Abstract
Free full text

The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes
Abstract
The cellular and molecular mechanisms that underlie species-specific membrane fusion between male and female gametes remain largely unknown. Here, by use of gene discovery methods in the green alga Chlamydomonas, gene disruption in the rodent malaria parasite Plasmodium berghei, and distinctive features of fertilization in both organisms, we report discovery of a mechanism that accounts for a conserved protein required for gamete fusion. A screen for fusion mutants in Chlamydomonas identified a homolog of HAP2, an Arabidopsis sterility gene. Moreover, HAP2 disruption in Plasmodium blocked fertilization and thereby mosquito transmission of malaria. HAP2 localizes at the fusion site of Chlamydomonas minus gametes, yet Chlamydomonas minus and Plasmodium hap2 male gametes retain the ability, using other, species-limited proteins, to form tight prefusion membrane attachments with their respective gamete partners. Membrane dye experiments show that HAP2 is essential for membrane merger. Thus, in two distantly related eukaryotes, species-limited proteins govern access to a conserved protein essential for membrane fusion.
Fusion of gametes of opposite sex (or mating type) to form a zygote is the defining moment in the life of a eukaryote. In the first phase of gamete interactions, cell adhesion molecules displayed on the surfaces of the gametes bring the two cells together. In animals, the sperm plasma membrane binds to the extracellular matrix of the egg (the zona pellucida in mammals and the jelly coat in many invertebrates). The interacting gametes use this first-phase adhesion step not only to bind to each other, but also to initiate a signal transduction cascade that activates the sperm and exposes new, fusogenic regions of the sperm plasma membrane. In the second phase of fertilization, the membrane fusion reaction, the plasma membranes of the two gametes come into intimate contact and then fuse, bringing about cytoplasmic continuity (Primakoff and Myles 2002; Rubinstein et al. 2006). Although these two steps—prefusion attachment of the plasma membranes of gametes and merger of their lipid bilayers—have been experimentally separated using in vitro bioassays, gene disruption studies to date have failed to distinguish the two, and no genes have been identified whose disruption allows prefusion attachment and disallows membrane merger. In mice, several proteins involved in gamete membrane interactions have been described, including ADAMS family members and CRISP proteins on sperm and integrins and tetraspanin family members CD9 and CD81 on eggs (for review, see Ellerman et al. 2006; Inoue et al. 2007; Primakoff and Myles 2007). Izumo, an immunoglobulin superfamily sperm protein that appears to be limited to mammals, is gamete-specific and shown by gene disruption to be essential at a late step in fertilization. Izumo is the best candidate to date for a role in the membrane fusion reaction in mice. Its specific function has yet to be determined, however, and the presence of conserved Ig superfamily domains predicts a role in membrane adhesion (Inoue et al. 2005, 2007; Primakoff and Myles 2007).
In nematodes, several genes have been identified whose disruption leads to sterility. The Caenorhabditis elegans gene SPE-9 is essential for gamete interactions, but it is proposed to be an adhesion and signaling molecule and probably is not involved in the membrane fusion reaction (Putiri et al. 2004). Other proteins are implicated in gamete interactions in worms, including EGG-1, EGG-2, SPE-38, and SPE-42 (Chatterjee et al. 2005; Kadandale et al. 2005; Kroft et al. 2005), but their precise roles in fertilization also are unclear, in large part because of the difficulty of studying sperm–egg interactions in vitro. The protein Prm1p has been implicated in cell–cell fusion of Saccharomyces cerevisiae. Its disruption reduces fusion of a and alpha cells by 50%, but only when the gene is disrupted in both cell types (Heiman and Walter 2000; Aguilar et al. 2007; Heiman et al. 2007); thus, it is not essential for fusion. In invertebrates, at least two sperm proteins have emerged as candidate adhesion/fusion molecules—an 18-kDa protein in abalone (Swanson and Vacquier 1995) and the sea urchin protein bindin (Kamei and Glabe 2003). Because of the difficulty of generating mutants in these organisms, the functions of the abalone and urchin proteins in gamete interactions remain unidentified.
A recent screen for sterile mutants in Arabidopsis identified the male-specific sterility gene HAP2 (Johnson et al. 2004). A HAP2 family member called GCS1 (for generative cell-specific) was subsequently identified in a screen for lily genes whose transcripts were up-regulated in sperm (generative cells) (Mori et al. 2006). In Arabidopsis, the only organism for which HAP2 mutants are available, the gene is involved in pollen tube guidance, is expressed in sperm, and also is essential for seed formation (Johnson et al. 2004; Mori et al. 2006; von Besser et al. 2006). Although the gene acts after sperm deposition in the female gametophyte, its cellular and molecular functions in seed formation are unknown, since the cellular events subsequent to sperm deposition—sperm migration, sperm–egg attachment, and sperm–egg fusion—have not been distinguished experimentally. Interestingly, HAP2 is conserved, and in addition to being in Arabidopsis and rice (Johnson et al. 2004), members were found in Chlamydomonas, a red alga, a slime mold, Plasmodium falciparum, and Leishmania major (Mori et al. 2006).
In contrast to fertilization in the organisms described above, fertilization in some protists is highly amenable to study. In the unicellular, biflagellated green alga Chlamydomonas reinhardtii, initial adhesion of the flagella of mating type minus and mating type plus gametes in the first phase of interactions triggers cilium-generated signaling (Wang et al. 2006) and gamete activation (for review, see Pan and Snell 2000; Goodenough et al. 2007). Gamete activation prepares the gametes for fusion and comprises a complex signaling pathway including a protein tyrosine kinase (Wang and Snell 2003), a cGMP-dependent protein kinase (Wang et al. 2006), a flagellar adenylyl cyclase (Saito et al. 1993; Zhang and Snell 1994), and a 10- to 20-fold increase in cAMP. Both gametes shed their glycoproteinaceous cell walls through the action of a metalloprotease released from each cell (Matsuda et al. 1985; Buchanan et al. 1989). And sites specialized for cell fusion—the mating structures—located on the apical cell membranes between the two flagella of both gametes are activated (Goodenough et al. 1982; Wilson et al. 1997). The activated plus mating structure is a microvillus-like cellular extension ~3 μm in length and ~0.15 μm in diameter. The activated minus mating structure lacks actin filaments and is shorter and more bulbous. Continued flagellar adhesion brings the activated mating structures into intimate contact, and within seconds after contact, the membranes of the two mating structure fuse, followed by complete coalescence of the two gametes into a single, quadriflagellated zygote. The entire process of fertilization, from initial flagellar adhesion of gametes through fusion, can occur within 30 sec or less.
Previously, we showed that the plus gamete-specific protein FUS1, which is not found in other species, is present on the plasma membrane of the mating structure, the fertilization tubule. Furthermore, in experiments with flagellar adhesion mutants, we demonstrated that wild-type plus gametes, but not fus1 plus gametes, were capable of adhering to minus gametes solely via their activated mating structures. Thus, during the membrane fusion reaction, plus-specific FUS1 is essential for prefusion attachment between the plus and minus mating structures (Ferris et al. 1996; Misamore et al. 2003).
Fertilization in the rodent malaria organism, Plasmodium berghei, is also highly amenable to study. Sexual precursor stages, the gametocytes, form in the vertebrate host inside infected erythrocytes, but remain quiescent until ingested by a susceptible Anopheles mosquito. In the bloodmeal, gametocytes emerge from their host cells and within minutes differentiate into gametes. Each female (macro) gametocyte gives rise to a single immotile female gamete, while male (micro) gametocytes generate up to eight flagellated male gametes in a process termed “exflagellation”; within minutes after release, the gametes meet, adhere for a few seconds, and then fuse to form a zygote (Sinden 1983). Male gamete adhesion to a female gamete requires the species-limited surface protein and transmission-blocking vaccine candidate P48/45 (van Dijk et al. 2001). P48/45 interacts physically with at least one other gametocyte protein, P230 (Kumar 1987), and in P. falciparum is required to retain the complex on the cell surface once gametes have emerged from their host cells (Eksi et al. 2006). The male-specific function of the P48/45–P230 complex is in contrast with its expression in both male and female gametes, and whether either protein on male gametes binds directly to a receptor on female gametes is unknown.
Within 15–20 h, the zygote transforms into a motile ookinete, which penetrates the midgut epithelium and establishes the infection in the mosquito by forming an oocyst between the midgut epithelial cells and their underlying basal lamina. Thus, gamete adhesion and fusion are obligate steps in mosquito transmission of malaria and attractive targets for transmission-blocking vaccines. In the rodent malaria parasite P. berghei, gametocytes respond efficiently to well-characterized developmental triggers (Billker et al. 1998) in vitro, and gametogenesis, fertilization, and ookinete formation are accessible to analysis in culture. Moreover, targeted gene disruption is now a routinely used method in Plasmodium (Janse et al. 2006).
To date, no widely conserved mechanism of gamete fusion has been identified (Chen and Olson 2005; Rubinstein et al. 2006; Primakoff and Myles 2007). Thus, it remains unknown for any organism whether adhesion of gamete membranes and fusion of the membranes are together accomplished by a single set of proteins, as with fusion of many viruses (Earp et al. 2005), or if the two functions are allocated to distinct sets of proteins. We also do not understand the molecular basis for the species specificity of gamete fusion in many organisms (Ferris et al. 1997; Swanson and Vacquier 2002; Vieira and Miller 2006).
Here, in coupled studies of fertilization in Chlamydomonas and Plasmodium, we show that gamete fusion requires the plant sterility gene HAP2. We genetically distinguish attachment of gamete fusogenic membranes from membrane merger, and show that the membrane fusion reaction is governed by species-limited proteins required for prefusion attachment. In both species, post-adhesion events resulting in membrane fusion depend on the conserved HAP2 protein.
Results
A screen for zygote formation mutants in Chlamydomonas identifies a homolog of an Arabidopsis male sterility gene
To identify proteins in Chlamydomonas minus gametes that are essential at a late stage of gamete interactions, we generated insertional mutants in Chlamydomonas minus cells (strain B215, mt−) by transformation with a paromomycin resistance gene (Sizova et al. 2001). Colonies that grew on paromomycin were induced to undergo gametogenesis, and the gametes were mixed with wild-type plus gametes and screened for their ability to undergo flagellar adhesion within minutes after mixing and to form the zygote aggregates that appear ~4 h after mixing. After screening ~2500 insertional mutants, we identified one transformant with the desired phenotype, 63B10. Gametes of 63B10 adhered via their flagella to wild-type plus gametes and formed small clusters visible at 10 min after mixing similarly to wild-type minus gametes (Fig. 1A, top panels). On the other hand, the 63B10 minus/wild-type plus mixtures failed to form the large zygote aggregates characteristic of wild-type/wild-type gamete mixtures at 4 h and remained in the small clusters (Fig. 1A, bottom panels). In contrast to this easily observed phenotype when they were gametes, the growth, motility, phototaxis, and morphology of 63B10 vegetative cells were indistinguishable from those of wild-type vegetative cells. When we examined wild-type minus/wild-type plus gamete mixtures and 63B10 minus/wild-type plus gamete mixtures soon after mixing, we found that, as expected, most wild-type cells had fused to form quadriflagellated zygotes, but 63B10 minus/wild-type plus mixtures failed to form quadriflagellated cells. Instead, the 63B10 minus gametes continued flagellar adhesion with the wild-type plus gametes, many visible as pairs of cells with their flagella entwined and their apical ends closely apposed (Fig. 1B).
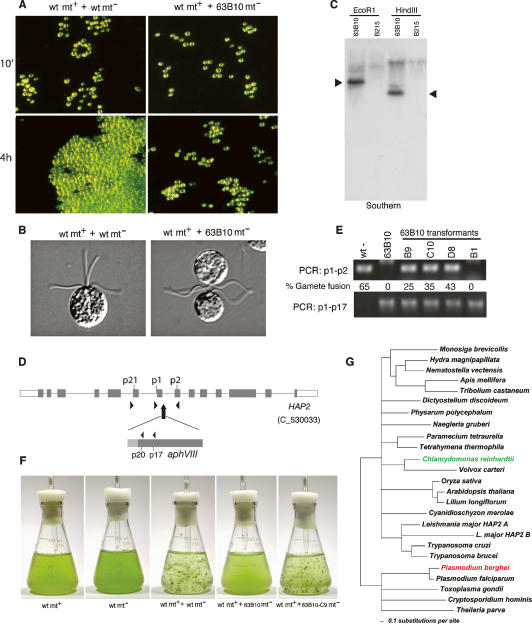
HAP2 is required for fertilization in Chlamydomonas and is phylogenetically conserved in many eukaryotes. (A) Dark-field images of wild-type plus gametes 10 min and 4 h after mixing with wild-type or 63B10 minus gametes. Clusters of gametes undergoing flagellar adhesion are present in both samples at 10 min. A large aggregate of zygotes is visible in the wild-type mixture at 4 h, whereas the 63B10 minus gametes failed to form zygote aggregates with wild-type plus gametes, and the small clusters persisted. (B) Differential interference contrast (DIC) microscopy images of a quadriflagellated zygote formed from fusion of a wild-type plus gamete with a wild-type minus gamete (left panel) and a wild-type plus gamete undergoing flagellar adhesion with a 63B10 minus gamete, but failing to fuse (right panel). (C) Southern blotting of EcoRI- and HindIII-digested Chlamydomonas genomic DNA from 63B10 and wild-type (B215) strains with a pSI103/PvuII DNA fragment as a probe. Arrowheads indicate the pSI103 plasmid in both samples. (D) Structure of the HAP2 gene and location of the aphVIII gene pSI103 plasmid. (E) Diagnostic PCR on genomic DNA showing the presence of disrupted HAP2 (primers p1–p17) in 63B10 gametes and the absence of intact HAP2 in 63B10 gametes and its reappearance in those 63B10 gametes that were rescued for fusion with the wild-type HAP2 gene (primers p1/p2). (F) Large aggregates of zygotes were present only in mixtures of wild-type plus and minus gametes and wild-type plus and 63B10 minus gametes rescued with the HAP2-HA construct (63B10-C9). (G) Phylogenetic tree based on trees generated using MOLPHY and TREE-PUZZLE illustrating the relationships of HAP2 proteins from several species (see Supplemental Material for accession numbers and methods).
After confirming by Southern blotting (Fig. 1C) that 63B10 contained a single insertion of the paromomycin resistance gene, we used thermal asymmetric interlaced PCR (TAIL–PCR) to identify 180 base pairs (bp) of genomic sequence adjacent to the inserted plasmid. Searches of version 2.0 of the Chlamydomonas reinhardtii genome sequence from the DOE Joint Genome Institute identified C_530033 as the adjacent gene (Fig. 1D). Transformation of 63B10 cells with BAC 20L3, which contained several putative genes in addition to C_530033, restored their ability to form zygotes (data not shown), thus confirming that we had identified the genomic region containing the disrupted gene. To confirm that C_530033 indeed was sufficient for rescue of zygote formation, we transformed 63B10 cells with a 20L3 restriction fragment whose only full-length gene was C_530033. As shown in the diagnostic PCR in Figure 1E, the transformants in which formation of quadriflagellated zygotes was restored contained the rescuing wild-type gene as well as the disrupted gene. Moreover, only wild-type gametes and rescued 63B10 gametes formed the macroscopic aggregates characteristic of zygotes 4 h after mixing wild-type minus and plus gametes (Fig. 1F). Thus, C_530033 was essential for gamete fusion. To our surprise, BLAST searches of NCBI databases (Altschul et al. 1997) showed that C_530033 encodes the Chlamydomonas homolog of HAP2, the Arabidopsis pollen tube guidance and male sterility gene first reported by Johnson et al. (2004), Mori et al. (2006), and von Besser et al. (2006).
HAP2 is present in multicellular animals and in P. berghei
Using PSI-BLAST (Altschul et al. 1997), we extended previous results (Johnson et al. 2004; Mori et al. 2006) on the species distribution of HAP2, and found HAP2 family members in many higher plants whose genome sequences are available including maize, wheat, and tomato (data not shown), as well as in many other nonpathogenic and pathogenic protists including Toxoplasma gondii, Cryptosporidium hominis, Theileria parva, Naegleria gruberi, and Trypanosoma brucei (Fig. 1G). The HAP2 gene is also present in a choanoflagellate (Monosiga brevicollis), one of the closest unicellular relatives of animals. Consistent with its presence in Monosiga, HAP2 family members were also present in multicellular animals including Hydra magnipapillata, the starlet sea anemone Nematostella vectensis, and insects (the honeybee and the flower beetle) (Fig. 1G). An alignment of predicted HAP2 proteins generated by PROMALS (Pei and Grishin 2007) and phylogenetic trees of HAP2 proteins generated by MOLPHY (Adachi and Hasegawa 1996) and TREE-PUZZLE (Jones et al. 1992) are shown in the Supplemental Material.
Previous studies on HAP2 in Arabidopsis showed that it was involved in pollen tube guidance (Johnson et al. 2004; von Besser et al. 2006) and that it had a second role in seed formation after release of sperm from the pollen tube (Johnson et al. 2004; Mori et al. 2006; von Besser et al. 2006). Our results showing that Chlamydomonas HAP2 mutants were fully motile and fully capable of flagellar adhesion demonstrated that the protein functions directly in the interactions between minus and plus gametes at a step in fertilization after initial gamete recognition. Moreover, we found that in addition to being present in the human malaria parasite P. falciparum (Mori et al. 2006), HAP2 was also present in the rodent malaria parasite P. berghei (Fig. 1G), in which sexual development is most amenable to experimentation. We therefore chose this species to ask if HAP2 functioned directly in gamete interactions in an organism that is only very distantly related to plants and green algae.
Plasmodium HAP2 is essential for mosquito transmission of malaria
To test for a function of HAP2 in P. berghei, we used targeted gene disruption. Cultured Plasmodium schizonts were electroporated with a targeting vector that contained an expression cassette conveying resistance to pyrimethamine and designed to replace all of the protein-coding sequence of the P. berghei HAP2 gene (GenBank accession no. XM_671808) (Fig. 2A). Following dilution cloning of drug-resistant parasites, genotyping by Southern blotting (Fig. 2B) of two independently produced hap2 clones and diagnostic PCR (Fig. 2C) documented that the replacement construct had integrated at the targeted site.
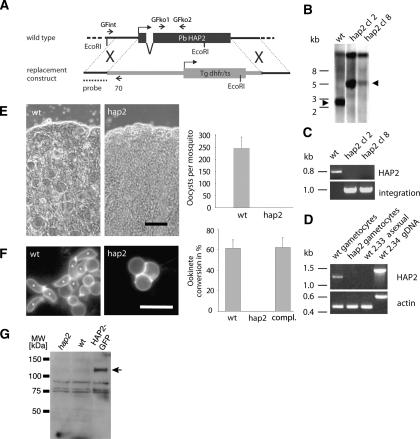
HAP2 is essential for sexual development and mosquito transmission of P. berghei. (A) Structure of the Plasmodium HAP2 gene and gene replacement construct. Short arrows indicate oligonucleotides used for PCR genotyping. (B) Southern hybridization of EcoRI-digested genomic DNA using the 5′ targeting sequence as a probe. Arrowheads indicate diagnostic 2.8-kb (wild-type) and 5.0-kb (HAP2) bands. (C) Diagnostic PCR with genomic DNA templates and primers GFko1/GFko2 to test for the presence of HAP2, and primers GFint/70 to detect a unique 1-kb product across the integration site. (D) RT–PCR detection of HAP2 transcript in parasite lines and stages (the expected larger product from genomic DNA includes one intron). (E) Representative images of midguts from A. stephensi mosquitoes 10 d after feeding on wild-type and hap2-infected mice (bar, 100 μm) and bar chart showing average numbers of oocysts per gut (error bar indicates SEM; n = 47 wild-type or hap2-exposed mosquitoes from three independent experiments). The overall prevalence of infection was 87% for wild type, and 0% for hap2. (F) Immunofluorescence images of live 20-h Plasmodium cultures immunostained for the female gamete/zygote marker P28. Elongate ookinetes (asterisks) were absent from the hap2 mutant (bar, 10 μm), which possessed only round female gametes. The bar chart shows ookinete conversion rates for wild type, hap2 clone 8, and the hap2 mutant that was complemented with HAP2-GFP. Conversion rate is expressed as the percentage of P28-positive parasites that had progressed to the ookinete stage (error bar indicates SD; n = 3). (G) Western blot analysis with anti-GFP, showing expression of a HAP2-GFP fusion protein (arrow) of the expected motility (120 kDa predicted) only in the complemented parasite line. Nonspecifically recognized bands confirm equal loading.
Analysis by RT–PCR showed that HAP2 transcripts were present in wild-type gametocytes, but not in hap2 gametocytes or in wild-type asexual erythrocytic stages of a gametocyte-deficient parasite strain (Fig. 2D). Consistent with this sexual stage-specific transcription, examination of mice infected with hap2 clones showed that the parasites underwent normal asexual development in erythrocytes. Neither the rate of gametocyte formation nor the sex ratio was affected, and gametocytes were able to emerge from their host cells and differentiate into gametes when exposed to activating conditions (data not shown). To test for a role of HAP2 in fertilization, we first allowed female Anopheles mosquitoes to feed on mice infected with hap2 parasites and 10 d later used phase contrast microscopy to examine the walls of midguts from the mosquitoes for the presence of oocysts. As shown in Figure 2E, whereas oocysts were plentiful in midguts of control mosquitoes allowed to feed on mice infected with wild-type P. berghei (Fig. 2E, left panel of photomicrograph and bar graph), we failed to detect oocysts in the mosquitoes that were fed on mice infected with hap2 parasites (Fig. 2E, right panel of photomicrograph and bar graph). Thus, HAP2 is required for transmission of P. berghei to mosquitoes.
The complete block of oocyst formation and thus of malaria transmission in vivo was similar to our finding that Chlamydomonas hap2 mutants failed to form zygotes. To determine if zygote formation per se was also blocked in P. berghei hap2 mutants, we examined ookinete formation in blood collected from mice infected with hap2 parasites. Ookinetes are differentiated, motile forms of Plasmodium zygotes. They are capable of invading the midgut epithelium, passing through the midgut cells, and taking up residence beneath the basement membrane, where they become static oocysts. Gametocytes in the infected blood were induced to escape from blood erythrocytes and allowed to undergo gametogenesis and gamete fusion by incubation in ookinete medium at 19°C. After ~24 h, the cultures were stained with an antibody against P28, a surface protein of activated female gametes, zygotes, and ookinetes. As shown in Figure 2F, in blood cultures from mice infected with wild-type parasites, most activated female gametes were fertilized and thus transformed into characteristically banana-shaped ookinetes. Blood cultures from mice infected with hap2 parasites failed to produce ookinetes (Fig. 2F), a finding we confirmed with another hap2 mutant clone from an independent transfection (data not shown). When we reintroduced a wild-type HAP2 gene fused to a coding sequence for green fluorescent protein (GFP) into the disrupted hap2 locus of the mutant, gametocytes expressed a fusion protein of the predicted size (Fig. 2G), ookinete formation was fully restored (Fig. 2F, bar graph), and the complemented parasites were able to complete their life cycle through the mosquito (data not shown). These results are consistent with a key role for HAP2 upstream of zygote formation during fertilization in Plasmodium.
HAP2 is a sex- and gamete-specific protein in Chlamydomonas and P. berghei and functions after gamete activation
To dissect the function of HAP2 in Chlamydomonas fertilization, we asked whether it was required in minus or plus gametes or both. Studies in Arabidopsis were consistent with a male-specific role of the protein in seed formation (Johnson et al. 2004; Mori et al. 2006; von Besser et al. 2006), whereas RT–PCR studies by Mori et al. (2006) indicated that HAP2 was expressed in mt+ and mt− gametes of a slime mold (Physarum polycephalum) and in Chlamydomonas minus and plus gametes, although expression was stronger in Chlamydomonas minus than plus. To generate plus gametes containing only the hap2 allele, we crossed wild-type plus gametes with 63B10 minus gametes that had been rendered fusion-competent by transformation with the wild-type HAP2 gene and selected plus progeny that contained only the mutant allele. Southern blotting confirmed that the plus cells contained only the hap2 allele (Fig. 3A, top panel). Plus strain cells that lacked a functional HAP2 gene exhibited no detectable mutant phenotype during vegetative growth or gametogenesis, and the hap2 plus gametes underwent gamete fusion to form quadriflagellated zygotes similarly to wild type (Fig. 3A, bottom panel). Thus, HAP2 protein is essential in fusion of Chlamydomonas minus gametes only.
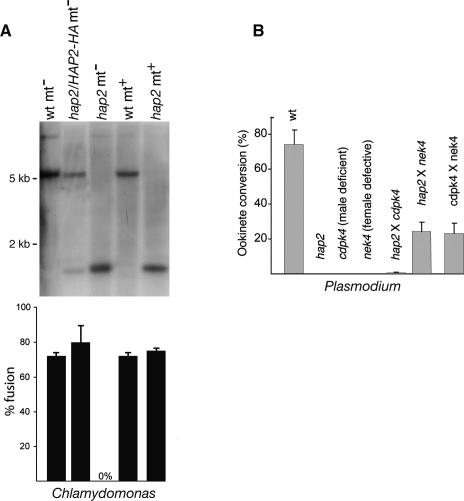
HAP2 has a sex-restricted function in both Chlamydomonas and Plasmodium. (A) Chlamydomonas hap2 plus gametes can fuse with wild-type minus gametes. (Top panel) Genotyping by Southern hybridization of one mating partner. The second mating partner was always wild type (not shown). The 5.3 kb Not1 fragment is diagnostic of wild-type HAP2 and the 1.3-kb fragment is diagnostic of the disrupted allele. (Bottom panel) Percentage of the indicated gametes that fused when mixed with wild-type gametes of the opposite sex. (B) In vitro malaria ookinete conversion analysis demonstrates that the Plasmodium hap2 mutant shows productive cross-fertilization with the nek4 sterility mutant, which produces functional males only, and not with cdpk4, which produces functional females only (error bar indicates SD; n = 3).
Malaria parasites always produce gametocytes of both sexes at the same time, and fertilization experiments with pure male or female gamete populations are not practical. Cross-fertilization experiments, however, with known gender-specific sexual development mutants, such as the male-deficient cdpk4 or the female-defective nek4 mutant (Billker et al. 2004; Reininger et al. 2005), make it possible to detect gender-specific sterility phenotypes. As shown in Figure 3B, neither cdpk4 nor nek4 strains produced ookinetes when cultured on their own, but when cultures containing both mutants were mixed, nek4 male gametes productively fertilized cdpk4 female gametes, restoring the capacity to form ookinetes (Fig. 3B). The reduced conversion rate compared with wild type was expected, because cultures also contained nek4 female gametes, which are unable to convert to ookinetes. In hap2/nek4 crosses the hap2 female gametes were productively fertilized by nek4 male gametes, but cdpk4 females remained unable to differentiate into ookinetes in cdpk4/hap2 crosses (Fig. 3B), showing that hap2 males are sterile. Thus, our results demonstrate that during fertilization in Chlamydomonas and Plasmodium, HAP2 is essential in gametes of only one sex, minus in Chlamydomonas and male in Plasmodium.
Unlike many organisms whose gametes possess an extracellular matrix that must be removed before fusion, Plasmodium’s gametes are “naked” (Sinden 1983). We reasoned, therefore, that HAP2 also would function at a step in Chlamydomonas fertilization when the gametes are “naked”; that is, after gamete activation and degradation of their glycoproteinaceous cell walls. One of the first events in the cilium-generated signaling pathway triggered by flagellar adhesion is release into the medium of an active metalloprotease that degrades the gamete cell walls. The experiments shown in Figure 1 demonstrated that HAP2 functioned after flagellar adhesion, but they did not address whether the hap2 gametes were capable of being activated when mixed with wild-type plus gametes, nor did they address whether hap2 minus gametes were capable of activating wild-type plus gametes during flagellar adhesion. To test whether hap2 mutants possessed the signaling pathway essential for cell wall degradation, we incubated them in dibutyrlyl cAMP, a cell-permeable analog of the second messenger that activates Chlamydomonas gametes (Pasquale and Goodenough 1987) and used a detergent sensitivity assay to assess cell wall loss (Snell 1982). To determine if hap2 cells responded to more physiological triggers of gamete activation, we also assessed wall loss in mixtures of hap2 gametes with wild-type plus gametes and with flagella isolated from wild-type plus gametes. As shown in Figure 4A, all of the gametes in such mixtures lost their cell walls in each of the three experimental conditions. The hap2 minus gametes released their walls when mixed with wild-type plus cells, indicating that at least one of the gametes was activated by the interaction; they released their walls when incubated with cAMP, indicating that the signaling pathway was intact in the absence of flagellar adhesion; and they released their cell walls when undergoing flagellar adhesion with isolated plus flagella (Snell 1982), indicating that they were capable of responding to flagellar adhesion per se and releasing their own metalloprotease. Thus, the flagellar adhesion-induced gamete activation pathway was fully functional in hap2 gametes.
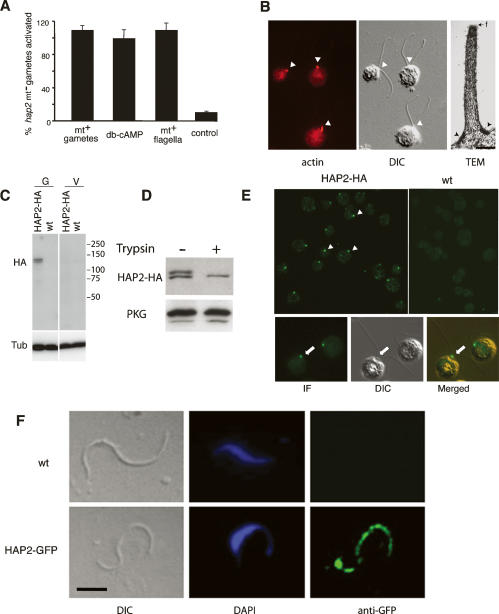
HAP2 functions after gamete activation, is present at the surface of the minus mating structure in Chlamydomonas (A–E), and is distributed along the length of the male gamete in Plasmodium (F). (A) 63B10 gametes were activated by incubation with wild-type plus gametes, flagella isolated from wild-type plus gametes, or db-cAMP. The percentage of cells that was activated was determined by measuring cell wall loss. (B, left panel) Fluorescence image of activated plus gametes in a mixture of wild-type plus gametes and 63B10 minus gametes. The arrowheads in the fluorescence (left panel) and DIC (middle panel) images indicate the site of the actin-filled mating structures. The right panel is an electron micrograph of an activated mating structure (fertilization tubule) on a wild-type plus that had been mixed with hap2 minus gametes. The actin filaments within the fertilization tubule originate from the cone-shaped, electron-dense doublet zone (arrowheads) at the base of the process (Detmers et al. 1983). Fringe (f; arrow) is visible on the surface of the fertilization tubule. Bar, 200 nm. (C) Immunoblotting with an anti-HA antibody documents that 63B10 cells rescued with HA-tagged HAP2 expressed HAP2-HA protein only in the gamete phase of their life cycle. (D) Immunoblotting with anti-HA antibody indicates that only a single form of HAP2-HA remained after treatment with 0.01% trypsin for 20 min at room temperature. (E) Anti-HA immunostaining combined with DIC microscopy of HAP2-HA gametes demonstrates that HAP2-HA is localized between the two flagella at the site of the minus mating structure. (F) Anti-GFP immunostaining and DIC microscopy of HAP2-GFP gametes of P. berghei shows HAP2-GFP localized along the length of the male gamete (bar, 2 μm).
The above experiments, nevertheless, would not have indicated whether the defect in hap2 minus gametes abrogated their ability to activate plus gametes, since it was possible, for example, that the wall loss we observed in the mixture of hap2 and wild-type plus gametes was due to release of the metalloprotease from the minus gametes only. When we examined the plus cells in such mixtures, however, we found that they formed the typical, actin-filled fertilization tubules that characterize activated plus gametes (Fig. 4B). In the electron micrograph (Fig. 4B, right panel), the actin filaments within the fertilization tubule are visible and can be seen arising from the submembranous doublet zone (Fig. 4B, arrowheads) at the base of the process. The fringe material (Fig. 4B, arrow) on the outer surface of this wild-type plus mating structure is reported to be a manifestation of the FUS1 protein and is absent in fus1 mutants (Goodenough et al. 1982). In an independent approach to confirm that the defect in the hap2 gametes was downstream from cAMP signaling, we found that addition of db-cAMP to samples of adhering wild-type plus gametes and 63B10 minus gametes failed to rescue the formation of zygote aggregates (data not shown). Thus, HAP2 is not required for activation of minus gametes, nor is it required for minus gametes to activate plus gametes to undergo the cytoskeletal reorganization required for formation of the fertilization tubule.
HAP2 is exposed on the external surface of the plasma membrane of Chlamydomonas and localized at the minus mating structure, a site specialized for cell fusion
To dissect further the function of HAP2 in gamete interactions, we introduced an HAP2-HA into 63B10 gametes. Immunoblotting with an anti-hemagglutinin (HA) antibody showed that HAP2-HA, which exhibited the predicted molecular mass of ~120 kDa, was expressed only in gametes and was not detectable in vegetative cells (Fig. 4C). The presence of two closely spaced isoforms of HAP2-HA in SDS-PAGE (Fig. 4D) suggested that the protein undergoes post-translational modification. If HAP2 were exposed on the external surface of the cell, as expected from the presence of a signal peptide and a single transmembrane domain near the C terminus of the predicted protein, then it should be sensitive to protease treatment of live cells. To determine if HAP2 is exposed at the cell surface, HAP2-HA-expressing minus gametes were incubated for 20 min at room temperature with 0.01% trypsin. Examination by phase contrast microscopy of the trypsin-treated gametes showed that the cells remained fully viable and motile, indicating that the protease treatment was not toxic to the cells. Analysis by immunoblotting indicated that the staining profile of HAP2-HA was modified by the trypsin treatment. The upper form was trypsin-sesitive and only a more rapidly migrating form remained. Although further experiments would be required to determine the relationships between the HAP2-HA isoforms present before and after trypsin treatment, this result demonstrated that at least one of the isoforms is exposed on the external surface of gametes (Fig. 4D). The presence on the immunoblots of equal amounts of cGMP-dependent protein kinase (PKG) (Wang et al. 2006), which is not exposed on the external cell surface, confirmed equal loading on the control and trypsin-treated lanes.
Immunofluorescence imaging of minus gametes expressing HAP2-HA showed that the protein was present at a single spot on each cell near the bases of the two flagella, the location of the mating structure (Fig. 4E). Control wild-type minus gametes did not stain with the antibody. Thus, the topology and the location of HAP2 were consistent with the cell biological properties of the protein, all of which pointed to a function of the protein in the membrane fusion reaction, either in prefusion attachment of the fusogenic membranes or in the membrane merger events that are downstream from attachment.
In contrast to studies in Chlamydomonas, ultrastructural studies from Plasmodium do not suggest that gametes possess mating structures or otherwise specialized cell surface areas to serve as preferred sites for adhesion and fusion (Sinden et al. 1976). We therefore hypothesized that in Plasmodium male gametes HAP2 might be more evenly distributed. Consistent with this notion, we detected a faint signal along the length of male gametes by fluorescence microscopy of live HAP2-GFP male gametes (data not shown), a result that was confirmed when fixed cells were stained with anti-GFP antibody (Fig. 4F). Fluorescence microscopy of live HAP2-GFP microgametocytes detected HAP2 on the cell periphery and additionally in one or a few dot-like intracellular locations of unknown identity (data not shown). Consistent with its stage- and gender-specific function in Plasmodium, HAP2-GFP expression from the endogenous HAP2 promoter occurred exclusively in Plasmodium male gametocytes and gametes, but neither in female or asexual erythrocytic stages, ookinetes, oocysts, nor salivary gland sporozoites (data not shown).
HAP2 functions downstream from gamete membrane attachment in Chlamydomonas and Plasmodium
Previously, we showed that the plus gamete-specific Chlamydomonas protein FUS1 is localized on the surface of the plus mating structure and is essential for prefusion attachment between the mating structures of activated minus and plus gametes. A simple model for mating structure interactions would be that HAP2 is a binding partner for FUS1, and that this interaction is required for prefusion attachment of the mating structures. To test this model, we examined whether HAP2 was essential for mating structure adhesion. In our initial characterization of the hap2 mutant, we found that hap2 minus gametes bound firmly to wild-type plus gametes, but we were unable to establish whether the overall cell adhesion we observed was due to adhesion of the mating structures or of the flagella, or if both were involved in binding the two cells together (Fig. 1). In previously published work, we had also determined that the FUS1 on unactivated plus gametes was nonfunctional in adhesion and that plus gamete activation was required to generate active FUS1 (Misamore et al. 2003).
To examine mating structure adhesion between plus gametes and hap2 minus gametes without the interference of flagellar adhesion, we used a plus mutant, sag1-1, which does not express SAG1, the plus flagellar agglutinin (Ferris et al. 2005). Furthermore, since flagellar adhesion is essential for gamete activation, we activated the hap2 and sag1-1 gametes for these experiments with dibutyryl cAMP before we mixed them. Such treatment ensured that the cell walls were removed and that the mating structures were exposed and in their activated states. The activated sag1-1 plus gametes were subsequently fixed and tagged with a fluorescent marker. To our surprise, when we mixed the activated hap2 and sag1-1 gametes together, they formed prefusion attachments with each other at their mating structures (Fig. 5A, right two sets of panels, arrowheads indicate the sag1-1 plus gametes), which were indistinguishable from the mating structure adhesions of wild-type minus gametes and sag1-1 plus gametes (Fig. 5A, left two sets of panels; Misamore et al. 2003). Even though the mating structure interactions were brought about by random collisions of the cells and not by flagellar adhesion, ~20% of the sag1-1 plus gametes formed pairs with both the wild-type and hap2 minus gametes (Fig. 5A). That the gametes remained as pairs as they were propelled through the medium by flagellar motility indicated that mating structures were tightly attached to each other. Thus, these experiments demonstrated that in the absence of HAP2, FUS1-dependent mating structure adhesion was not perturbed, and that unlike FUS1, HAP2 is dispensable for tight prefusion attachment during the membrane fusion reaction. These data indicate that gamete membrane adhesion and fusion are separate molecular events in which FUS1 and HAP2 function sequentially, rather than through direct interaction.
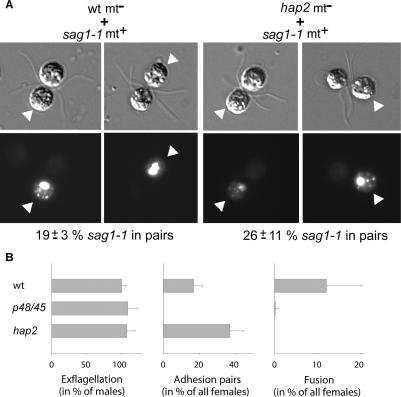
HAP2 functions in the gamete fusion reaction downstream from gamete membrane adhesion. (A) Activated live 63B10 gametes, like activated live wild-type minus gametes, adhered via their mating structures to activated, fixed, fluorescently tagged sag1-1 plus gametes, which are incapable of flagellar adhesion (top panel shows differential interference microscopy; bottom panel shows fluorescence; arrowheads indicate the sag1-1 plus gametes). The percent (±SEM) of sag1-1 gametes forming pairs when mixed with an excess of activated 63B10 or wild-type minus gametes is shown below the figure (average from two independent experiments; n = 150–200 sag1-1 cells examined in each). Similar results were obtained when the agglutinin mutant sag1-2 was used (not shown). Between 0 and 6% pairs were detected in controls in which activated live sag1-1 gametes were mixed with the fixed sag1-1 gametes (not shown). (B) Efficiency of exflagellation, gamete adhesion, and gamete fusion in wild-type, p48/45, and hap2 clones of Plasmodium (error bar indicates SD; n = 3 experiments, each examining 100 gametocytes).
To determine whether HAP2 functioned at a similar step in the membrane fusion reaction of Plasmodium gametes, we used phase contrast microscopy to examine the interactions of hap2 male gametes with female gametes. As with Chlamydomonas, binding of wild-type male gametes to wild-type females is followed within seconds by fusion. Microscopic examination of fertilization in vitro showed that in the absence of HAP2 or in the absence of P48/45, exflagellation occurred normally as expected. On the other hand, the incidence of male/female Plasmodium gamete pairs in the hap2 mutant was approximately doubled compared with wild type (Fig. 5B); the failure to detect fertilization indicated that hap2 pairs formed and persisted but failed to progress from adhesion to membrane fusion. In marked contrast, in fertilization experiments with a p48/45 mutant, a complete lack of gamete binding explained fully the absence of fertilization (Fig. 5B), confirming the importance of the Plasmodium-specific P48/45-containing complex in gamete adhesion (van Dijk et al. 2001). Thus, as with Chlamydomonas, fertilization in Plasmodium can be experimentally divided into an adhesion step that relies on at least one species-limited membrane molecule, here P48/45, and a subsequent fusion step requiring HAP2.
Post-attachment membrane merger in Chlamydomonas requires HAP2
Although the above experiments pointed to a role for HAP2 in membrane merger per se, it was possible that HAP2 functioned downstream from membrane merger. For example, even though in Chlamydomonas the hap2 minus gametes did not form quadriflagellated zygotes, membrane merger could have occurred, and HAP2 could have been required to bring about the cellular events that accomplish complete coalescence of the two cells into a single quadriflagellated zygote after cytoplasmic continuity was established. Studies of membrane fusion reactions in other systems have established that fusion can initiate through opening of a proteinaceous pore followed by lipid merger (Han et al. 2004) or, alternatively, with the merger of the outer leaflets of the interacting membranes to form a hemifusion connection, followed by formation of a pore lined by the fused lipid bilayers (Chernomordik and Kozlov 2005; Lu et al. 2005; Reese and Mayer 2005; Xu et al. 2005). To test whether HAP2 functioned downstream from either partial or complete membrane merger in Chlamydomonas, we tested for redistribution of a membrane dye (PKH26) (Podbilewicz et al. 2006) from the plasma membranes of labeled wild-type plus gametes to wild-type and hap2 minus gametes. When the labeled wild-type plus gametes (Fig. 6A, top panel, arrowheads) were mixed with unlabeled wild-type minus gametes, we detected initiation of redistribution of dye from the plus gametes to the minus gametes within seconds after their mating structures came into close contact, and complete redistribution occurred immediately thereafter (Fig. 6A, top panels). Interestingly, even the flagellar membranes of the minus gametes acquired the dye very quickly (Hunnicutt et al. 1990). On the other hand, when labeled wild-type plus gametes were mixed with hap2 minus gametes, redistribution of label was never detected in the hundreds of wild-type plus/63B10 pairs examined in several independent experiments (Fig. 6A, bottom panels; data not shown).
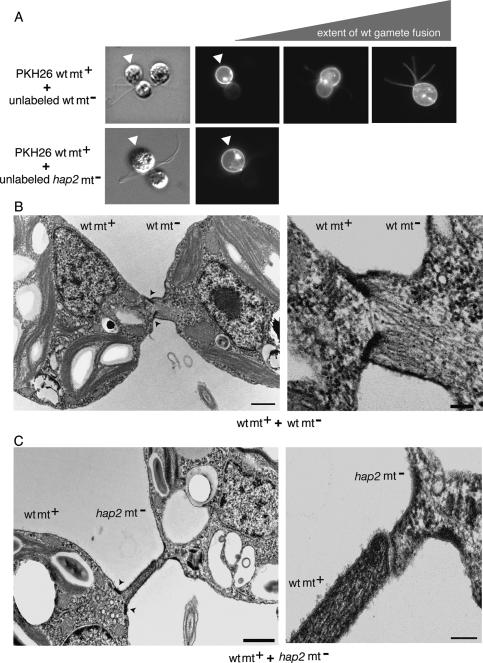
HAP2 is essential for membrane merger in Chlamydomonas. (A) The plasma membranes of activated plus gametes were labeled with the fluorescent lipid PKH26 (arrowheads), mixed with wild-type minus gametes (top panels) or 63B10 minus gametes (bottom panels), and the live cells were examined by DIC (left top and bottom panels) and epifluorescence microscopy. (B) A pair of wild-type plus and minus gametes just after fusion is shown in transmission electron microscopy at low magnification (left panel; bar, 200 nm) and at higher magnification (right panel; bar, 50 nm). The actin filaments of the fertilization tubule of the plus gamete (cell on the left in both images) have become incorporated into the minus gamete as the two cells merge. (C, left panel) In a mixture of wild-type plus gametes and hap2 minus gametes, the tip of the fertilization tubule on a wild-type plus gamete (cell on the left) is tightly associated with the apex of the minus mating structure (bar, 200 nm; arrowheads show the doublet zone). (Right panel) A higher-magnification view (bar, 50 nm) shows that the membranes of the two mating structures are separated from each other by ~10 nm.
Transmission electron microscopy confirmed the absence of fusion in the wild-type/hap2 mixtures. The top panels of Figure 6B show a wild-type plus (cell on right) and wild-type minus gamete whose mating structures have just fused (bars in left panel, 200 nm; bar in right panel, 50 nm). The actin filaments within the foreshortened plus mating structure were still attached to the doublet zone and had penetrated into the cytoplasm of the minus gamete as fusion progressed (see also Detmers et al. 1983). Attesting to the rapidity of the fusion process, even after examining 500–600 wild-type gametes fixed 3 min after mixing, we were unable to capture wild-type gametes in the attachment stage of mating structure interactions or at a stage in fusion earlier than that shown in these images. On the other hand, and consistent with the absence of dye transfer described above, we never detected mating structure fusion in mixtures of wild-type plus and hap2 minus gametes. Instead, we observed many pairs in which the tip of the mating structure of the wild-type plus gamete was closely apposed to the apex of the shorter minus mating structure. Figure 6C shows one such pair at low magnification (left panel, bar, 200 nm; arrowheads point to the doublet zone in the plus) and higher magnification (right panel, bar, 50 nm). The membranes of the mating structures appeared to be adherent to each other by the FUS1 fringe and were separated by ~10 nm. Taken together, these results demonstrated that HAP2 is essential at a step in the gamete membrane fusion reaction that occurs within seconds after species-specific, tight, prefusion attachment of the fusogenic membranes and that is concomitant with or immediately precedes membrane fusion.
Discussion
By taking advantage of gene discovery methods in Chlamydomonas, targeted gene disruption in P. berghei, and distinctive features of fertilization in both organisms, we uncovered a novel mechanism for gamete membrane fusion in which species-limited proteins are essential for binding the male and female gamete membranes together and a broadly conserved protein is essential—either directly or indirectly—for fusion per se. Each of these distantly related organisms depends on a species-limited protein—FUS1 in Chlamydomonas and a P48/45-containing protein complex in P. berghei—for attachment of the plasma membranes of gametes of opposite sex. And each requires a member of the broadly conserved HAP2 protein family to accomplish the subsequent membrane merger step of the membrane fusion reaction. Chlamydomonas is particularly amenable to use of gene discovery methods to identify fertilization mutants because it is haploid for most of its life, and thus mutant phenotypes are detectable in the immediate progeny of the cell that received the mutagenizing plasmid (in these experiments, an antibiotic resistance gene), and zygotes form aggregates of a distinctive morphology that allows for rapid screening of mutants. Plasmodium offers the power of targeted gene disruption methods. The many steps that compose fertilization in both organisms are experimentally accessible, and genes essential at several steps in fertilization have already been identified.
HAP2 is not required for gamete migration or species-limited gamete recognition and signaling
The ability to use mutants and cell bioassays to dissect several steps in fertilization in both organisms allowed us to position HAP2 in the gamete interaction pathway with much more precision than is possible in plants. In vitro fertilization between isolated sperm and eggs has been achieved in rice and maize (Faure et al. 1994; Kranz and Lorz 1994; Khalequzzaman and Haq 2005; Peng et al. 2005), but not yet in Arabidopsis. Previous studies had established that in addition to its role in pollen tube guidance in Arabidopsis, HAP2 also was essential in seed formation at a step after deposition of sperm in the synergid cell in the general region of the egg (Johnson et al. 2004; Mori et al. 2006; von Besser et al. 2006). Unknown, however, was the step in fertilization that required HAP2. Mori et al. (2006) proposed that HAP2 might play a role in gamete recognition. It was also possible that the protein was essential for chemotaxis or for motility to bring the sperm and egg together (von Besser et al. 2006).
In our studies, we established that HAP2 in Chlamydomonas is dispensable for the flagellar adhesion and signaling necessary for release of cell walls and activation of the fusogenic membranes of gametes. Presumably, these events are not required for the “naked” gametes of Arabidopsis or P. berghei. Furthermore, the results that the hap2 mutants in both Chlamydomonas and P. berghei were fully capable of motility ruled out the possibility that HAP2 functioned in migration. Finally, our observations that hap2 gametes in both Chlamydomonas and Plasmodium recognized and adhered to gametes of the opposite sex indicated that HAP2 is not essential for species-limited membrane recognition or the tight, prefusion attachment of the fusogenic membranes that precedes membrane fusion.
Although the working model for gamete fusion has been that prefusion attachment and membrane fusion per se depend on separate sets of gene products, the model was not supported by genetic evidence because no mutants were available that allowed adhesion and blocked fusion in any organism. Our results assigning HAP2 function to a step in the gamete membrane fusion reaction after close (10-nm) prefusion attachment is the first gene disruption-based evidence that the gamete membrane fusion reaction depends on at least two separate sets of proteins that function at discrete steps in the reaction. The 10-nm gap between the attached membranes is similar to the gap between Drosophila myoblast membranes at the prefusion attachment step of myoblast fusion (Doberstein et al. 1997) and similar to that between C. elegans epithelial cells just before EFF-1-dependent cell fusion (Shemer et al. 2004). Presumably, the two membranes are close enough for a fusion protein or protein complex to bridge the gap and cause membranes to merge.
Mechanisms of gamete fusion
Molecular mechanisms of membrane fusion have been best studied in viral fusion and fusion of intracellular vesicles. A complex series of protein-binding and -folding reactions drives both sets of reactions. After an initial adhesion step, the fusion proteins link the opposing membranes, followed by protein folding that pulls the membranes in close proximity. Fusion pores form, and the process culminates in the complete merging of the lipid bilayers (Sollner 2004; Earp et al. 2005). Viruses use a single protein for both specific contact and for fusion itself, and the several classes of viral fusion proteins apparently evolved independently (Earp et al. 2005). Intracellular vesicle fusion employs distinct sets of conserved protein families for each step—rabs and their effectors for specific adhesion and SNARES for membrane merger (Jahn et al. 2003). Examination of the HAP2 protein sequence fails to offer any hints as to its possible role in the membrane fusion reaction. The HAP2 sequence predicts a single-pass transmembrane protein (Mori et al. 2006), and our results document that it is present on the external surface of the plasma membrane of Chlamydomonas. On the other hand, HAP2 lacks a predicted fusion peptide and the coiled-coil domains present in many viral fusion proteins.
Formation of syncytia in C. elegans depends on a single protein (EFF-1 or AFF-1) (Mohler et al. 2002; del Campo et al. 2005; Sapir et al. 2007), but the fusogen must be present on both of the fusing membranes (Podbilewicz et al. 2006). And myoblast fusion requires a mix of conserved and species-limited proteins for prefusion attachment, signaling, and cytoskeletal rearrangements (Chen and Olson 2005). The proteins in myoblasts that mediate the membrane merger step per se have yet to be identified, although recent studies on the MARVEL domain protein, Singles Bar, showed that it is required for progression past the prefusion attachment stage of myoblast fusion (Estrada et al. 2007).
Whether HAP2 functions directly as a fusogen or has a more indirect, broadly conserved role in hypothetical events after prefusion attachment and before union of the two lipid bilayers remains to be investigated. Membrane fusion reaction mechanisms were invented only infrequently during evolution (Jahn et al. 2003), and the conserved function of HAP2 in the gamete membrane fusion reaction in two widely disparate organisms would thus be consistent with a direct role for HAP2 in the final event of fertilization.
HAP2 is located specifically at the mating structure of Chlamydomonas minus gametes, yet in P. berghei it is distributed along the length of the microgamete, suggesting that in the Plasmodium microgametes, no organelle or membrane area specialized for gamete fusion may exist. HAP2-GFP was notably not concentrated at the anterior end of the microgamete, where a juxta-kinetosomal sphere and granule of unknown function are located (Sinden et al. 1976). Future work will have to examine the expression and cellular localization of HAP2 on the microgametes of human malaria species, as well as its accessibility to antibodies, which may be able to block malaria transmission by interfering with gamete fusion within the infected blood meal. Consistent with its mosquito stage-specific expression in P. berghei, a low frequency of single nucleotide polymorphisms in HAP2 sequences from different P. falciparum isolates (Jeffares et al. 2007; Mu et al. 2007; Volkman et al. 2007) suggests that PfHAP2 (accession no. PF10_0139) is under no diversifying immune selection, an important property it shares with current transmission-blocking vaccine candidates (Saul 2007).
Gamete fusion, species specificity, and a conserved gene family
Because gamete fusion in many organisms is a species-specific event, it has been unclear whether the elucidation of mechanisms of the membrane fusion reaction in one species will be broadly applicable. The functional conservation of a role for HAP2 at the membrane merger step of fertilization in Chlamydomonas and P. berghei makes it likely that close to the unresolved root of eukaryote evolutionary history the last common ancestor of both species and of the higher plants would have used HAP2 for gamete fusion. The presence of HAP2 homologs in choanoflagellates and other protists, cnidarians, and some bilaterian animals further suggests that dissecting the molecular function of HAP2 could offer insights into fundamental mechanisms of gamete fusion for a large portion of the Earth’s species.
The model that species-specific proteins govern access to a conserved, HAP2-dependent process of membrane fusion is attractive because it provides an explanation for the observation that many genes involved in gamete recognition diverge rapidly (Ferris et al. 1997; Swanson and Vacquier 2002; Vieira and Miller 2006), although the process of fertilization is fundamental to all eukaryotes. Divergence of the prefusion attachment genes could contribute to establishment of barriers to fertilization that might lead to speciation. The functional separation of membrane adhesion and subsequent events resulting in fusion between two different membranes may thus be the way in which many eukaryotes reconcile two opposite evolutionary needs, on the one hand, to ensure reproductive isolation through rapidly changing gamete recognition mechanisms, and, on the other hand, to preserve the machinery for the biophysically complex process of membrane fusion.
Materials and methods
Chlamydomonas
Unless stated otherwise, strains were from the Chlamydomonas Genetics Center, Duke University, Durham, NC. Cell culture, induction of gametogenesis, flagellar isolation, gamete activation by db-cAMP, measurement of cell wall loss, SDS-PAGE, and immunoblotting were as described previously (Wang et al. 2006).
Insertional mutagenesis: We screened 2500 insertional mutants generated in the nitrate reductase-deficient B215 mt− strain (Greg Pazour, University of Massachusetts) for clones whose gametes underwent flagellar adhesion for 12–18 h but failed to fuse. Insertional mutants were generated using the plasmid pSI103 linearized with PvuII and transformed into B215 cells using the glass bead method with selection on agar plates containing 10 μg/mL paromomycin (Sigma) in M medium (Kindle et al. 1989; Fang et al. 2006). Transformed colonies were induced to undergo gametogenesis by transferring them into 96-well plates containing M-N medium. After agitation on a reciprocal shaker for 2 h, 5 μL from each well were transferred into a duplicate 96-well plate containing M media to maintain a stock of the cells in vegetative growth. After continued agitation overnight, samples from each well of the plate with M-N were mixed with wild-type plus gametes. Each well was scored on an inverted microscope for flagellar agglutination at 10 min and 4 h. Zygote formation, as determined by the presence of large aggregates of zygotes visible in the inverted microscope, was assessed at 4 h. To confirm a single insertion of plasmid pSI103 in 63B10 cells, Southern blotting was carried out with genomic DNA from wild-type and 63B10 cells digested separately with EcoRI and HindIII. The probe was a pSI103/PvuII DNA fragment labeled using a Random Primed DNA labeling kit (Roche Applied Science).
PCR and TAIL–PCR: We used TAIL–PCR (Liu et al. 2005) to determine that gene model C_530033 (Protein ID 166688; version 2.0 of the Chlamydomonas genome sequence at http://genome.jgipsf.org/cgi-bin/dispGeneModel?db=chlre2&tid=166688) (Grossman et al. 2003) had been disrupted by insertion of the aphVIII plasmid pSI103 mutagen (a paromomycin resistance gene) (Sizova et al. 2001) in mutant 63B10. The specific, nested primers to identify genomic sequence in the 5′-flanking region of the inserted aphVIII plasmid in clone 63B10 cells were the following: primary, Aph.p22 (5′-GCGCCCTCATAGCCCGCCAAATC-3′); secondary, Aph.p21 (5′-CCGCCAAATCAGTCCTGTAGCTTC-3′); and tertiary, Aph.p20 (5′-TGCGCGCTTGGCGTAATCATGGTC-3′). The arbitrary degenerate primer was Ad.p24 [(G/C)TAGA(G/C)T(G/C)A(G/C)C(A/T)CA(G/C)] (C. Silflow, pers. comm.). The PCR product from the tertiary reaction, which was cloned and sequenced, is the following (the underlined sequence is C_530033, the lowercase sequence is an Escherichia coli cytosine methylase presumably from the plasmid host bacterium, and the nonunderlined sequence is from the aphVIII plasmid): (5′-CCGCCAAATCAGTCCTGTAGCTTCCATATCT GATTCGCAATCTTGCCTTGCACCTGCCTGCCACGCTCAT ACCATGTCGCCGTGACCCCAAAACAGGCCTGTCTGTCCG GCCAGCTCAAGGACCTGTGGGAGGCGGACCTGGCGCGT ACCGCGGACGGCCGGGTGCCGCTGTACATGATCACCAG GTTCACTGGCGGCAGCGAGGGCTAATCGCGCCGGAAAA TATATCAGTAACCGATTCATACAGCACCGGGAATGCCGCACAGGCAATGCTGGAGAA ACTGCTGCAAATTTATGATGTTAAAACGTTGGTGGCGCAGCTTAATGGTGTAGGTGAGAATCACTGG AGCGCGGCAATTTTAAAACGTGCGCTGGCGAATGACTCGGCATGGCACCGTTTAAGTGAGAAAGAG TTCGCCCATCTGCAAACGTTATTACCCAAACCACCGGCACATCATCCGCATTATGCGTTTCGCTTTA TCGATCTATTCGCCGGAATTGGCGGCATCCGTCGCGGTTTTGAATCGATTGGCGGACAGTGCGTGTTT TCCAGCGAATGGAACAAACATGCGGTACGCACTTATAAAGCCAACCATTATTGCGATCCGGCGAC GCATCATTTTAATGAAGATATCCGCGACATCACCCTCAGCCATAAAGAAGGCGTGAGTGATGAGGC GGCGGCGGAACATATTCGTCAACAATTTCACACAGGAAACAGCTATG ACCATGATTACGCCAAGCGCGCA-3′). Other primers used for PCR were the following: Hap2.p1 (5′-ATGTCGCCGTGAC CCCAAAACAG-3′); Hap2.p2 (5′-CTGGCTGGTGACAGGCAGC GCGAA-3′); and Aph.p17 (5′-TTGGCTGCGCTCCTTCTGGCGC-3′).
Transformation of Chlamydomonas with HAP2 constructs: HAP2-HA: The 8.3-kb SstI fragment from DNA BAC clone 20L3 obtained from the Clemson University Genomics Institute, Clemson University, containing gene model C_530033 was inserted into the SstI site of pUC119 to generate pYJ36. Standard methods were used to insert a PCR product encoding three copies of the 9-amino-acid HA epitope (Silflow et al. 2001) into the NheI site of pYJ36 to generate pYJ58. To obtain 63B10 cells containing the HAP2-HA construct, we carried out cotransformation with the glass bead method using pYJ58 and plasmid pmn56 encoding the nitrate reductase gene (Tam and Lefebvre 1993). For the experiment shown in Figure 1B, 63B10 cells were cotransformed with the gel-purified 8.3-kb Sst1 fragment of BAC clone 20L3 and pmn56. Transformants were selected for their ability to undergo fusion with wild-type plus gametes.
Generation of a plus strain containing only disrupted HAP2: 63B10 minus gametes rescued for fusion with transgenic HAP2-HA were crossed with 21gr plus gametes, and the progeny were grown using standard procedures (Harris 1988). Colonies formed by germinated zygotes on 2% agar plates were pooled and inoculated into a growth flask containing M media. Progeny cells were subcloned on agar selection plates containing 10 μg/mL paromomycin and screened for plus progeny that contained the disrupted hap2 allele from the 63B10 cells and lacked both the wild-type HAP allele and the HAP2-HA insert. To confirm the genotype of the transformant, Southern blotting was carried out with genomic DNA digested with NotI. The probe was a cloned PCR product generated using Hap2.p21 (CTGATGGTGCCG CAGCCGCCAGC) and Aph.p20 primers with 63B10 genomic DNA as template and labeled using a Random Primed DNA labeling kit (Roche Applied Science).
Indirect immunofluorescence: Gametes were washed with MT buffer (30 mM Tris acetate at pH 7.3, 5 mM MgSO4, 5 mM EDTA, 25 mM KCl, 1 mM dithiothreitol) and loaded onto eight-well slides coated with 0.1% polyethylenimine for 10 min (Mahjoub et al. 2004). Cells were fixed in 100% ice-cold methanol for 20 min at −20°C, washed three times for 5 min in PBS, and blocked for 30 min with blocking serum (1% cold-water fish gelatin, 0.1% bovine serum albumin [BSA], 5% goat serum in PBS). The slides with fixed cells were incubated with rat monoclonal anti-HA antibody (Roche Applied Science; diluted 100-fold) for 2 h, rinsed three times in PBS, and then incubated for 1 h with fluorescein-conjugated goat anti-rat IgG (ICN/CAPPEL; 1:400 dilution) in blocking serum. The slides were rinsed in PBS and mounted in Fluoromount-G (Southern Biotech). Fluorescence microscopy was performed using an Ultraview ERS spinning disk confocal microscope (Perkin Elmer). Final composite images were constructed using Image J (NIH) and Adobe Photoshop (Adobe Systems).
Assessing gamete activation: To test whether 63B10 gametes were capable of gamete activation, 250 μL of 63B10 gametes at 1.6 × 107 cells per milliliter were mixed for 30 min with an equal number of 21gr plus gametes, with dibutyryl cAMP, or with flagella isolated from plus gametes. For the experiment with isolated flagella, 10 cell equivalents of flagella were added at 5-min intervals, and cell wall loss was determined as described previously (Snell 1982). The data shown are averages from three independent experiments, each done in duplicate, and the error bars are SEM. To evaluate activation of plus gametes by 63B10 minus gametes, the two types of gametes were mixed together for 30 min, and fertilization tubules, an indicator of plus gamete activation, were visualized with the actin-specific fluorochrome Alexa 546-phalloidin (Molecular Probes) as described previously (Wilson et al. 1997).
Prefusion attachment assay: Activated sag1-1 gametes were incubated in 2.5% glutaraldehyde in N-free medium for 10 min, washed twice in 1% glycine in 10 mM phosphate-buffered M-N medium for 5 min, incubated with the live-cell impermeant, nucleic acid fluorochrome SYTOX-green (1 mM in M-N medium) for 10 min, and washed twice in M-N medium. To carry out the attachment assay, fixed, SYTOX-labeled sag1-1 plus gametes were mixed with live, activated 63B10 or wild-type minus gametes and allowed to interact for 30 min. The mixed samples were placed on a microscope slide, fixed in 0.1% paraformaldehyde, covered with a coverslip supported by Vaseline posts, and viewed by fluorescence and differential interference contrast (DIC) microscopy.
Assessing membrane fusion: The plasma membranes of activated plus gametes (2 × 107 cells per milliliter in M-N medium) were labeled by mixing the cells in M-N with an equal volume of Staining Solution containing PKH26 red fluorescent dye (Sigma; 4 μM in M-N) for 10 min at 23°C. The reaction was stopped by addition of BSA to a final concentration of 1% for 1 min, and cells were washed three times with M-N medium by centrifugation. The labeled gametes were mixed with unlabeled wild-type or 63B10 minus gametes and examined by epifluorescence and DIC microscopy.
Electron microscopy: Plus wild-type gametes were mixed with wild-type or hap2 minus gametes for 3 min (wild-type plus gametes and wild-type minus gametes) or 30 min (wild-type plus gametes and hap2 minus gametes) and fixed in 1% glutaraldehyde and 0.2% tannic acid following the procedure of Goodenough et al. (1982) with some modifications (Begg et al. 1978; Detmers et al. 1983). Concentrated fixative (1.5 mL of 5% glutaraldehyde, 1% tannic acid in M-N medium) was added, one drop at a time with gentle mixing, to 6 mL of cells in M-N medium. Cells were fixed for 2 h and were then allowed to settle onto poly-L-lysine-coated coverglasses. Cells on coverglasses were rinsed in M-N medium, post-fixed in 0.5% osmium tetroxide, en bloc stained in 2% uranyl acetate, dehydrated in a graded ethanol series, and transferred through propylene oxide into EMbed-812 resin. After overnight polymerization, coverglasses were removed by immersion in liquid nitrogen. Thin sections (55 nm) were cut on a Leica EM UC6 and post-stained with 2% uranyl acetate and lead citrate. Sections were viewed in a FEI Tecnai G2 Spirit electron microscope operating at 120 kV. Images were acquired with a Gatan UltraScan 1000 camera and DigitalMicrograph software.
Plasmodium
The P. berghei ANKA wild-type strain 2.34 was used throughout this study. Strain 2.33 is a gametocyte-deficient strain derived from 2.34 by continued blood passage. cdpk4, nek4, and p48/45 mutant clones were described previously (van Dijk et al. 2001; Billker et al. 2004; Reininger et al. 2005). P. berghei was maintained in Theiler’s Original outbred mice and transmitted to Anopheles stephensi, strain SDA 500, as described previously (Sinden et al. 2002). Mice were infected by intraperitoneal injection of infected blood containing ~106 parasites. The course of infections and gametocyte production were monitored on Giemsa-stained blood films. To trigger exflagellation, 5 μL of gametocyte-infected blood was mixed with 400 μL of exflagellation medium (RPMI1640 containing 25 mM HEPES, 20% fetal bovine serum [FBS], 10 mM sodium bicarbonate, 50 μM xanthurenic acid at pH 7.6). Between 15 and 25 min later, exflagellation centers per 104 red blood cells (RBCs) were counted in a hemocytometer. To determine the relative ability of microgametocytes to exflagellate, numbers of exflagellation centers were compared with numbers of morphologically mature microgametocytes per 104 RBCs on Giemsa-stained blood films prepared in parallel from the same infected mouse. The proportion of female gametes undergoing either adhesion or fusion with a male gamete was quantified by phase contrast microscopy in cultures undergoing gametogenesis under Vaseline-rimmed coverslips on glass microscope slides. Starting 15 min after activation, when mature microgametocytes had started to release male gametes, ~100 female gametes per experiment were observed and scored. Gamete adhesion was recorded when one or more male gametes were persistently associated with the surface of the female gamete, but when there was no sign of gamete fusion. Gamete fusion was recorded when adhesion resulted in the male gamete entering the female, or when characteristic motions in the zygote’s cytoplasm caused by the continued intermittent beating of the axoneme of the male gamete (Sinden and Croll 1975) indicated that fertilization had occurred. Since axoneme motility ceases ~1 min after fusion, our analysis underestimates the true incidence of fusion in wild type, as evidenced by the higher efficiency of ookinete formation in the same cultures (see Fig. 2). Ookinete conversion rates for different mutants and genetic crosses were determined in 20-h in vitro cultures of infected blood by immunolabeling the macrogamete/zygote/ookinete marker P28, as described previously (Reininger et al. 2005). For mosquito transmission experiments, batches of 50 mosquitoes were allowed to feed on anaesthetized mice on day 3–4 of a blood-induced infection for 20 min at 19°C. Unfed mosquitoes were removed the following day, and oocysts were counted on dissected midguts on day 10 after feeding.
Deletion of the HAP2 gene: To replace all protein-coding sequence of the HAP2 gene (GenBank accession no. XM_671808) with a T. gondii dhfr/ts expression cassette conveying resistance to pyrimethamine, a targeting vector was constructed in plasmid pBS-DHFR. A 736-bp fragment comprising 5′-flanking sequence immediately upstream of the start codon was amplified from P. berghei genomic DNA using primers GF-1 (5′-CCCCGGGCCCGCGCGTTATTATTATTCGGGC-3′, restriction site underlined) and GF-2 (5′-GGGGAAGCTTTTTTTC TAAATGAAATATTAAAGAATGGC-3′) and inserted into ApaI and HindIII restriction sites upstream of the dhfr/ts cassette of pBS-DHFR. A 967-bp fragment of 3′-flanking sequence was then generated using primers GF-3 (5′-CCCCGAATTCAT TACATGGAATAGTATTTGCAAATTTG-3′) and GF-4 (5′-GGGGTCTAGACAATATACATGCTGATAACCTCC-3′) and inserted downstream from the dhfr/ts cassette using EcoRI and XbaI restriction sites. The replacement construct was excised as an ApaI/XbaI fragment and used for the electroporation of cultured P. berghei schizonts as described (Janse et al. 2006). Following dilution cloning of drug-resistant parasites, genotyping of two hap2 clones was done by Southern blot hybridization on EcoRI-digested genomic DNA using the ApaI/HindIII fragment of 5′ targeting sequence as a probe. Diagnostic PCR analysis used primers GFko1 (5′-CTCGAATATGTAGATATATCCA GATG-3′) and GFko2 (5′-CAGAGATGTTATAGCTAGT GATATAAC-3′) specific for HAP2, and primers GFint (5′-CTAAGTAGCAACTATTTTGTAAAATTATATC-3′) and 70 (1) to span the predicted 5′ integration site.
Complementation of a hap2 mutant: A 4.2-kb fragment comprising 1.5 kb of upstream sequence and the complete PbHAP2 genomic sequence, except the stop codon, was amplified by PCR from genomic DNA using primers 485 (5′-ATATGGTAC CACGCTACTTATATATAGTGATAACC-3′) and 482 (5′-ATA TGGGCCCTCGCAATGGGGGTATTTTACTTTTAC-3′), inserted into KpnI and ApaI restriction sites of vector p277, upstream of and in frame with an EGFP gene, which was followed by 0.5 kb of 3′ untranslated region (UTR) derived from the PbDHFR/TS gene. The protein-coding sequence of the complementation construct was confirmed by sequencing. The vector was linearized in a unique EcoRV site in the putative HAP2 promoter and introduced into a hap2 knockout clone, and resistant parasites were selected with WR99210. Integration of the complementation construct into the disrupted hap2 locus was verified by PFGE and Southern blot analysis. HAP2-GFP expression was verified by Western blot analysis of purified wild-type and HAP2-GFP gametocytes, which were lysed by the addition of SDS-PAGE sample buffer (3% SDS, 3% mercaptoethanol, 20% glycerol, 60 mM Tris at pH 8, 0.1% bromphenol blue) and thereafter separated on a 10% SDS-PAGE gel. Proteins were immunoblotted onto PVDF filters (Millipore) and incubated with anti-GFP antibody (Molecular Probes; diluted 1:5000) followed by incubation with HRP-conjugated anti-rabbit antibodies (GE healthcare; diluted 1:20,000). Detection was enabled with Immobilon Western chemiluminescent HRP substrate (Millipore).
RT–PCR analysis of HAP2 expression: P. berghei RNA was isolated from equivalent numbers of purified wild-type and hap2 gametocytes and strain 233 asexual parasites using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Any residual gDNA was removed by treatment with RQ1 RNase-free DNase (Promega), and the resulting RNA was extracted with phenol/chloroform, precipitated with ethanol, resuspended in DEPC-treated water, and quantified by 0.8% agarose gel electrophoresis. First-strand cDNA synthesis from 1 μg of total RNA was done with M-MLV Reverse Transcriptase (Invitrogen) for 50 min at 37°C. Following heat inactivation for 15 min at 70°C, 2 μL of cDNAs were used per PCR reaction. Primers selected to amplify sections of the HAP2 ORF (spanning the 209-bp intron) were Forward, 5′-GCATAAGATTCACAAATA CAAAAAGG-3′; and Reverse, 5′-GGTCTTCCTCTAAGTAT T-3′. The expected RT amplicon was 1203 bp, whereas the gDNA amplicon was 1412 bp. The ubiquitously expressed α-tubulin gene PB300720.00.0 was amplified for each sample to ensure amplifiability of cDNA from respective RNA samples (Forward, 5′-CCAGATGGTCAAATGCCC-3′; Reverse, 5′-CT GTGGTGATGGCCATGAAC-3′). The expected products were 432 bp (cDNA) and 592 bp (gDNA). Thirty RT–PCR cycles were carried out with denaturation for 1 min at 94°C, annealing for 45 sec at 50°C, and extension for 1.5 min at 68°C, and products were visualized on a 0.8% agarose gel.
Indirect immunofluorescence: Immunofluorescence analysis was done with enriched male gametes prepared essentially as described by Carter and Chen (1976). Blood containing wild-type or HAP2-GFP gametocytes was activated by the addition of exflagellation media for 30 min at 19°C and thereafter centrifuged twice at 500g for 10 min. The resulting supernatant was then centrifuged at 13,000g for 10 min and left for 15 min at room temperature. The supernatant was again centrifuged at 20,000g for 10 min. Thick smears from the pellet, containing mainly male gametes, were fixed in ice-cold methanol, blocked in 10% FBS in PBS, and incubated with rabbit anti-GFP antibodies (Molecular Probes; diluted 1:1000) followed by incubation with Alexa 488-conjugated anti-rabbit antibodies (Molecular Probes; diluted 1:2000). Fluorescence microscopy was performed using a Leica DMR microscope. Images were constructed using AxioVision (Carl Zeiss imaging solutions) and Microsoft Office Picture Manager (Microsoft).
Acknowledgments
We thank Meredith Williams (University of Texas Southwestern) for assistance with the trypsin experiments; Carolyn Silflow (University of Minnesota, St. Paul, MN) for guidance with the TAIL–PCR; Nicole King (University of California at Berkeley, Berkeley, CA) for providing prepublication access to the Monosiga genome sequences; Dr. Kate Luby-Phelps (Director, University of Texas Southwestern Live Cell Imaging Core) and Laurie Mueller for enlightened guidance with microscopy; and Mike Misamore (Texas Christian University, Ft. Worth, TX), Pete Lefebvre (University of Minnesota, St. Paul, MN), John Abrams (University of Texas Southwestern), and Fred Grinnell (University of Texas Southwestern) for helpful discussions. This work was supported by a National Institutes of Health grant (GM056778 to W.J.S.) and by grants from the UK Medical Research Council (to O.B.), the Wellcome Trust (to R.E.S., R.T., and O.B.), the Lister Institute of Preventive Medicine (to O.B.), the Swedish Research Council (to S.G.), and the BBSRC (to R.E.S.), and also received support from the European Union through the BioMalPar Network of Excellence (to R.E.S. and O.B.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1656508.
References
- Adachi J., Hasegawa M. MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 1996;28:72–76. [Google Scholar]
- Aguilar P.S., Engel A., Walter P. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol. Biol. Cell. 2007;18:547–556. [Europe PMC free article] [Abstract] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. [Europe PMC free article] [Abstract] [Google Scholar]
- Begg D.A., Rodewald R., Rebhun L.I. The visualization of actin filament polarity in thin sections. Evidence for the uniform polarity of membrane-associated filaments. J. Cell Biol. 1978;79:846–852. [Europe PMC free article] [Abstract] [Google Scholar]
- Billker O., Lindo V., Panico M., Etienne A.E., Paxton T., Dell A., Rogers M., Sinden R.E., Morris H.R. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392:289–292. [Abstract] [Google Scholar]
- Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004;117:503–514. [Abstract] [Google Scholar]
- Buchanan M.J., Imam S.H., Eskue W.A., Snell W.J. Activation of the cell wall degrading protease, lysin, during sexual signalling in Chlamydomonas: The enzyme is stored as an inactive, higher relative molecular mass precursor in the periplasm. J. Cell Biol. 1989;108:199–207. [Europe PMC free article] [Abstract] [Google Scholar]
- Carter R., Chen D.H. Malaria transmission blocked by immunisation with gametes of the malaria parasite. Nature. 1976;263:57–60. [Abstract] [Google Scholar]
- Chatterjee I., Richmond A., Putiri E., Shakes D.C., Singson A. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132:2795–2808. [Abstract] [Google Scholar]
- Chen E.H., Olson E.N. Unveiling the mechanisms of cell–cell fusion. Science. 2005;308:369–373. [Abstract] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Membrane hemifusion: Crossing a chasm in two leaps. Cell. 2005;123:375–382. [Abstract] [Google Scholar]
- del Campo J.J., Opoku-Serebuoh E., Isaacson A.B., Scranton V.L., Tucker M., Han M., Mohler W.A. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr. Biol. 2005;15:413–423. [Abstract] [Google Scholar]
- Detmers P.A., Goodenough U.W., Condeelis J. Elongation of the fertilization tubule in Chlamydomonas: New observations on the core microfilaments and the effect of transient intracellular signals on their structural integrity. J. Cell Biol. 1983;97:522–532. [Europe PMC free article] [Abstract] [Google Scholar]
- Doberstein S.K., Fetter R.D., Mehta A.Y., Goodman C.S. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 1997;136:1249–1261. [Europe PMC free article] [Abstract] [Google Scholar]
- Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. [Europe PMC free article] [Abstract] [Google Scholar]
- Eksi S., Czesny B., van Gemert G.J., Sauerwein R.W., Eling W., Williamson K.C. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol. Microbiol. 2006;61:991–998. [Abstract] [Google Scholar]
- Ellerman D.A., Cohen D.J., Da Ros V.G., Morgenfeld M.M., Busso D., Cuasnicu P.S. Sperm protein ‘DE’ mediates gamete fusion through an evolutionarily conserved site of the CRISP family. Dev. Biol. 2006;297:228–237. [Abstract] [Google Scholar]
- Estrada B., Maeland A.D., Gisselbrecht S.S., Bloor J.W., Brown N.H., Michelson A.M. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev. Biol. 2007;307:328–339. [Europe PMC free article] [Abstract] [Google Scholar]
- Fang S.C., de los Reyes C., Umen J.G. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2006;2:e167. 10.1371/journal.pgen.0020167. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Faure J.E., Digonnet C., Dumas C. An in vitro system for adhesion and fusion of maize gametes. Science. 1994;263:1598–1600. [Abstract] [Google Scholar]
- Ferris P.J., Woessner J.P., Goodenough U.W. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell. 1996;7:1235–1248. [Europe PMC free article] [Abstract] [Google Scholar]
- Ferris P.J., Pavlovic C., Fabry S., Goodenough U.W. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. 1997;94:8634–8639. [Europe PMC free article] [Abstract] [Google Scholar]
- Ferris P.J., Waffenschmidt S., Umen J.G., Lin H., Lee J.H., Ishida K., Kubo T., Lau J., Goodenough U.W. Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell. 2005;17:597–615. [Abstract] [Google Scholar]
- Goodenough U.W., Detmers P.A., Hwang C. Activation for cell fusion in Chlamydomonas: Analysis of wild-type gametes and nonfusing mutants. J. Cell Biol. 1982;92:378–386. [Europe PMC free article] [Abstract] [Google Scholar]
- Goodenough U., Lin H., Lee J.H. Sex determination in Chlamydomonas. Semin. Cell Dev. Biol. 2007;18:350–361. [Abstract] [Google Scholar]
- Grossman A.R., Harris E.E., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., et al. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell. 2003;2:1137–1150. [Europe PMC free article] [Abstract] [Google Scholar]
- Han X., Wang C.T., Bai J., Chapman E.R., Jackson M.B. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. [Abstract] [Google Scholar]
- Harris E.H. The Chlamydomonas sourcebook. Academic Press; San Diego: 1988. [Google Scholar]
- Heiman M.G., Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. [Europe PMC free article] [Abstract] [Google Scholar]
- Heiman M.G., Engel A., Walter P. The Golgi-resident protease Kex2 acts in conjunction with Prm1 to facilitate cell fusion during yeast mating. J. Cell Biol. 2007;176:209–222. [Europe PMC free article] [Abstract] [Google Scholar]
- Hunnicutt G.R., Kosfiszer M.G., Snell W.J. Cell body and flagellar agglutinins in Chlamydomonas reinhardtii: The cell body plasma membrane is a reservoir for agglutinins whose migration to the flagella is regulated by a functional barrier. J. Cell Biol. 1990;111:1605–1616. [Europe PMC free article] [Abstract] [Google Scholar]
- Inoue N., Ikawa M., Isotani A., Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. [Abstract] [Google Scholar]
- Inoue N., Yamaguchi R., Ikawa M., Okabe M. Sperm– egg interaction and gene manipulated animals. Soc. Reprod. Fertil. Suppl. 2007;65:363–371. [Abstract] [Google Scholar]
- Jahn R., Lang T., Sudhof T.C. Membrane fusion. Cell. 2003;112:519–533. [Abstract] [Google Scholar]
- Janse C.J., Ramesar J., Waters A.P. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 2006;1:346–356. [Abstract] [Google Scholar]
- Jeffares D.C., Pain A., Berry A., Cox A.V., Stalker J., Ingle C.E., Thomas A., Quail M.A., Siebenthall K., Uhlemann A.C., et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat. Genet. 2007;39:120–125. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. Arabidopsis hapless mutations define essential gametophytic functions. Genetics. 2004;168:971–982. [Europe PMC free article] [Abstract] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. [Abstract] [Google Scholar]
- Kadandale P., Stewart-Michaelis A., Gordon S., Rubin J., Klancer R., Schweinsberg P., Grant B.D., Singson A. The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr. Biol. 2005;15:2222–2229. [Abstract] [Google Scholar]
- Kamei N., Glabe C.G. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes & Dev. 2003;17:2502–2507. [Europe PMC free article] [Abstract] [Google Scholar]
- Khalequzzaman M., Haq N. Isolation and in vitro fusion of egg and sperm cells in Oryza sativa. Plant Physiol. Biochem. 2005;43:69–75. [Abstract] [Google Scholar]
- Kindle K.L., Schnell R.A., Fernandez E., Lefebvre P.A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 1989;109:2589–2601. [Europe PMC free article] [Abstract] [Google Scholar]
- Kranz E., Lorz H. In vitro fertilisation of maize by single egg and sperm cell protoplast fusion mediated by high calcium and high pH. Zygote. 1994;2:125–128. [Abstract] [Google Scholar]
- Kroft T.L., Gleason E.J., L’Hernault S.W. The spe-42 gene is required for sperm–egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev. Biol. 2005;286:169–181. [Abstract] [Google Scholar]
- Kumar N. Target antigens of malaria transmission blocking immunity exist as a stable membrane bound complex. Parasite Immunol. 1987;9:321–335. [Abstract] [Google Scholar]
- Liu Y.G., Chen Y., Zhang Q. Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. Methods Mol. Biol. 2005;286:341–348. [Abstract] [Google Scholar]
- Lu X., Zhang F., McNew J.A., Shin Y.K. Membrane fusion induced by neuronal SNAREs transits through hemifusion. J. Biol. Chem. 2005;280:30538–30541. [Abstract] [Google Scholar]
- Mahjoub M.R., Qasim Rasi M., Quarmby L.M. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol. Biol. Cell. 2004;15:5172–5186. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuda Y., Saito T., Yamaguchi T., Kawase H. Cell wall lytic enzyme released by mating gametes of Chlamydomonas reinhardtii is a metalloprotease and digests the sodium perchlorate-insoluble component of cell wall. J. Biol. Chem. 1985;260:6303–6377. [Abstract] [Google Scholar]
- Misamore M.J., Gupta S., Snell W.J. The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol. Biol. Cell. 2003;14:2530–2542. [Europe PMC free article] [Abstract] [Google Scholar]
- Mohler W.A., Shemer G., del Campo J.J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J.G., Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2002;2:355–362. [Abstract] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. Generative Cell Specific 1 is essential for angiosperm fertilization. Nat. Cell Biol. 2006;8:64–71. [Abstract] [Google Scholar]
- Mu J., Awadalla P., Duan J., McGee K.M., Keebler J., Seydel K., McVean G.A., Su X.Z. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat. Genet. 2007;39:126–130. [Abstract] [Google Scholar]
- Pan J., Snell W.J. Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 2000;3:596–602. [Abstract] [Google Scholar]
- Pasquale S.M., Goodenough U.W. Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 1987;105:2279–2292. [Europe PMC free article] [Abstract] [Google Scholar]
- Pei J., Grishin N.V. PROMALS: Towards accurate multiple sequence alignments of distantly related sequences. Bioinformatics. 2007;23:802–808. [Abstract] [Google Scholar]
- Peng X.B., Sun M.X., Yang H.Y. A novel in vitro system for gamete fusion in maize. Cell Res. 2005;15:734–738. [Abstract] [Google Scholar]
- Podbilewicz B., Leikina E., Sapir A., Valansi C., Suissa M., Shemer G., Chernomordik L.V. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev. Cell. 2006;11:471–481. [Abstract] [Google Scholar]
- Primakoff P., Myles D.G. Penetration, adhesion, and fusion in mammalian sperm–egg interaction. Science. 2002;296:2183–2185. [Abstract] [Google Scholar]
- Primakoff P., Myles D.G. Cell–cell membrane fusion during mammalian fertilization. FEBS Lett. 2007;581:2174–2180. [Abstract] [Google Scholar]
- Putiri E., Zannoni S., Kadandale P., Singson A. Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev. Biol. 2004;272:448–459. [Abstract] [Google Scholar]
- Reese C., Mayer A. Transition from hemifusion to pore opening is rate limiting for vacuole membrane fusion. J. Cell Biol. 2005;171:981–990. [Europe PMC free article] [Abstract] [Google Scholar]
- Reininger L., Billker O., Tewari R., Mukhopadhyay A., Fennell C., Dorin-Semblat D., Doerig C., Goldring D., Harmse L., Ranford-Cartwright L., et al. A NIMA-related protein kinase is essential for completion of the sexual cycle of malaria parasites. J. Biol. Chem. 2005;280:31957–31964. [Abstract] [Google Scholar]
- Rubinstein E., Ziyyat A., Wolf J.P., Le Naour F., Boucheix C. The molecular players of sperm–egg fusion in mammals. Semin. Cell Dev. Biol. 2006;17:254–263. [Abstract] [Google Scholar]
- Saito T., Small L., Goodenough U.W. Activation of adenylyl cyclase in Chlamydomonas reinhardtii by adhesion and by heat. J. Cell Biol. 1993;122:137–147. [Europe PMC free article] [Abstract] [Google Scholar]
- Sapir A., Choi J., Leikina E., Avinoam O., Valansi C., Chernomordik L.V., Newman A.P., Podbilewicz B. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev. Cell. 2007;12:683–698. [Europe PMC free article] [Abstract] [Google Scholar]
- Saul A. Mosquito stage, transmission blocking vaccines for malaria. Curr. Opin. Infect. Dis. 2007;20:476–481. [Abstract] [Google Scholar]
- Shemer G., Suissa M., Kolotuev I., Nguyen K.C., Hall D.H., Podbilewicz B. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr. Biol. 2004;14:1587–1591. [Abstract] [Google Scholar]
- Silflow C.D., LaVoie M., Tam L.W., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P.A. The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 2001;153:63–74. [Europe PMC free article] [Abstract] [Google Scholar]
- Sinden R.E. Sexual development of malarial parasites. Adv. Parasitol. 1983;22:153–216. [Abstract] [Google Scholar]
- Sinden R.E., Croll N.A. Cytology and kinetics of microgametogenesis and fertilization in Plasmodium yoelii nigeriensis. Parasitology. 1975;70:53–65. [Abstract] [Google Scholar]
- Sinden R.E., Canning E.U., Spain B. Gametogenesis and fertilization in Plasmodium yoelii nigeriensis: A transmission electron microscope study. Proc. R. Soc. Lond. B. Biol. Sci. 1976;193:55–76. [Abstract] [Google Scholar]
- Sinden R.E., Butcher G., Beetsma A. Maintenance of the Plasmodium berghei life cycle. In: Doolan D.L., editor. Malaria methods and protocols. Methods in molecular medicine. Vol. 72. Humana; Totowa, NJ: 2002. pp. 25–40. [Abstract] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. [Abstract] [Google Scholar]
- Snell W.J. Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp. Cell Res. 1982;138:109–119. [Abstract] [Google Scholar]
- Sollner T.H. Intracellular and viral membrane fusion: A uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. [Abstract] [Google Scholar]
- Swanson W.J., Vacquier V.D. Liposome fusion induced by a M(r) 18,000 protein localized to the acrosomal region of acrosome-reacted abalone spermatozoa. Biochemistry (Mosc.) 1995;34:14202–14208. [Abstract] [Google Scholar]
- Swanson W.J., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. [Abstract] [Google Scholar]
- Tam L.W., Lefebvre P.A. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. [Europe PMC free article] [Abstract] [Google Scholar]
- van Dijk M.R., Janse C.J., Thompson J., Waters A.P., Braks J.A., Dodemont H.J., Stunnenberg H.G., van Gemert G.J., Sauerwein R.W., Eling W. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. [Abstract] [Google Scholar]
- Vieira A., Miller D.J. Gamete interaction: Is it species-specific? Mol. Reprod. Dev. 2006;73:1422–1429. [Abstract] [Google Scholar]
- Volkman S.K., Sabeti P.C., DeCaprio D., Neafsey D.E., Schaffner S.F., Milner D.A., Daily J.P., Sarr O., Ndiaye D., Ndir O., et al. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 2007;39:113–119. [Abstract] [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. [Abstract] [Google Scholar]
- Wang Q., Snell W.J. Flagellar adhesion between mating type plus and mating type minus gametes activates a flagellar protein–tyrosine kinase during fertilization in Chlamydomonas. J. Biol. Chem. 2003;278:32936–32942. [Abstract] [Google Scholar]
- Wang Q., Pan J., Snell W.J. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 2006;125:549–562. [Abstract] [Google Scholar]
- Wilson N.F., Foglesong M.J., Snell W.J. The Chlamydomonas mating type plus fertilization tubule, a prototypic cell fusion organelle: Isolation, characterization, and in vitro adhesion to mating type minus gametes. J. Cell Biol. 1997;137:1537–1553. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu Y., Zhang F., Su Z., McNew J.A., Shin Y.K. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2005;12:417–422. [Abstract] [Google Scholar]
- Zhang Y., Snell W.J. Flagellar adhesion-dependent regulation of Chlamydomonas adenylyl cyclase in vitro: A possible role for protein kinases in sexual signaling. J. Cell Biol. 1994;125:617–624. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.1656508
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/22/8/1051.full.pdf
Free to read at www.genesdev.org
http://www.genesdev.org/cgi/content/abstract/22/8/1051
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/content/full/22/8/1051
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/reprint/22/8/1051
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gad.1656508
Article citations
Plasmodium NEK1 coordinates MTOC organisation and kinetochore attachment during rapid mitosis in male gamete formation.
PLoS Biol, 22(9):e3002802, 10 Sep 2024
Cited by: 1 article | PMID: 39255311 | PMCID: PMC11414920
The C-terminal region of the Plasmodium berghei gamete surface 184-kDa protein Pb184 contributes to fertilization and male gamete binding to the residual body.
Parasit Vectors, 17(1):304, 13 Jul 2024
Cited by: 0 articles | PMID: 39003498 | PMCID: PMC11246575
Novel requirements for HAP2/GCS1-mediated gamete fusion in Tetrahymena.
iScience, 27(6):110146, 28 May 2024
Cited by: 0 articles | PMID: 38904066 | PMCID: PMC11187246
Transcriptional control of the Cryptosporidium life cycle.
Nature, 630(8015):174-180, 29 May 2024
Cited by: 5 articles | PMID: 38811723
From gametes to zygote: Mechanistic advances and emerging possibilities in plant reproduction.
Plant Physiol, 195(1):4-35, 01 Apr 2024
Cited by: 2 articles | PMID: 38431529
Review
Go to all (197) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas.
Development, 137(9):1473-1481, 24 Mar 2010
Cited by: 34 articles | PMID: 20335357 | PMCID: PMC2853849
Plasmodium berghei HAP2 induces strong malaria transmission-blocking immunity in vivo and in vitro.
Vaccine, 27(38):5187-5194, 09 Jul 2009
Cited by: 71 articles | PMID: 19596419
Review
Species-specific gamete recognition initiates fusion-driving trimer formation by conserved fusogen HAP2.
Nat Commun, 12(1):4380, 19 Jul 2021
Cited by: 15 articles | PMID: 34282138 | PMCID: PMC8289870
Is HAP2-GCS1 an ancestral gamete fusogen?
Trends Cell Biol, 20(3):134-141, 18 Jan 2010
Cited by: 67 articles | PMID: 20080406
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council
Medical Research Council (1)
A systematic analysis of protein kinases in malaria parasites to identify, validate and prioritise novel drug targets
Professor Oliver Billker, Wellcome Trust Sanger Institute
Grant ID: G0501670
NIGMS NIH HHS (2)
Grant ID: GM056778
Grant ID: R01 GM056778





