Abstract
Free full text

The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2
Abstract
Cancer progression has similarities with the process of epithelial-to-mesenchymal transition (EMT) found during embryonic development, during which cells down-regulate E-cadherin and up-regulate Vimentin expression. By evaluating the expression of 207 microRNAs (miRNAs) in the 60 cell lines of the drug screening panel maintained by the Nation Cancer Institute, we identified the miR-200 miRNA family as an extraordinary marker for cells that express E-cadherin but lack expression of Vimentin. These findings were extended to primary ovarian cancer specimens. miR-200 was found to directly target the mRNA of the E-cadherin transcriptional repressors ZEB1 (TCF8/δEF1) and ZEB2 (SMAD-interacting protein 1 [SIP1]/ZFXH1B). Ectopic expression of miR-200 caused up-regulation of E-cadherin in cancer cell lines and reduced their motility. Conversely, inhibition of miR-200 reduced E-cadherin expression, increased expression of Vimentin, and induced EMT. Our data identify miR-200 as a powerful marker and determining factor of the epithelial phenotype of cancer cells.
MicroRNAs (miRNAs) are small 20- to 22-nucleotide (nt)-long noncoding RNAs that inhibit gene expression at the post-transcriptional level. Each of the ~400 miRNAs known to exist in mammalian cells has multiple targets, making them powerful regulators of complex processes such as differentiation and cancer progression. Indeed, a strong link between miRNA and human cancers has been established, as miRNAs have been demonstrated to act as either oncogenes (e.g., miR-155, miR-17-5p, and miR-21) (He et al. 2005; Voorhoeve et al. 2006) or tumor suppressors (e.g., miR-15a, miR-16-1, and let-7) (Calin et al. 2002; Takamizawa et al. 2004; Johnson et al. 2005; Akao et al. 2006; Yanaihara et al. 2006).
Epithelial-to-mesenchymal transition (EMT)-like processes occur as part of embryonic development and wound healing (Savagner 2001), and during carcinogenesis (Dvorak 1986) when cancer cells undergo a change from a differentiated to a more invasive dedifferentiated tumor (Savagner 2001; Fuchs et al. 2002). After EMT induction, cells lose epithelial features and acquire mesenchymal characteristics, including Vimentin filaments and a flattened phenotype. They become more invasive by expressing proteases that allow them to pass through the underlying basement membrane and migrate, both being crucial steps in the multistep process of metastasis.
As part of EMT, cancer cells express various transcription factors—such as the E-box-binding factors SNAI1 (Snail), SNAI2 (Slug), and the basic helix–loop–helix (bHLH) factor Twist (Peinado et al. 2007)—in response to stimulation with a number of soluble factors that are present in the tumor environment, such as TGFβ or TNFα. In addition, two other E-box-binding transcription factors—ZEB1 (also known as TCF8 and δEF1) and ZEB2 (also known as ZFXH1B and SMAD-interacting protein 1 [SIP1])—are important regulators in the complex network of transcriptional repressors that regulate the expression of E-cadherin and EMT through repression of a number of master regulators of epithelial polarity (Comijn et al. 2001; Eger et al. 2005; Aigner et al. 2007; Shirakihara et al. 2007).
The 60 cell lines of the drug screening panel of human cancer cell lines at the National Cancer Institute (NCI60) represent nine different cancers (http://dtp.nci.nih.gov). However, they can be genetically divided into two major branches: Supercluster 1 (SC1) cells have a mesenchymal gene signature, and SC2 cells were described to express an epithelial gene profile (Ross et al. 2000), suggesting that SC1 and SC2 cells might represent two stages of tumor progression often equated with EMT. We previously identified let-7 as a marker for SC2 cells and loss of let-7 as a marker for more advanced cancer (Shell et al. 2007). The expression of let-7 and concomitant up-regulation of its main target, HMGA2, was found to be a characteristic of early neoplastic transformation in ovarian cancer patients. However, neither in the NCI60 cell lines nor in the primary patient tumors did let-7 or HMGA2 correlate well with expression of E-cadherin or Vimentin. While E-cadherin expression is known to be lost during cancer progression, let-7 was unlikely to have a role in preventing loss of E-cadherin expression and EMT, leaving open the question of whether miRNAs are involved in the regulation of EMT. We have now identified the miR-200 family of miRNAs as a general marker for E-cadherin-positive and Vimentin-negative cancer cells in both the NCI60 cells as well as ovarian cancer patients. We report an extraordinarily tight control of E-cadherin/Vimentin expression by miR-200 miRNAs through targeting the mRNAs of ZEB1 and ZEB2. Taken together, our data identify miR-200 as a marker for E-cadherin-positive cancer cells and as a powerful regulator of EMT.
Results
The NCI60 cells can be divided into three groups according to the ratio of E-cadherin to Vimentin expression
To identify miRNAs that might be important for regulating E-cadherin expression, we set out to identify miRNAs whose expression correlated with the expression of E-cadherin and Vimentin in the NCI60 cells. We first determined the expression levels of E-cadherin and Vimentin in 59 of the NCI60 cells using Western blot analysis followed by densitometry (Fig. 1A). We then sorted all 59 NCI60 cell lines from highest to lowest E-cadherin/Vimentin ratio (Fig. 1B). Cells fell into three groups: an epithelial group with very high E-cadherin expression and no detectable Vimentin expression, a group that had either both markers expressed at the same level or no expression of either of these two markers, and finally, a mesenchymal group with high Vimentin and no detectable E-cadherin expression. We reasoned that the comparison of the cells with an epithelial phenotype with cells of the mesenchymal group would allow us to identify miRNAs whose expression correlates with either of the two stages and that potentially could be regulators of EMT.
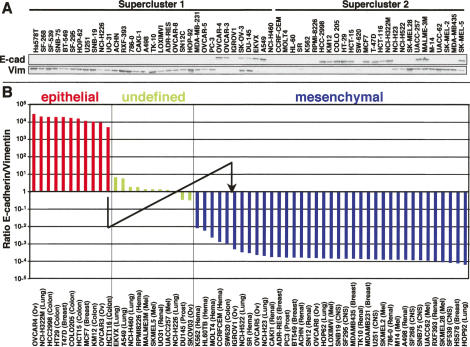
Identification of E-cadherin-positive and -negative cells among the NCI60 cell lines. (A) Western blot analysis of 59 of the NCI60 cell lines for E-cadherin and Vimentin expression grouped into superclusters. (B) NCI60 cells ranked according to highest to lowest ratio of E-cadherin/Vimentin expression. Epithelial cells express only E-cadherin and mesenchymal cells express only Vimentin. Undefined cell lines express either both markers or neither of them. The E-cadherin/Vimentin ratio was determined using densitometry of the data shown in A. Expression was normalized to actin bands run twice on different gels (not shown). The arrow indicates the change in E-cadherin/Vimentin ratio in HCT116 cells after inhibition of miR-200 as shown in Figure 5A.
Identification of the miR-200 family as a general marker for E-cadherin-positive and Vimentin-negative cancer cell lines
To identify miRNAs expression that could separate epithelial from mesenchymal cells, we interrogated a data set containing the expression levels of 207 miRNAs determined by real-time PCR in all NCI60 cells (Gaur et al. 2007). All miRNAs were ranked according to the difference in expression between the epithelial and mesenchymal cells (Figure 2A; Supplemental Fig. S1). Five miRNAs were selectively expressed in the epithelial cells, four of them with extraordinarily low P-values between 10−12 and 10−20, suggesting a very strong association with the epithelial and mesenchymal phenotype, respectively. These four miRNAs were miR-200a, miR-200b, miR-200c, and miR-141, all of which are part of the family of miR-200 miRNAs that also includes miR-429 (Fig. 2B). miR-429 could not be detected because it was not part of the data set available to us. The miR-200 family can be grouped into two subfamilies in two ways. The first way is functionally, according to the presence of two types of seed sequences (Fig. 2B): miR-200a/miR-141 and miR-200b/miR-200c/miR-429. The second way is genetically, according to the location in two gene clusters on two different chromosomes: miR-200b/miR-200a/miR-429 on chr. 1, and miR-200c/miR-141 on chr. 12 (Fig. 2C). In our study, we performed most experiments with two miR-200 family members—miR-200a and miR-200c—because they represent members of both subfamilies. For simplicity, we will refer to the members of the miR-200 family collectively as miR-200. Expression of all tested miR-200 family members showed an inverse correlation with the Vimentin protein expression in almost all 59 cell lines (Fig. 2D; data not shown). We also plotted the expression of miR-200c versus the expression of Vimentin mRNA (Fig. 2E). Consistent with transcriptional regulation, we found a similar inverse correlation between all four miR-200 miRNAs and Vimentin mRNA expression (Fig. 2E; data not shown). Interestingly, the same outlier, DU145, was found with moderate expression of both miR-200c and Vimentin on both the protein and the mRNA level. miR-200c was positively correlated with the expression of E-cadherin, as there was not a single cell line with high or intermediate miR-200 expression that lacked E-cadherin expression on both the protein and the mRNA level (Fig. 2F,G; data not shown). The best correlation was found between the expression of miR-200 and the ratio of E-cadherin and Vimentin mRNA (Supplemental Fig. S2), suggesting that miR-200 was a powerful marker for cell lines with an epithelial nature. Our data suggested a strong association between miR-200 expression and the epithelial nature of cancer cell lines.
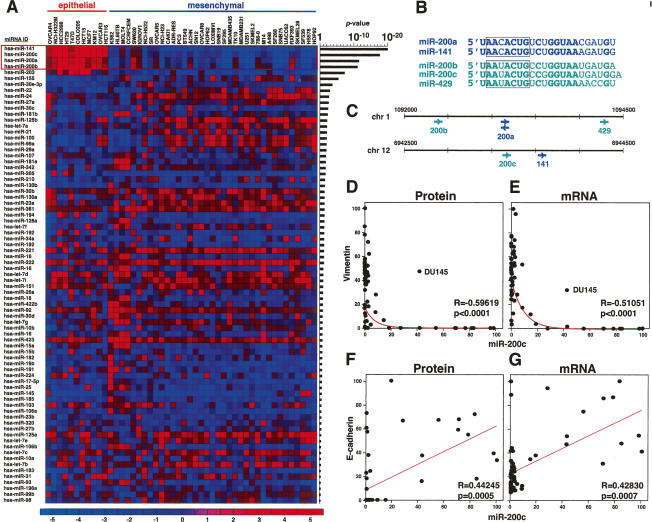
The miR-200 family of miRNAs is selectively expressed in only E-cadherin-positive and Vimentin-negative cells. (A) Expression of the 77 miRNAs with the highest expression in the NCI60 cells (Shell et al. 2007) ranked according to expression in either epithelial or mesenchymal cell lines. P-values are the result of a two-sample t-test analysis. High expression is indicated in red and low expression is in blue. Fold change (log2) is indicated. (B) The miR-200 family of miRNAs consists of two closely related subfamilies. Identical positions among all five family members are shown in bold. Seed sequences are boxed. (C) The five miR-200 family members are located on two different genetic loci. The correlation to the two functional subfamilies is indicated by using different colors. (D–G) Comparison of Vimentin protein expression (D), Vimentin mRNA expression (E), E-cadherin protein expression (F), and E-cadherin mRNA expression (G) with that of miR-200c in the NCI60 cells. Pearson correlation coefficients (R) and P-values (p) are reported.
The mRNAs of ZEB1 and ZEB2 are targeted by miR-200, and the two factors are inversely expressed with E-cadherin/Vimentin in the NCI60 cells
The E-box-binding transcription factors ZEB1 and ZEB2 have both been suggested to be targets of miR-200 (Hurteau et al. 2006; Christoffersen et al. 2007). According to TargetScan 4.1 (http://www.targetscan.org), ZEB2 has two predicted miR-200a/141 and five miR-200b/200c/429 seed matches in its 3′ untranslated region (UTR) (Fig. 3A). Remarkably, five of these miR-200 recognition sites are conserved 8mers. ZEB1 carries five putative miR-200b/200c/429 and three miR-200a/141 sites in its 3′UTR (Fig. 3A). To determine whether endogenous miR-200 in general could target the 3′UTR of ZEB2 in NCI60 cells, we transfected a ZEB2 reporter construct in which the murine ZEB2 3′UTR was fused to luciferase into the miR-200 low-expressing cell line IGROV-1 and the miR-200 high-expressing cell lines HCT116 and KM12 (Fig. 3B). Luciferase activity was strongly repressed in HCT116 and KM12 cells and was only slightly affected in the IGROV-1 cells, suggesting that endogenous levels of miR-200 may be sufficient to target the ZEB2 3′UTR. Using both wild-type and mutant constructs of human ZEB1 and ZEB2 3′UTRs carrying mutations in all miR-200b/c/429 targeting sites, we now demonstrate that ectopic miR-200c suppresses by directly affecting the predicted miR-200c targeting sites (Fig. 3C). Both mutant ZEB 3′UTRs were completely resistant to the suppressing activity of miR-200c. To determine whether endogenous miR-200 suppresses luciferase activity through silencing the 3′UTR of ZEB2 and ZEB1, we transfected HCT116 cells with either the murine ZEB2 3′UTR (used in Fig. 3B) or the human ZEB2 3′UTR (used in Fig. 3C) or human ZEB1 3′UTR together with a miR-200 inhibitor. To inhibit all members of the miR-200 family, we designed a locked nucleic acid (LNA)-based general anti-miR-200 inhibitor following a strategy described recently for mir-29 (Mott et al. 2007). The luciferase 3′UTR-expressing cells were either left untreated or cotransfected with either scrambled LNA oligo or LNA-200 (Fig. 3D). Luciferase activity of all constructs increased in LNA-200-treated cells. Consistent with the 3′UTRs of human and mouse ZEB2 being 94.2% identical in sequence, luciferase activity increased to about the same level with these two constructs (Fig. 3D). Our data confirm that endogenous miR-200 targets the ZEB1 and ZEB2 3′UTRs. Expression of most proteins regulated by miRNAs is regulated through a silencing mechanism that only affects expression of the proteins and not their mRNA levels. Interestingly, for both ZEB genes, mRNA degradation was suggested to be the mechanism of suppression rather than translational silencing (Hurteau et al. 2006; Christoffersen et al. 2007). However, this assumption was based on experiments involving overexpression of miR-200, which can force degradation of 3′UTRs. Using the same murine ZEB2 luciferase 3′UTR construct used by Christoffersen et al. (2007), we therefore determined whether the ZEB2 mRNA can be targeted by endogenous levels of miR-200 expressed in a cancer cell line. Using real-time PCR, we detected reduced expression of firefly luciferase mRNA in HCT116 cells cotransfected with the vector carrying the ZEB2 3′UTR and scrambled oligo when compared with just the empty luciferase vector (Fig. 3E). Inhibition of miR-200 using the LNA miR-200 eliminated the mRNA reduction, suggesting that endogenous miR-200—at least in these cells—was able to cause reduction in the level of mRNA that carries the ZEB2 3′UTR. To determine whether the actual ZEB1 and ZEB2 mRNAs are generally under the control of miR-200, we plotted the expression of ZEB1 and ZEB2 mRNAs versus the expression of miR-200a, miR-200b, miR-200c, or miR-141 in the NCI60 cell lines on a cell-by-cell basis (Fig. 3F,G; data not shown). A strong inverse correlation between the expression of miR-200 and ZEB1 and ZEB2 mRNA was detected. The correlation between miR-200 and ZEB2 mRNA was especially striking, with not a single cell line expressing both miR-200 and ZEB2 (Fig. 3G; data not shown). Taken together, these data strongly suggest that in the 59 cancer cell lines, both ZEB1 and ZEB2 mRNA are targeted by endogenous miR-200.
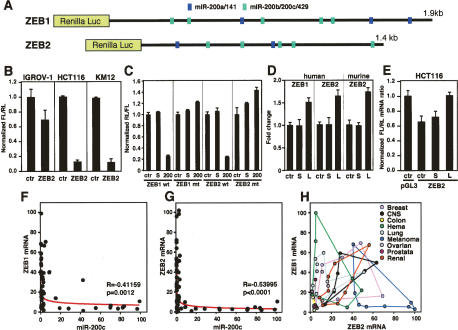
Endogenous miR-200 targets the mRNAs of ZEB1 and ZEB2. (A) Schematic of the ZEB1 and ZEB2 3′UTRs with the location of the predicted miR-200a/141 (blue) and miR-200b/c/429 (green) target sites. Shown are the luciferase constructs with the human ZEB 3′UTRs. (B) Activity of the firefly luciferase gene linked to the 3′UTR of murine ZEB2 72 h after transfection into cell lines with different endogenous miR-200 levels (IGROV-1, low; HCT116 and KM12, high). (C) 293T cells were transfected with the ZEB Renilla luciferase constructs shown in A or corresponding mutants in which all five miR-200b/c/429 sites were mutated and either left untreated (ctr) or cotransfected with scrambled pre-miR (S) or miR-200c (200). (D) HCT116 cells were transfected with either 10 nM scrambled LNA oligo (S) or LNA-200 (L) or left untreated and cotransfected with luciferase constructs containing either human ZEB1, human ZEB2, or murine ZEB2 3′UTR. (E) Real-time PCR to quantify luciferase mRNA in HCT116 cells transfected with either empty luciferase vector (pGL3) or the murine ZEB2 luciferase construct. Cells were either left untreated or cotransfected with either 50 nM scrambled LNA oligo (S) or LNA-miR-200 (L). (F) Comparison of the ZEB1 mRNA expression with that of miR-200c in the NCI60 cells. (G) Comparison of the ZEB2 mRNA expression with that of miR-200c in the NCI60 cells. Pearson correlation coefficients (R) and P-values (p) are reported. (H) Comparison of the ZEB1 mRNA expression with that of the mRNA of ZEB2 in the NCI60 cells. Cells from nine different cancer origins are labeled in different colors.
Our analysis suggests that both ZEB mRNAs are under the tight control of miR-200. It has been shown that ZEB proteins are expressed in a tissue-specific manner, making it difficult to assign a unique function to either protein (Postigo and Dean 2000). To determine whether ZEB proteins are expressed at different levels in different human cancer cells, we plotted the expression of ZEB1 mRNA against the expression of ZEB2 mRNA in all NCI60 cell lines. Interestingly, every cell line had a characteristic ratio of ZEB1/ZEB2 expression, with all cell lines expressing significant levels of miR-200 almost negative in ZEB expression (data not shown). To determine whether this ratio corresponds to different cancer origins, we labeled the nine cancer types represented in the NCI60 cells in different colors. Every cancer type occupied a different and characteristic area in the two-dimensional space of ZEB mRNA distribution (Fig. 3H). Colon-derived cancer cells lack ZEB expression, whereas CNS- and kidney-derived tumor cells express both factors. In contrast, most melanoma cells only express ZEB2. ZEB proteins are expressed during development in various organs. Our analysis of the NCI60 cells representing nine different human tissues indicates that cancer cells express various combinations of the two ZEB proteins. The fact that there was not a single NCI60 cell line that expresses high levels of both ZEB1 and ZEB2 suggests that while different cancers may use different ratios of ZEB proteins, the overall concentration of both factors is tightly regulated by miR-200, suggesting that ZEB1 and ZEB2 have similar functions, and differences in their activities are mainly due to tissue-specific expression.
We did not find a correlation between the ratio of expression between the miR-200a/miR141 and the miR-200b/miR-200c functional subfamilies and the cell type among the NCI60 cells (data not shown), suggesting that the ratio of ZEB1/ZEB2 expression in the different tissues is determined by other mechanisms than expression of different miR-200 genes. Interestingly, however, the ratio of combined expression of miR-200a and miR-141 to the combined expression of miR-200b and miR-200c (representing the two miR-200 families with slightly different targeting activities) was constant across all NCI60 cells (Supplemental Fig. S3A), suggesting that expression of miR-200 members from both gene loci is regulated in a way that the net outcome target coverage is very similar in all cells. In contrast, the contribution of the two gene clusters to the miR-200 content in the NCI60 cell lines varies significantly (Supplemental Fig. S3B). The R2 of the ratio of combined expression of miR-200a and miR-200b to the combined expression of miR-200c and miR-141 (representing the two different gene clusters) was much less significant than between the two functional families. This analysis suggests expression of miRNAs from the two miR-200 gene loci is regulated in a way that maintains equal expression of the two different functional miR-200 classes, most likely due to the fact that both gene loci contain members of each functional subclass.
Increasing miR-200 levels induces mesenchymal-to-epithelial transition (MET) in established human cancer cell lines, reducing their aggressiveness
Many established tumor cells, such as the NCI60 cell lines, are relatively resistant to many EMT-inducing reagents (C. Feig, S.-M. Park, and M.E. Peter, unpubl.). We therefore determined whether manipulation of miR-200 levels in a number of NCI60 cell lines could influence marker expression in a way consistent with EMT or MET induction. To determine whether miR-200 could cause induction of E-cadherin expression, we transfected a mixture of miR-200a and miR-200c into miR-200 low-expressing cell lines MDA-MB-231, A549, and ACHN. miR-200a and miR-200c were chosen because they represent members of the two functional subfamilies of miR-200 (Fig. 2B). Repeated transfection of 50 nM miR-200a/c caused up-regulation of E-cadherin in all three cell lines (Fig. 4A). Consistent with a regulation of ZEB1 and ZEB2 by miR-200, the expression of these two miR-200 targets was reduced. In the MDA-MB-231 cells, up-regulation of E-cadherin protein was evident already 3 d after transfection (Fig. 4B), and morphological changes consistent with the cells undergoing MET were seen after 6 d (Fig. 4C). miR-200 was highly potent in inducing these changes since they were already observed in cells 3 d after transfection with only 5 nM miR-200a/c (Supplemental Fig. S4). Reintroduction of ZEB2 into miR-200a/c-treated cells reversed the expression of E-cadherin, demonstrating that ZEB2 alone was able to suppress miR-200-induced E-cadherin (Fig. 4D). To test whether increasing the level of miR-200 in the highly metastatic MDA-MB-231 cells affected their motility, these cells were transfected with miR-200a/c and subjected to an in vitro motility assay (Fig. 4E). miR-200c-treated cells showed significantly reduced motility, demonstrating that miR-200 levels affect the aggressiveness of cancer cells. This was not due to reduced growth of the miR-200-transfected cells (data not shown).
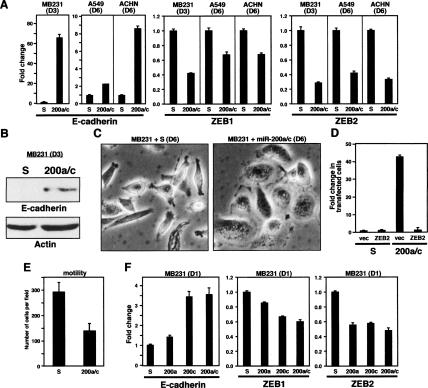
Induction of E-cadherin expression and MET in established cancer cell lines by introducing exogenous miR-200. (A) Cells were transfected with either scrambled pre-miRNA (S) or a mixture of pre-miR-200a and pre-miR-200c (200a/c). Expression of E-cadherin, ZEB1, and ZEB2 was determined by real-time PCR either 3 d (D3) or 6 d (D6) after the first transfection. (B) Detection of E-cadherin by Western blot analysis in MDA-MB-231 cells 3 d after transfecting scrambled oligo (S) or miR-200a/c. (C) Morphological changes of the cells after treatment as in B after 6 d. (D) MDA-MB-231 cells that had received six cycles of transfection were first transfected with either scrambled pre-miR oligo or miR-200a/c to further induce E-cadherin expression, and after 24 h were transfected with control vector or ZEB2 expression plasmid. Three days later, E-cadherin expression was quantified by real-time PCR. Numbers were normalized to a transfection efficiency of 23%. (E) Twenty-two-hour in vitro motility assay of MDA-MB-231 cells transfected three times (D9) with either 50 nM scrambled oligo (S) or miR-200a/c. (F) MDA-MB-231 cells were transfected with either scrambled oligo (S), miR-200a (200a), or miR-200c (200c) individually or in combination (200a/c), and after 24 h the expression of E-cadherin, ZEB1, and ZEB2 was quantified by real-time PCR.
miR-200 family members target a similar and highly overlapping set of genes
According to two miRNA target prediction algorithms—TargetScan and TargetRank (http://hollywood.mit.edu/targetrank)—miR-200a and miR-200c target a different but overlapping set of targets. TargetScan predicts 429 miR-200a and 602 miR-200c targets, with 75 shared targets (12.4% of the predicted targets for miR-200c). TargetRank predicts 1115 miR-200a and 1316 miR-200c targets, with an overlap of 314 targets (23.9% of the predicted miR-200c targets). To determine whether members of the two families have different potencies in suppressing expression of ZEB1 or ZEB2, and hence different activities in inducing MET, MDA-MB-231 cells were transfected with either miR-200a or miR-200c individually. To minimize the secondary effects, E-cadherin and ZEB1/2 mRNA levels were determined 1 d after transfection (Fig. 4F). Both miR-200 family members reduced expression of ZEB2, but miR-200c was more potent in reducing ZEB1 levels and in inducing E-cadherin expression. To determine the efficiency of miR-200a and miR-200c to target genes genome-wide, we performed a gene chip analysis of MDA-MB-231 cells transfected with either miR-200a or miR-200c. It has been shown that, 24 h after transfection of a miRNA, expression of genes that carry predicted seed matches of the transfected miRNA in their 3′UTR is preferentially reduced (Johnson et al. 2007). Of the ~45,000 probe sets represented on the U133 chip set, only 323 genes were decreased in their expression in cells transfected with miR-200c, and only 108 genes were decreased in their expression in cells transfected with miR-200a compared with the scrambled control (only >0.5-fold down-regulation was considered) (data not shown). Supplemental Table S1 lists all genes that were down-regulated by both miR-200a and miR-200c. In the top half of mRNAs detected with reduced expression, many were predicted miR-200 targets, whereas in the bottom half of the list of genes barely any predicted targets were found, consistent with many of the highly down-regulated genes being direct targets of either miR-200a or miR-200c. Of the top 20 probe sets with reduced expression in both the cells transfected with miR-200a or with miR-200c, nine were predicted by either or both target prediction algorithms (Table 1). In contrast, genes that were down-regulated by either miR-200a or miR-200c alone contained only a negligible number of predicted targets (Supplemental Tables S2, S3). Our data are consistent with miR-200a and miR-200c having very similar sets of targets and causing similar functional changes in cells.
Table 1.
The top 20 genes most down-regulated by both miR-200a and miR-200c
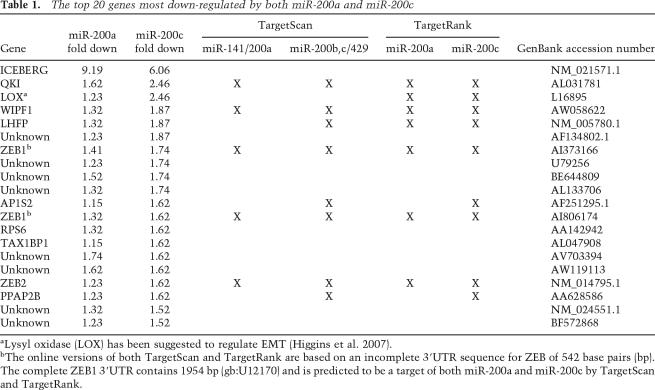
aLysyl oxidase (LOX) has been suggested to regulate EMT (Higgins et al. 2007).
bThe online versions of both TargetScan and TargetRank are based on an incomplete 3′UTR sequence for ZEB of 542 base pairs (bp). The complete ZEB1 3′UTR contains 1954 bp (gb:U12170) and is predicted to be a target of both miR-200a and miR-200c by TargetScan and TargetRank.
Reducing miR-200 levels induces EMT in an established human cancer cell line
We chose HCT116, among NCI60 cell lines with very high miR-200 expression, to test whether reduction of miR-200 levels could induce EMT. Transfection of HCT116 cells with the LNA-miR-200 inhibitor caused a strong reduction of E-cadherin mRNA and protein expression (Fig. 5A–C), and a concomitant induction of Vimentin (mRNA and protein) and ZEB1 and ZEB2 mRNAs (Fig. 5A,C). These changes were associated with a change from a cobblestone to a spindle-like morphology (Fig. 5D), a classical marker of EMT induction. Analysis of the E-cadherin and Vimentin expression before and after inhibition of miR-200 revealed that HCT116 cells had moved from the epithelial to the mesenchymal group (Fig. 1B, arrow). To determine whether the changes observed in HCT116 cells after inhibition of endogenous miR-200 were due to the up-regulation of ZEB proteins or other known inhibitors of E-cadherin expression such as Snail or Twist, we sought to selectively suppress the expression of ZEB1 and ZEB2 using two siRNA pools and tested how that affected expression of E-cadherin. Cells were first treated for six cycles with either LNA scrambled oligo or LNA-200 to induce expression of ZEB1 and ZEB2, and after 24 h the same cells were transfected with either the two siRNA pools individually or a combination of both pools. After 2 d, the expression levels of ZEB1, ZEB2, and E-cadherin were quantified by real-time PCR (Fig. 5E). The siRNA against ZEB2 reduced ZEB2 expression ~70% without changing expression of ZEB1 (Fig. 5E). In contrast, the siRNA against ZEB1 completely blocked the expression of ZEB1 to background levels and also affected the expression of ZEB2 ~50%, a phenomenon described previously in another report (Chua et al. 2007). The partial reduction of ZEB2 expression alone did not have an effect on the expression of E-cadherin. In contrast, the complete reduction of ZEB1 expression together with the substantial reduction in ZEB2 expression detected in the siZEB1-transfected cells caused a significant re-expression of E-cadherin, and so did the treatment with both siZEB1 and siZEB2 (Fig. 5E). These data suggest that only when both expressed ZEB proteins are suppressed can E-cadherin be expressed in cells with inhibited miR-200, demonstrating that ZEB proteins are indeed responsible for the suppression of E-cadherin in HCT116 cells with inhibited miR-200.
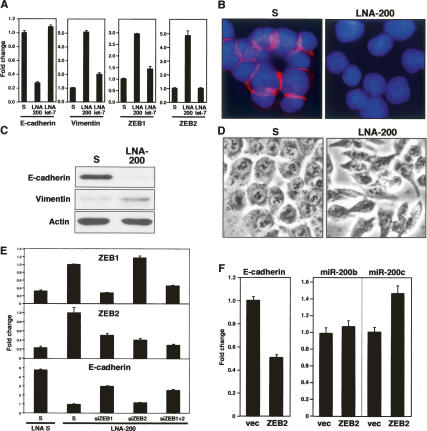
Reducing endogenous miR-200 expression induces EMT in HCT116 cells. (A) Cells were transfected repeatedly every 3 d with either scrambled LNA (S), LNA-200, or LNA-let-7 oligo. Expression of mRNAs was quantified by real-time PCR 18 d after the first transfection. (B) Immunofluorescence analysis of E-cadherin expression (pink) in cells repeatedly transfected for 22 d. Cells were counterstained with DAPI. (C) Western blot analysis of cells treated for 15 d. (D) Phase contrast picture showing cells treated for 15 d. (E) Cells were first transfected with either 50 nM LNA scrambled or LNA-200, and after 24 h, once E-cadherin was up-regulated, they were again transfected with either scrambled siRNA pool, a ZEB1-specific siRNA pool, a ZEB2-specific siRNA pool, or both. After 72 h, expression of ZEB1, ZEB2, and E-cadherin was quantified by real-time PCR. (F) Cells were transfected with empty vector (vec) or with HA-ZEB2, and after 3 d expression of E-cadherin, miR-200a, and miR-200c was quantified by real-time PCR. Numbers were normalized to a transfection efficiency of 48%. Expression of HA-ZEB2 was confirmed by Western blot analysis (not shown).
MiR-200 family members are expressed from two different loci that seem to be coregulated. To test whether ZEB proteins are responsible in their coregulation by repressing their activity in a positive feedback loop, we transfected HCT116 cells with human ZEB2, and 3 d after transfection determined the expression of miR-200b and miR-200c, which are expressed from the two different loci (Fig. 5F). Ectopically expressed ZEB2 caused down-regulation of E-cadherin, consistent with the established function of ZEB2. However, rather then causing a down-regulation of miR-200, miR-200b was not affected by ZEB2 and miR-200c was up-regulated in the transfected cells, suggesting that miR-200 family members on the different loci are not codown-regulated when ZEB2 levels drop. Collectively, the data demonstrate that the miR-200 family is a powerful regulator of EMT/MET by targeting ZEB1 and ZEB2, which control the expression of E-cadherin. Loss of these miRNAs may therefore have the potential to promote tumor metastasis by initiating EMT.
MiR-200 but not let-7 is a predictor of the epithelial nature of cancer cells
One of the main activities of let-7 may be to control expression of early embryonic genes that could be re-expressed during cancer progression (Park et al. 2007). We now provide evidence that the miR-200 family of miRNAs is selectively expressed in epithelial cells and is absent in all cells with a low E-cadherin to Vimentin ratio. A shift in this ratio is also often found during tumor progression. Therefore, we hypothesize that both miRNA families let-7 and miR-200 may be lost during cancer progression. Consistent with this assumption is the observation that while let-7 is a much better marker for Type I/SC1 and Type II/SC2 cells, miR-200 also shows a significantly higher expression in Type II/SC2 cells (Supplemental Fig. S5A). Interestingly, while let-7 shows a graded expression across all cell lines, the expression of miR-200 is switch-like, with many cell lines not expressing any detectable miR-200, a property consistent with the switch-like expression of epithelial markers. Supplemental Figure S5, B and C, directly compares the expression of let-7d and miR-200c in the different groups of NCI60 cells with different ratios in the expression of E-cadherin and Vimentin. It is obvious that let-7 expression does not vary among the different groups. In contrast, miR-200a, miR-200b, miR-200c, and miR-141 are expressed almost exclusively in the epithelial group I cells that do not express ZEB1 or ZEB2 (Supplemental Figs. S2, S5C). Interestingly, most of these cell lines are SC2 cells (Supplemental Fig. S5C, labeled in red), and the SC2 cells tend to express more miR-200 than the SC1 cells. These data suggest that in at least ~20% of the NCI60 cells the expression of let-7 and miR-200 might be linked.
MiR-200 expression correlates with E-cadherin expression in tumors from ovarian cancer patients
To determine whether the expression of E-cadherin is also under control of miR-200 in human tumor cells, we quantified miR-200c expression in a set of ovarian cancers. KM12, a cell line with high miR-200 expression, was used as positive control and IGROV-1 cells were used as negative controls for miR-200 expression (Fig. 6A). Consistent with the role of miR-200 in mesenchymal cells, RNA isolated from two patient tumor stroma samples did not express detectable miR-200c levels. We tested two sets of patient samples derived from primary serous papillary ovarian cancer: one isolated according to the highest expression of HMGA2 (characterizing the most aggressive tumors) (Fig. 6B), and one isolated according to highest tumor content as determined by H&E staining (Fig. 6C). We found a significant correlation (P = 0.013 and P = 0.046) between E-cadherin and miR-200c expression in both sample sets, confirming the association of miR-200 with the E-cadherin status in primary human cancer samples.
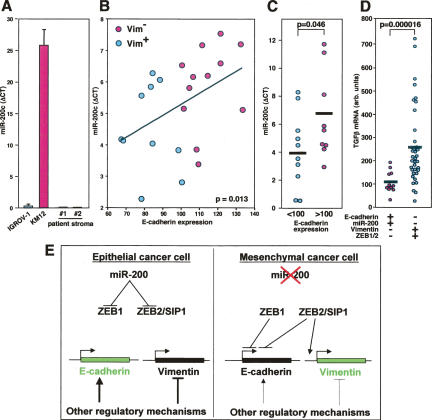
The expression of miR-200c and E-cadherin correlates in tumor tissue samples of ovarian cancer patients. (A) Real-time PCR analysis of miR-200c in IGROV-1 and KM12 cells and two patient stroma samples. (B,C) Correlation between the expression of miR-200c and E-cadherin mRNA in two sets of patient samples. Tumors were enriched using method 1 (B) or method 2 (C) as described in Materials and Methods. Linear regression is shown in B, as well as the P-value of the significance of difference in expression of miR-200c in E-cadherin high/Vimentin low (red symbols) and E-cadherin low/Vimentin high (blue symbols) specimens. (D) TGFβ1 mRNA expression in the two groups among the NCI60 cells with the indicated expression pattern. The P-value of the difference in expression between the two groups is shown. (E) Model to explain the function of miR-200 in maintaining an epithelial phenotype of cancer cells. See the text for details.
Our work complements a recent study that identified all members of the miR-200 family as regulators of TGFβ-induced EMT in MDCK cells (Gregory et al. 2008). Since human cancer cells often up-regulate TGFβ late during tumor progression (Jakowlew 2006), TGFβ produced by the cancer cells or the tumor microenvironment may induce reduction of miR-200 in the tumor, in part driving progression through inducing EMT. To test whether cancer cells could be driving miR-200 reduction through up-regulation of TGFβ, we compared the expression level of TGFβ1 mRNA between the two groups of NCI60 cells that either express miR-200 and E-cadherin or ZEB1/2 and Vimentin (Fig. 6D). Interestingly, TGFβ1 expression was significantly higher in the mesenchymal group of cancer cells, and could therefore be involved in regulating miR-200 expression during cancer progression.
Discussion
EMT is a complex differentiation process important in processes as different as embryonic development, wound healing, and tumor progression. It has been shown to involve changes in the expression of at least 4000 genes (Zavadil et al. 2001), suggesting that it is highly regulated at the transcriptional level. It can be induced by a bewildering number of intracellular and extracellular factors feeding into a large number of signaling pathways (for review, see Hay 1995). While the upstream signals and the types of tissues can be different, the final effectors controlling the cell phenotype—including the cytoskeleton and the cell–cell and the cell–substratum adhesion systems—appear to be remarkably similar in all forms of the EMT-related differentiation processes (Savagner 2001). This suggests the existence of a master regulator of EMT. However, to this date none of the described regulators of EMT have the same regulatory power in all cells. Finding a universal factor that would induce EMT or MET in established human cancer cells has been difficult (C. Feig, S.-M. Park, and M.E. Peter, unpubl.), presenting an obstacle in developing new treatment modalities that are aimed at reversing EMT in human cancers. We now report the identification of the miR-200 family of miRNAs as a powerful marker for epithelial cells and a regulator of EMT. By using the NCI60 cells, we could investigate the direct relation between endogenous miRNAs and EMT markers, such as E-cadherin and Vimentin, in a large number of cell lines derived from nine different human cancers. We did not find a single cell line among 59 NCI60 cell lines that had an E-cadherin high and Vimentin low phenotype and did not express miR-200. This remarkable level of correlation between the expression of miR-200 and the E-cadherin/Vimentin ratio across all NCI60 cells strongly suggests that miR-200 is a universal regulator of the epithelial nature of cancer cells. Multiple functional assays confirmed that miR-200 is actively involved in maintaining the epithelial nature of cells, and altering miR-200 levels in established cancer cell lines caused changes consistent with either EMT or MET induction.
We recently identified another family of miRNAs, let-7, as a specific marker for well-differentiated cancers. We found that SC2 cells expressed higher amounts of let-7 (in particular, let-7d and let-7g) than SC1 cells (Shell et al. 2007). It had been reported that the SCs found in the NCI60 cell lines are defined by the difference in expression of epithelial and stromal markers (Ross et al. 2000). However, we concluded that let-7 is not a regulator of EMT for the following reasons (Park et al. 2007): (1) In an hierarchical cluster analysis, many of the established “epithelial” cell lines among the NCI60 cells separated into the SC1 “stromal” group (Ross et al. 2000). (2) E-cadherin and Vimentin did not separate well into SC1 from SC2 cells (Shell et al. 2007). (3) Let-7 levels did not inversely correlate with levels of established EMT markers such as E-cadherin, Vimentin, or Snail (Park et al. 2007; Shell et al. 2007). (4) The expression pattern of let-7 during development is not consistent with its involvement in EMT, but rather with a role of promoting and/or arresting general differentiation pathways (Schulman et al. 2005). While let-7 is unlikely to regulate EMT, it may contribute to tumor progression by regulating a number of early embryonic genes that are re-expressed during the early stages of neoplastic transformation (Park et al. 2007).
In addition to being essential for embryonic development, EMT has been implicated in the process of metastasis formation through down-regulation of E-cadherin (Thiery 2002; Thiery and Sleeman 2006). ZEB1, ZEB2, and the transcription factors Snail, Slug, E47, and Twist are all able to initiate EMT through binding to two bipartite E-box motifs within the E-cadherin promoter, repressing its transcription (Peinado et al. 2004). Both ZEB proteins have been implicated in human malignancies. ZEB2 is up-regulated in advanced stages of ovarian, gastric, and pancreatic cancer (Rosivatz et al. 2002; Elloul et al. 2005, 2006; Imamichi et al. 2007), and serves as a prognostic marker for squamous cell carcinoma (Maeda et al. 2005). ZEB1 has been associated with aggressive behavior of colorectal tumors and uterine cancers (Pena et al. 2005, 2006; Spoelstra et al. 2006). Together, these findings point to ZEB1 and ZEB2 as critical promoters of cancer progression. Our findings suggest that ZEB1 and ZEB2 expression is controlled by the miR-200 family, and that down-regulation of these miRNAs contributes to the early stage in tumor metastasis.
ZEB1 and ZEB2 have been suggested recently to be targets of miR-200c and miR-200b, respectively (Christoffersen et al. 2007; Hurteau et al. 2007), and miR-200 was demonstrated to cause changes in E-cadherin expression (Christoffersen et al. 2007; Hurteau et al. 2007). However, these studies did not conclusively demonstrate that the 3′UTRs of the two ZEBs are directly targeted. Furthermore, neither of these studies demonstrated that inhibition of endogenous miR-200 can induce EMT. Our work now provides a detailed analysis of the network of the two ZEBs and their regulation by different miR-200 family members. The repression of the ZEB proteins by miR-200 represents an interesting model of regulation. Both ZEB proteins are expressed in human cancer cell lines, albeit with different ratios. Interestingly, the total amount of both ZEBs never exceeds the highest amount of either ZEB protein found in cells that only express one of the two factors, suggesting that the total amount of ZEBs must not exceed a certain threshold. This is accomplished by a network of miR-200 miRNAs that can be grouped into two families in two ways. The first way is according to their specificity, as the two groups differ in one nucleotide in their seed sequences. Target prediction algorithms predicted a significant difference in the spectrum of genes targeted by the two subfamilies. However, a gene chip analysis of cells transfected with individual members of these subfamilies indicated a much wider overlap in targets than expected from the results given by the prediction algorithms. Coregulation of ZEB1 and ZEB2 is also suggested by the fact that both have five miR-200b/c/429 and three and two miR-200a/141 sites in their 3′UTRs, respectively. The second way in which the five miR-200 miRNAs can be grouped is according to their chromosomal location. MiR-200a/b and miR-429 are located in one cluster on chr. 1, while miR-200c and miR-141 are part of another cluster on chr. 12. MiRNAs in each cluster are likely coregulated, and indeed we found variation of >100-fold in the expression of members of one cluster versus the other cluster among the NCI60 cell lines. Interestingly, that did not translate into a major difference in the ratio of expression of miR-200 family members of the two functional subclasses, presumably because each genetic locus expresses members of either functional class. Our data lend support to the view that the two ZEB proteins have similar functions (Postigo and Dean 2000), and our data suggest that both can be regulated by all members of the miR-200 family. This network of redundant activities likely contributes to the switch-like nature of the process of EMT, and could be one of the reasons for the extraordinarily tight connection between miR-200 and the E-cadherin/Vimentin ratio observed in the NCI60 cell lines.
Based on our data, we propose the following model for the function of the miR-200 family in EMT and cancer progression (Fig. 6E): In epithelial cancer cells and well-differentiated cancers, miR-200 is highly expressed, controlling the expression of ZEB1 and ZEB2. Other mechanisms cause induction of E-cadherin expression and suppression of Vimentin expression. In contrast, when miR-200 expression is down-regulated (either in mesenchymal cancer cells or in tissues undergoing EMT), ZEB1 and ZEB2 proteins are expressed. Both ZEB1 and ZEB2 are strong suppressors of E-cadherin expression. At the same time, ZEB2 has been shown to directly activate the Vimentin promoter by an unknown mechanism (Bindels et al. 2006), causing Vimentin to be expressed. Our data identified miR-200 as a powerful master regulator of EMT in cancer cells, and suggest that introducing miR-200 into cancer cells could be a novel way of reversing tumor progression.
Materials and methods
Western blot analysis and quantification of protein expression in the NCI60 cells
For the analysis of protein expression in the NCI60 cell lines, frozen cell pellets of 107 cells were obtained from Dr. Susan Holbeck (National Cancer Institute) and lysed directly in RIPA buffer. Twenty-five micrograms of cell lysates were analyzed on 12% SDS-PAGE as described recently (Shell et al. 2007). Quantification of protein bands on Western blots was performed using ScanAnalysis (Biosoft). The antibodies used were E-Cadherin (610182, Pharmingen), Vimentin (550513, Pharmigen), β-actin (A-5441, Sigma), anti-mouse HRP (sc-2005, Santa Cruz Biotechnology), and anti-rabbit HRP (4030-05, Southern Biotechnology). To determine the ratio of E-cadherin to Vimentin expression in the NCI60 cells, band intensities were normalized to that of the mean band intensity of actin (run twice on two separate gels). For both E-cadherin and Vimentin, the band with the highest expression was set to 100%, and for each cell line the ratio of E-cadherin to Vimentin band intensity was determined. To identify epithelial and mesenchymal cells among the NCI60 cell lines, cells were sorted according to their E-cadherin/Vimentin ratio.
Identification of miRNAs expressed in epithelial cells
The data set including expression levels of 207 individual miRNA determined by real-time PCR was described recently (Gaur et al. 2007). The expression of each miRNA was normalized to the mean of all other miRNAs in the data set. Fold change was calculated, and a two-tailed t-test with unequal variance was conducted to test for differences in the expression between the 11 epithelial and the 37 mesenchymal cell lines. MiRNAs were sorted in ascending order by P-value. A heat map was generated as described recently (Park et al. 2008).
Correlating protein and mRNA expression with miRNA expression in the NCI60 cell lines
The expression levels of miR-200a, miR-200b, miR-200c, and miR-141 were plotted against the expression levels of E-cadherin protein, Vimentin protein, the ratio of E-cadherin protein to Vimentin protein, E-cadherin mRNA (gene array analysis GC34162 at the developmental therapeutics program of the NCI; http://dtp.nci.nih.gov), Vimentin mRNA (GC37090), ZEB1 mRNA (GC156730), or ZEB2 mRNA (GC38696). All analyses were normalized by setting the value in the cell line with the highest expression as 100%. In addition, the expression of TGFβ1 mRNA (GC33890) was determined in the epithelial and mesenchymal NCI60 cells, and the significance of difference in expression was determined using the Student’ two-tailed t-test.
Cell culture and transfections
A549, ACHN, MDA-MB-231, IGROV-1, HCT116, KM12, and 293T cells were maintained in RPMI 1640 (Cellgro), supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. Cells were grown at a humidified atmosphere of 5% CO2 at 37°C. Transfections of miRNAs were carried out with siPORT NeoFX transfection reagent (Ambion) according to the manufacturer’s instructions. Cells with low miR-200 expression such as A549, ACHN, and MDA-MB-231 were transfected with either pre-miR-200a or pre-miR-200c (Ambion) or a mixture of both at a final concentration of 5 nM or 50 nM, or an equivalent amount of negative control #2 precursor miRNA (Ambion), whereas a cell line with high miR-200, HCT116, was transfected with either 50 nM LNA-let-7 (Boyerinas et al. 2008) or LNA-miR-200 or control mercury knockdown LNA (Exiqon). In all cases, cells were either transfected once and tested after 1 d (D1) or 3 d (D3) or transfected repeatedly every 3 d for the indicated times (D6, D15, D18, or D22). LNA-miR-200 is a chimeric LNA/DNA oligonucleotide designed to hybridize with all mir-200 family members. The sequence is 5′-CCATCATTAC CCGGCTGTATTA-3′ (Proligo), where the underlined nucleotides indicate LNA bases. The On-target siRNA SMART pools against ZEB1 and ZEB2 were purchased from Dharmacon RNA Technology. Cells were transfected with 25 nM siRNA using Lipofectamine 2000 (Invitrogen).
Luciferase assays
For luciferase assays, 0.75 × 105 cells were seeded in 12-well plates 1 d prior to transfection. IGROV-1, HCT116, or KM12 cells were transfected with 100 ng of pGL3 containing the 3′UTR of murine ZEB2 (pGL3 S1-5) (Christoffersen et al. 2007) (kindly provided by Dr. Anders Lund, Biotech Research and Innovation Center, Copenhagen, Denmark), together with 50 ng of pRL-SV40 (Promega) using Lipofectamine 2000 (Invitrogen). Seventy-two hours later, luciferase activity was measured according to the manufacturer’s instructions (Promega). All experiments were performed in triplicate and normalized to Renilla luciferase activity. For quantitative real-time PCR, HCT116 cells were seeded 105 per well of a six-well plate 1 d before transfection. Cells were transfected with 100 ng of pGL3 containing the 3′UTR of murine ZEB2, 50 ng of pRL-SV40, and either scrambled or LNA-miR-200. After 72 h, total RNA was extracted from cells using the Trizol reagent (Invitrogen Life Technologies). To test the effects of mutated miR-200b/c/429 targeting sites in the 3′UTRs of ZEB1 and ZEB2, Renilla luciferase fusion constructs carrying either the entire ZEB1 or ZEB2 3′UTR and corresponding mutants (1 ng each) with all miR-200b/c/429 sites mutated as described (Gregory et al. 2008) were transfected into 293T cells together with 200 ng of pGL3 firefly luciferase plasmid and 0.4 nM scrambled or miR-200c. Normalized luciferase activity was determined 48 h after transfection as described previously (Shell et al. 2007).
Immunohistochemistry
Cells grown in chambered slides (Lab-Tek, Nalgen Nunc International) were washed twice with cold PBS and fixed with ice-cold methanol/acetone (1:1) for 4 min at −20°C. Slides were air-dried and rehydrated with PBS for 5 min. After blocking with 2% BSA/PBS for 1 h at room temperature, cells were incubated with anti-E-Cadherin (Pharmingen) overnight at 4°C. Cells were washed with PBS three times and incubated with Alexa 594-conjugated goat anti-mouse antibody (1:200; Molecular Probes) for 1 h at room temperature. Cells were washed three times and mounted with Prolong Gold anti-fade reagent with DAPI (Invitrogen).
In vitro motility assay
In vitro motility assay was performed as described previously (Barnhart et al. 2004). In short, Transwell insert chambers with 8-μm porous membrane (Costar) were used for the assay. Cells were washed three times with PBS and 50,000 cells were added to the top chamber in serum-free media. The bottom chamber was filled with media containing 10% FBS. Cells were incubated for 22 h at 37°C in a 5% CO2 humidified incubator. To quantify migrating cells, cells on the top chamber were removed by using a cotton-tipped swab, and the migrated cells were fixed in methanol and stained with Giemsa stain. Five random fields were counted.
Quantitative real-time PCR
Total RNA was DNase I-digested. cDNA was generated from 2 μg of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). mRNA expression levels were measured by using the TaqMan Gene Expression Assay kit (Applied Biosystems) containing specific primers and TaqMan MGB probe for each gene (E-cadherin, Vimentin, ZEB1, ZEB2, and GAPDH) on a 7500 quantitative Real-time PCR Machine and SDS software (Applied Biosystems). The relative amount of gene transcripts was normalized to GAPDH. For Renilla and firefly luciferase genes, the primers were used as described elsewhere (Christoffersen et al. 2007); the TaqMan MGB probe was designed using the Firebuilder software (Applied Biosystems). For miRNA real-time PCR, miRNA and U6-specific cDNA was generated from 20 ng of RNA extracted from formalin-fixed paraffin-embedded tissue (FFPET) as described recently (Shell et al. 2007) or total RNA using the TaqMan MicroRNA Reverse Transcription Kit with RNA-specific RT primer from the TaqMan MicroRNA Assay (Applied Biosystems). miRNA levels were measured by using the miRNA-specific TaqMan MGB probe provided in the MicroRNA Assays and performed in triplicate on the 7500 quantitative Real-time PCR Machine, and standard errors were determined. miRNA levels were normalized to U6 using the manufacturer’s software (Applied Biosystems).
Ovarian cancer specimens
Tissue blocks from a total of 36 patients with FIGO stage III–IV advanced ovarian cancer who had undergone tumor debulking by a gynecologic oncologist at the Section of Gynecologic Oncology, University of Chicago, between 1994 and 2004 were used for the study after institutional review board approval was obtained, and clinicopathologic parameters were collected as described (Sawada et al. 2007). The paraffin plugs were enriched for tumor tissue using two different methods. Method 1 involved enrichment of tumors from 22 patients according to highest staining for HMGA2 as reported previously (Shell et al. 2007). Method 2 involved enrichment of tumors from 21 patients (seven patients overlap with the samples isolated using method 1) by microscope control using H&E-stained sections of the same sample as guidance. RNA was extracted from FFPET. E-cadherin and Vimentin staining intensities were available from a previous study (Shell et al. 2007).
Statistical analysis
Two sample t-tests for differences in mean miRNA expression between epithelial and mesenchymal cell types were conducted. Pearson’s correlation coefficients were calculated for protein or mRNA expression and miR-200 expression in the NCI60 cell lines. Nonparametric rank tests to give an estimate for R were performed. For miR-200 expression of ovarian cancer samples, low and high E-cadherin-expressing groups were defined by dichotomizing at the median intensity. Significance in expression between groups was determined by performing a two-tailed t-test.
Gene chip analysis
MDA-MB-231 cells were transfected with 50 nM scrambled, pre-miR-200a, or pre-miR-200c, and after 24 h, total RNA was extracted using Trizol reagent. The RNA was further purified using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. The gene chip analysis was performed as described recently (Park et al. 2008) using the U133 chip from Affymetrix.
Acknowledgments
We thank Dr. S. Holbeck for providing the NCI60 cell lines and Dr. A. Lund for providing the murine ZEB2 3′UTR luciferase vector and the pcDNA3-HA-ZEB2 expression plasmid. We also thank Dr. G. Goodall for sharing unpublished data and for providing the two wild-type and mutant ZEB1 and ZEB2 3′UTR luciferase constructs. We are grateful to Dr. E. Kistner for performing statistical analyses and R. Schickel for performing one of the initial miR-200 transfection assays. S.M.P. was supported by a University of Chicago Women’s Board fellowship. This work was supported by the Ovarian Cancer Research Fund (Liz Tilberis Scholars Program) and NIH grants R01 CA111882 (to E.L.) and R01 GM61712 (to M.E.P).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1640608.
References
- Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M., Mikulits W., Brabletz T., Strand D., Obrist P., et al. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. [Europe PMC free article] [Abstract] [Google Scholar]
- Akao Y., Nakagawa Y., Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. [Abstract] [Google Scholar]
- Barnhart B.C., Legembre P., Pietras E., Bubici C., Franzoso G., Peter M.E. CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. EMBO J. 2004;23:3175–3185. [Europe PMC free article] [Abstract] [Google Scholar]
- Bindels S., Mestdagt M., Vandewalle C., Jacobs N., Volders L., Noel A., van Roy F., Berx G., Foidart J.M., Gilles C. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006;25:4975–4985. [Abstract] [Google Scholar]
- Boyerinas B., Park S.M., Shomron N., Hedegaard M.M., Vinther J., Andersen J.A., Feig C., Xu J., Burge C.B., Peter M.E. Identification of Let-7-regulated oncofetal genes. Cancer Res. 2008 (in press) [Abstract] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 2002;99:15524–15529. [Europe PMC free article] [Abstract] [Google Scholar]
- Christoffersen N.R., Silahtaroglu A., Orom U.A., Kauppinen S., Lund A.H. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. [Europe PMC free article] [Abstract] [Google Scholar]
- Chua H.L., Bhat-Nakshatri P., Clare S.E., Morimiya A., Badve S., Nakshatri H. NF-κB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: Potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. [Abstract] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. [Abstract] [Google Scholar]
- Dvorak H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. [Abstract] [Google Scholar]
- Eger A., Aigner K., Sonderegger S., Dampier B., Oehler S., Schreiber M., Berx G., Cano A., Beug H., Foisner R. δEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. [Abstract] [Google Scholar]
- Elloul S., Elstrand M.B., Nesland J.M., Trope C.G., Kvalheim G., Goldberg I., Reich R., Davidson B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103:1631–1643. [Abstract] [Google Scholar]
- Elloul S., Silins I., Trope C.G., Benshushan A., Davidson B., Reich R. Expression of E-cadherin transcriptional regulators in ovarian carcinoma. Virchows Arch. 2006;449:520–528. [Abstract] [Google Scholar]
- Fuchs I.B., Lichtenegger W., Buehler H., Henrich W., Stein H., Kleine-Tebbe A., Schaller G. The prognostic significance of epithelial–mesenchymal transition in breast cancer. Anticancer Res. 2002;22:3415–3419. [Abstract] [Google Scholar]
- Gaur A.B., Jewell D.A., Liang Y., Ridzon D., Moore J.H., Chen C., Ambros V.R., Israel M. Characerization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. [Abstract] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshild G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The microRNA-200 family and mir-205 regulate epithelial–mesenchymal transition by targeting the E-cadherin repressors, ZEB1 and SIP1. Nature Cell Biol. 2008 (in press) [Abstract] [Google Scholar]
- Hay E.D. An overview of epithelio–mesenchymal transformation. Acta Anat. (Basel) 1995;154:8–20. [Abstract] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. [Europe PMC free article] [Abstract] [Google Scholar]
- Higgins D.F., Kimura K., Bernhardt W.M., Shrimanker N., Akai Y., Hohenstein B., Saito Y., Johnson R.S., Kretzler M., Cohen C.D., et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J. Clin. Invest. 2007;117:3810–3820. [Europe PMC free article] [Abstract] [Google Scholar]
- Hurteau G.J., Spivack S.D., Brock G.J. Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT–PCR. Cell Cycle. 2006;5:1951–1956. [Abstract] [Google Scholar]
- Hurteau G.J., Carlson J.A., Spivack S.D., Brock G.J. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. [Abstract] [Google Scholar]
- Imamichi Y., Konig A., Gress T., Menke A. Collagen type I-induced Smad-interacting protein 1 expression downregulates E-cadherin in pancreatic cancer. Oncogene. 2007;26:2381–2385. [Abstract] [Google Scholar]
- Jakowlew S.B. Transforming growth factor-β in cancer and metastasis. Cancer Metastasis Rev. 2006;25:435–457. [Abstract] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. [Abstract] [Google Scholar]
- Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. [Abstract] [Google Scholar]
- Maeda G., Chiba T., Okazaki M., Satoh T., Taya Y., Aoba T., Kato K., Kawashiri S., Imai K. Expression of SIP1 in oral squamous cell carcinomas: Implications for E-cadherin expression and tumor progression. Int. J. Oncol. 2005;27:1535–1541. [Abstract] [Google Scholar]
- Mott J.L., Kobayashi S., Bronk S.F., Gores G.J. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. [Europe PMC free article] [Abstract] [Google Scholar]
- Park S.M., Shell S., Radjabi A.R., Schickel R., Feig C., Boyerinas B., Dinulescu D.M., Lengyel E., Peter M.E. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. [Abstract] [Google Scholar]
- Park S.M., Rajapaksha T.W., Zhang M., Sattar H.A., Fichera A., Ashton-Rickardt P.G., Peter M.E. CD95 signaling deficient mice with a wild-type hematopoietic system are prone to hepatic neoplasia. Apoptosis. 2008;13:41–51. [Abstract] [Google Scholar]
- Peinado H., Portillo F., Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 2004;48:365–375. [Abstract] [Google Scholar]
- Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. [Abstract] [Google Scholar]
- Pena C., Garcia J.M., Silva J., Garcia V., Rodriguez R., Alonso I., Millan I., Salas C., de Herreros A.G., Munoz A., et al. E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: Clinicopathological correlations. Hum. Mol. Genet. 2005;14:3361–3370. [Abstract] [Google Scholar]
- Pena C., Garcia J.M., Garcia V., Silva J., Dominguez G., Rodriguez R., Maximiano C., Garcia de Herreros A., Munoz A., Bonilla F. The expression levels of the transcriptional regulators p300 and CtBP modulate the correlations between SNAIL, ZEB1, E-cadherin and vitamin D receptor in human colon carcinomas. Int. J. Cancer. 2006;119:2098–2104. [Abstract] [Google Scholar]
- Postigo A.A., Dean D.C. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc. Natl. Acad. Sci. 2000;97:6391–6396. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosivatz E., Becker I., Specht K., Fricke E., Luber B., Busch R., Hofler H., Becker K.F. Differential expression of the epithelial–mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am. J. Pathol. 2002;161:1881–1891. [Europe PMC free article] [Abstract] [Google Scholar]
- Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P., Iyer V., Jeffrey S.S., Van de Rijn M., Waltham M., et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. [Abstract] [Google Scholar]
- Savagner P. Leaving the neighborhood: Molecular mechanisms involved during epithelial–mesenchymal transition. Bioessays. 2001;23:912–923. [Abstract] [Google Scholar]
- Sawada K., Radjabi A.R., Shinomiya N., Kistner E., Kenny H., Becker A.R., Turkyilmaz M.A., Salgia R., Yamada S.D., Vande Woude G.F., et al. c-Met overexpression is a prognostic factor in ovarian cancer and an effective target for inhibition of peritoneal dissemination and invasion. Cancer Res. 2007;67:1670–1679. [Abstract] [Google Scholar]
- Schulman B.R., Esquela-Kerscher A., Slack F.J. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 2005;234:1046–1054. [Europe PMC free article] [Abstract] [Google Scholar]
- Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A., Feig C., Lengyel E., Peter M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. 2007;104:11400–11405. [Europe PMC free article] [Abstract] [Google Scholar]
- Shirakihara T., Saitoh M., Miyazono K. Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β Mol. Biol. Cell. 2007;18:3533–3544. [Europe PMC free article] [Abstract] [Google Scholar]
- Spoelstra N.S., Manning N.G., Higashi Y., Darling D., Singh M., Shroyer K.R., Broaddus R.R., Horwitz K.B., Richer J.K. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. [Abstract] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. [Abstract] [Google Scholar]
- Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. [Abstract] [Google Scholar]
- Thiery J.P., Sleeman J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell. Biol. 2006;7:131–142. [Abstract] [Google Scholar]
- Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A., et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. [Abstract] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. [Abstract] [Google Scholar]
- Zavadil J., Bitzer M., Liang D., Yang Y.C., Massimi A., Kneitz S., Piek E., Bottinger E.P. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β Proc. Natl. Acad. Sci. 2001;98:6686–6691. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.1640608
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/22/7/894.full.pdf
Free to read at www.genesdev.org
http://www.genesdev.org/cgi/content/abstract/22/7/894
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/content/full/22/7/894
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/reprint/22/7/894
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
GRHL2 regulates keratinocyte EMT-MET dynamics and scar formation during cutaneous wound healing.
Cell Death Dis, 15(10):748, 14 Oct 2024
Cited by: 0 articles | PMID: 39402063 | PMCID: PMC11473813
Epithelial-Mesenchymal Plasticity and Epigenetic Heterogeneity in Cancer.
Cancers (Basel), 16(19):3289, 27 Sep 2024
Cited by: 0 articles | PMID: 39409910 | PMCID: PMC11475326
Review Free full text in Europe PMC
Relationship of Signaling Pathways between RKIP Expression and the Inhibition of EMT-Inducing Transcription Factors SNAIL1/2, TWIST1/2 and ZEB1/2.
Cancers (Basel), 16(18):3180, 17 Sep 2024
Cited by: 0 articles | PMID: 39335152 | PMCID: PMC11430682
Review Free full text in Europe PMC
miR-200c targeting GLI3 inhibits cell proliferation and promotes apoptosis in non-small cell lung cancer cells.
Medicine (Baltimore), 103(38):e39658, 01 Sep 2024
Cited by: 0 articles | PMID: 39312343 | PMCID: PMC11419521
The effect of ribociclib on the expression levels of miR-141 and CDK4/6-USP51 signaling pathway genes in MCF-7 and MDA-MB-231 cells.
PLoS One, 19(8):e0309289, 28 Aug 2024
Cited by: 0 articles | PMID: 39196911 | PMCID: PMC11355560
Go to all (1,563) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2.
J Biol Chem, 283(22):14910-14914, 14 Apr 2008
Cited by: 1116 articles | PMID: 18411277 | PMCID: PMC3258899
The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1.
Nat Cell Biol, 10(5):593-601, 30 Mar 2008
Cited by: 2617 articles | PMID: 18376396
MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma.
Oncol Rep, 31(5):2021-2028, 21 Mar 2014
Cited by: 56 articles | PMID: 24677166 | PMCID: PMC4020617
Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition.
Trends Cell Biol, 18(8):357-359, 26 Jun 2008
Cited by: 88 articles | PMID: 18585040
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA111882
NIGMS NIH HHS (2)
Grant ID: R01 GM061712
Grant ID: R01 GM61712





