants and, in addition, ethics approval was obtained from the participating institutions.
FUSION GWAS
The Finland-United States Investigation of NIDDM Genetics (FUSION) study GWAS included 1,161 Finnish type 2 diabetes (T2D) cases, 1,174 normal glucose tolerant (NGT) controls, and 122 offspring of case/control pairs6. Cases and controls were approximately frequency matched, taking into account age, sex, and birth province within Finland6. For the height analysis, our sample consisted of 1,084 T2D individuals and 1,287 NGT individuals with height measurements from clinical exams. Samples were genotyped with the Illumina Infinium II HumanHap300 BeadChip6 and with an Illumina GoldenGate Custom Panel designed to improve genomic coverage around T2D candidate genes. The two imputed SNPs selected for additional follow-up were subsequently genotyped using the TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA).
SardiNIA GWAS
The SardiNIA GWAS examined a total of 4,305 related individuals participating in a longitudinal study of aging-related quantitative traits in the Ogliastra region of Sardinia, Italy. These individuals are distributed across 1,133 inter-related sibships, each with an average of 3.9 phenotyped individuals. For this study, we analyzed phenotypes for 4,298 individuals, excluding 4 cases of short stature due to Morquio Syndrome (MIM 253000) and 3 individuals for whom height measurements were not available. Among the individuals examined, 1,412 were genotyped with the Affymetrix Mapping 500K Array Set. Taking advantage of the relatedness among individuals in the SardiNIA sample, we conducted a second round of computational analysis to impute genotypes for analysis in an additional 2,893 individuals who were genotyped only with the Affymetrix Mapping 10K Array. In this second round, we identified large stretches of chromosome shared within each family and probabilistically “filled-in” genotypes within each stretch whenever one or more of its carriers was genotyped with the 500K Array Set9,21. These 2,893 individuals were mostly offspring and siblings of the 1,412 individuals genotyped at high density. For computational efficiency, the second round of imputation was applied to sub-pedigrees, each including no more than 20–25 individuals.
Follow-up Samples and Genotyping
Based on analysis of the combined SardiNIA and FUSION results, SNPs rs6060369 and rs143383 were each examined in up to 28,801 additional individuals. These included individuals of European ancestry from the FUSION study stage 2 samples (N = 2,466), the Diabetes Genetics Initiative22 (N = 2,985), the Old Order Amish23,24 (N = 2,711), the Atherosclerosis Risk in Communities (ARIC) Study25 (N = 11,370), the Caerphilly Study26,27 (1,389 men), and the British Women’s Heart and Health Study28 (3,685 women). 4,195 African American individuals from the ARIC study were also examined. Additional details of follow-up samples and genotyping methods are included in Supplementary Methods. Within each follow-up sample, SNP genotype success rates were >90% and genotype counts were consistent with Hardy-Weinberg equilibrium (p>.05).
Genotype Imputation
Our initial screen was based on the meta-analysis of two genome-wide association studies. Because the studies used two different genotyping platforms, we imputed genotypes for all polymorphic HapMap SNPs in each study, using a Hidden Markov Model programmed in MACH6,8. Details are provided in the Supplementary Methods.
GWAS Analysis
Within the FUSION and SardiNIA study samples, we carried out association analyses to relate observed and estimated genotypes to height. At each SNP, height was related to allele counts for a reference allele in a regression model that also included sex, age, and age2 as covariates; FUSION covariates also included birth province and study6. For SNPs genotyped in the laboratory, allele counts were discrete (0, 1, or 2), whereas for imputed SNPs, allele counts were fractional (between 0.0 and 2.0, depending on the expected number of copies of the allele for each individual). For FUSION, T2D and control individuals were analyzed separately and results combined using the meta-analytic techniques described below. To allow for relatedness, regression coefficients were estimated in the context of a variance components model that can handle imputed genotypes and accounts for background polygenic effects9. When evaluating significance, we applied quantile normalization to trait values (SardiNIA) or to residuals after adjustment for covariates (FUSION), by ranking all height values and then converting them to z-scores according to quantiles of the standard normal distribution. In parallel to the analysis of quantile normalized data, we also analyzed untransformed height (in centimeters) to estimate effect sizes.
Meta-Analysis
To summarize results for the three initial scans (1,084 T2D cases and 1,287 controls from FUSION, and 4,298 individuals from Sardinia), we carried out a meta-analysis. We used meta-analysis rather than an analysis of pooled data to avoid an increase in false positive rates due to population stratification. The Sardinian and Finnish populations are genetically and geographically distinct, with an average Fst of .025 among the 45,284 autosomal SNPs genotyped in both samples, and clear differences in height. The genomic control parameter for our meta-analysis, which estimates inflation in test statistics due to the combined effects of population stratification, cryptic relatedness, and genotyping error29, was 1.02, suggesting both that population stratification and unmodeled relatedness had a negligible impact on our association results and that our meta-analysis of imputed genotypes resulted in appropriate control of false-positive rates.
For each marker, we selected an arbitrary reference allele and calculated a z-statistic characterizing the evidence for association in each study (summarizing both the p-value, in its magnitude, and the direction of effect, in its sign). We then calculated an overall z-statistic as a weighted average of the three individual statistics and calculated the corresponding p-value20. Weights were proportional to the square-root of the number of individuals examined in each sample and were selected such that the squared weights summed to 1.0. An analogous strategy was used to summarize results of follow-up genotyping.
Acknowledgements
The authors thank the many individuals who generously participated in this study. We thank Yun Li for computational analysis, Kyle Gaulton, Amy Swift, Michael Erdos, and Narisu Narisu for FUSION genotyping and technical expertise, Center for Inherited Disease Research investigators for FUSION GWAS genotyping, the staff and participants of the ARIC study for their important contributions, and the Amish Research Clinic Staff for their energetic efforts in study subject recruitment and characterization. The SardiNIA team thanks Monsignore Piseddu, Bishop of Ogliastra, the mayors and citizens of the 4 Sardinian towns (Lanusei, Ilbono, Arzana, and Elini), and the head of the Public Health Unit ASL4 for their volunteerism and cooperation; the team also thanks Andrea Maschio, Fabio Busonero, Antonella Mulas, Natascia Sestu and Maria Grazia Piras for genotyping and technical expertise, and all the physicians, nurses and the recruitment personnel of the ProgeNIA group in Lanusei. This research was supported (in part) by the intramural Research Program of the NIH, National Institute on Aging [contracts NO1-AG-1-2109 to the SardiNIA (“Progenia”) team and 263-MA-410953 to the University of Michigan (G.R.A)]. Other support was from NIH grants DK072193 (K.L.M.), HL084729 (G.R.A.), HG002651 (G.R.A.), DK062370 (M.B.), DK54361 (A.R.S.), HL72515 (A.R.S.), AG18728 (A.R.S.), AR046838 (A.R.S.); National Human Genome Research Institute intramural project number 1 Z01 HG000024 (F.S.C.); the American Diabetes Association, including a post-doctoral fellowship award (C.J.W.); and March of Dimes research grant 6-FY04-61 (J.N.H). K.L.M. and G.R.A are Pew Scholars in the Biomedical Sciences. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. FUSION GWAS genotyping was performed with support from CIDR NIH Contract Number N01-HG-65403 and the JHU GRCF SNP Center. The whole-genome genotyping and analysis in the DGI genome scan was supported by Novartis Institutes for BioMedical Research (to D. Altshuler), with additional support from The Richard and Susan Smith Family Foundation/American Diabetes Association Pinnacle Program Project Award (to D. Altshuler, J.N.H. and M.J. Daly). Funding and support were also provided by the University of Maryland General Clinical Research Center (M01 RR 16500), the NIDDK Clinical Nutrition Research Unit of Maryland (NIH P30 DK072488), the Department of Veterans Affairs, and Veterans Affairs Medical Center Baltimore Geriatric Research, Education and Clinical Center (GRECC). The Caerphilly study was funded by the Medical Research Council (UK). The Caerphilly study was undertaken by the former MRC Epidemiology Unit (South Wales) and was funded by the Medical Research Council of the United Kingdom. The data archive is maintained by the Department of Social Medicine, University of Bristol. We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
Footnotes
Accession numbers. GenBank mRNA sequence for growth differentiation factor 5 (GDF5), NM_000557 and for basic FGF-repressed Zic binding protein (BFZB, UQCC, C20orf44) is NM_018244.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ng.74
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2914680?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101881817
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ng.74
Article citations
Weight, height, waist circumference: association with knee osteoarthritis findings from the osteoarthritis initiative.
Pain Rep, 9(5):e1187, 20 Sep 2024
Cited by: 0 articles | PMID: 39315116 | PMCID: PMC11419520
Analysis of Runs of Homozygosity in Aberdeen Angus Cattle.
Animals (Basel), 14(15):2153, 24 Jul 2024
Cited by: 0 articles | PMID: 39123679 | PMCID: PMC11311081
A genome-wide association study of neutrophil count in individuals associated to an African continental ancestry group facilitates studies of malaria pathogenesis.
Hum Genomics, 18(1):26, 15 Mar 2024
Cited by: 1 article | PMID: 38491524 | PMCID: PMC10941368
Enhancing Genomic Prediction Accuracy for Body Conformation Traits in Korean Holstein Cattle.
Animals (Basel), 14(7):1052, 29 Mar 2024
Cited by: 0 articles | PMID: 38612291 | PMCID: PMC11011013
Decreased Risk of Knee Osteoarthritis with Taller Height in an East Asian Population: A Nationwide Cohort Study.
J Clin Med, 13(1):92, 23 Dec 2023
Cited by: 0 articles | PMID: 38202099 | PMCID: PMC10779701
Go to all (271) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (Showing 13 of 13)
- (1 citation) OMIM - 185800
- (1 citation) OMIM - 112600
- (1 citation) OMIM - 262400
- (1 citation) OMIM - 154700
- (1 citation) OMIM - 200700
- (1 citation) OMIM - 134934
- (1 citation) OMIM - 312870
- (1 citation) OMIM - 228900
- (1 citation) OMIM - 186500
- (1 citation) OMIM - 253000
- (1 citation) OMIM - 201250
- (1 citation) OMIM - 113100
- (1 citation) OMIM - 604831
Show less
RefSeq - NCBI Reference Sequence Database (2)
- (1 citation) RefSeq - NM_000557
- (1 citation) RefSeq - NM_018244
SNPs (3)
- (11 citations) dbSNP - rs6060369
- (5 citations) dbSNP - rs143383
- (1 citation) dbSNP - rs725908
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Positive selection on the osteoarthritis-risk and decreased-height associated variants at the GDF5 gene in East Asians.
PLoS One, 7(8):e42553, 14 Aug 2012
Cited by: 19 articles | PMID: 22905146 | PMCID: PMC3419199
A functional polymorphism in the 5' UTR of GDF5 is associated with susceptibility to osteoarthritis.
Nat Genet, 39(4):529-533, 25 Mar 2007
Cited by: 305 articles | PMID: 17384641
A common variant of HMGA2 is associated with adult and childhood height in the general population.
Nat Genet, 39(10):1245-1250, 02 Sep 2007
Cited by: 265 articles | PMID: 17767157 | PMCID: PMC3086278
Genetic regulation of adult stature.
Curr Opin Pediatr, 21(4):515-522, 01 Aug 2009
Cited by: 43 articles | PMID: 19455035
Review
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z01 HG000024
Medical Research Council (2)
Centre for molecular-based causal analyses in health and disease
Professor George Davey Smith, University of Bristol
Grant ID: G0600705
Assessment of biomedical risk factors and disease outcomes in the British 1958 cohort
Professor Christine Power, Institute of Education
Grant ID: G0000934
NCRR NIH HHS (2)
Grant ID: M01 RR 16500
Grant ID: M01 RR016500
NHGRI NIH HHS (4)
Grant ID: R01 HG002651-05S2
Grant ID: N01HG65403
Grant ID: R01 HG002651
Grant ID: HG002651
NHLBI NIH HHS (19)
Grant ID: HL084729
Grant ID: N01-HC-55018
Grant ID: HL72515
Grant ID: N01-HC-55015
Grant ID: N01HC55022
Grant ID: N01-HC-55019
Grant ID: N01HC55018
Grant ID: N01HC55020
Grant ID: N01-HC-55016
Grant ID: N01-HC-55020
Grant ID: N01-HC-55022
Grant ID: N01HC55016
Grant ID: N01HC55019
Grant ID: N01HC55021
Grant ID: N01-HC-55021
Grant ID: N01HC55015
Grant ID: U01 HL072515
Grant ID: U01 HL084729
Grant ID: U01 HL084729-03
NIA NIH HHS (2)
Grant ID: AG18728
Grant ID: R01 AG018728
NIAMS NIH HHS (2)
Grant ID: AR046838
Grant ID: R01 AR046838
NIDDK NIH HHS (10)
Grant ID: DK062370
Grant ID: DK072193
Grant ID: R01 DK062370
Grant ID: U01 DK062370
Grant ID: DK54361
Grant ID: P30 DK072488
Grant ID: R01 DK029867
Grant ID: R01 DK072193
Grant ID: R01 DK072193-04
Grant ID: R56 DK062370
Wellcome Trust (2)
A national DNA control series for genetic case-control studies based on the British 1958 birth cohort
Professor Paul Burton, University of Leicester
Grant ID: 068545
Grant ID: 068545/Z/02




