Abstract
Free full text

Isolation and characterization of chromosomal promoters of Streptococcus salivarius subsp. thermophilus.
Abstract
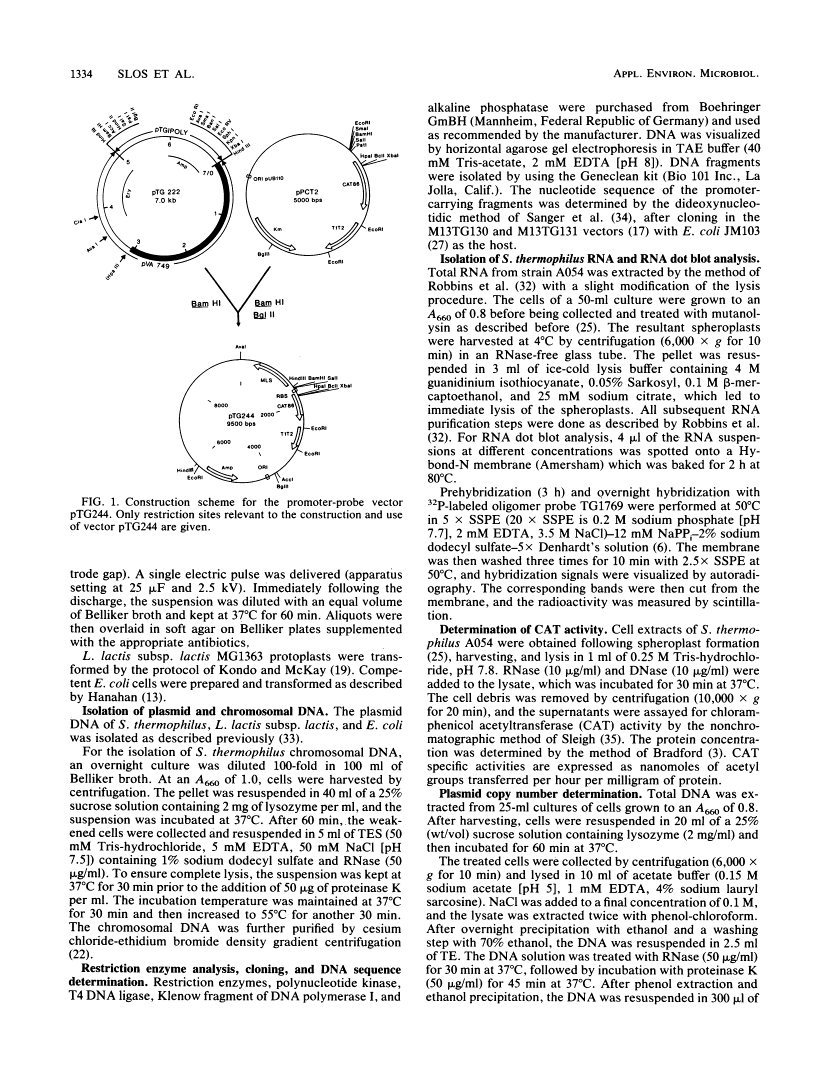
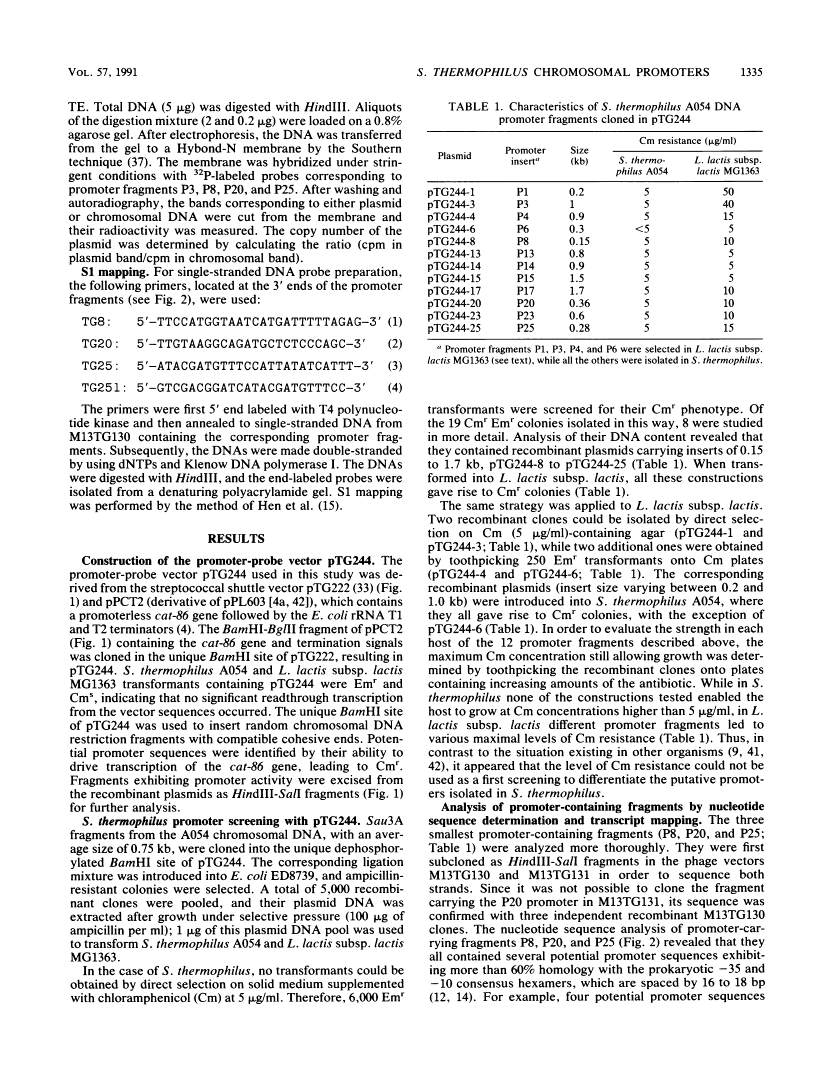
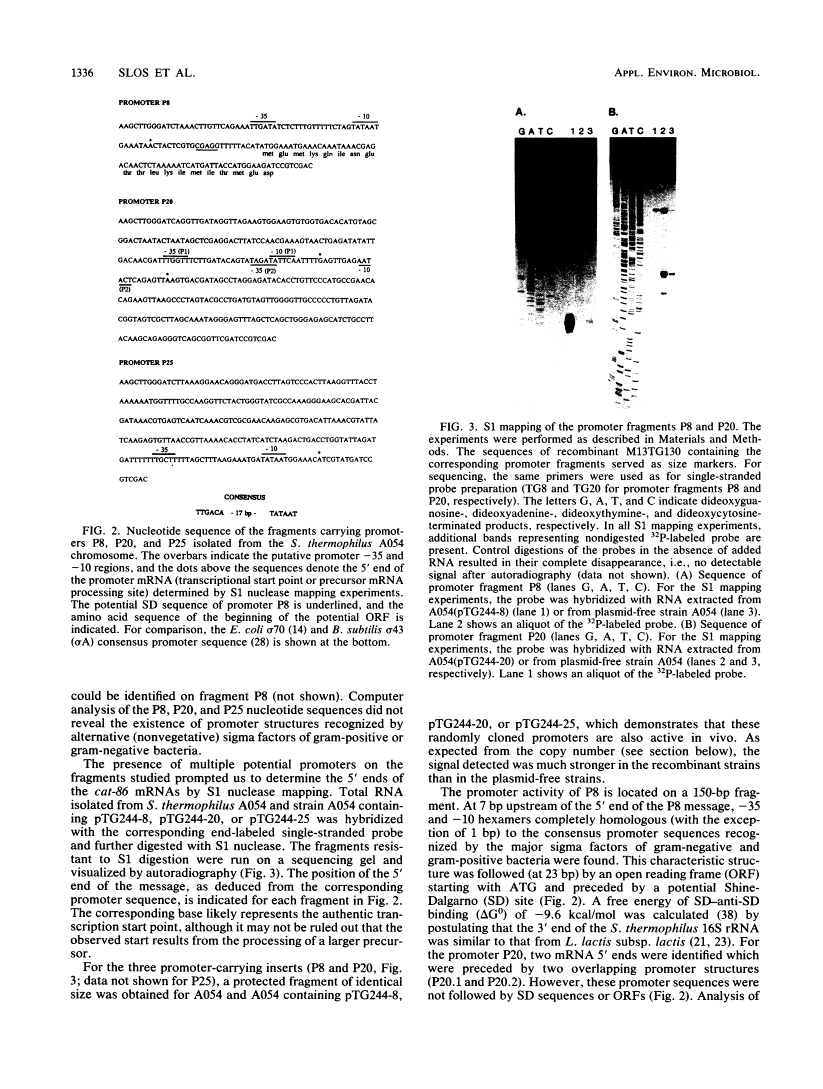
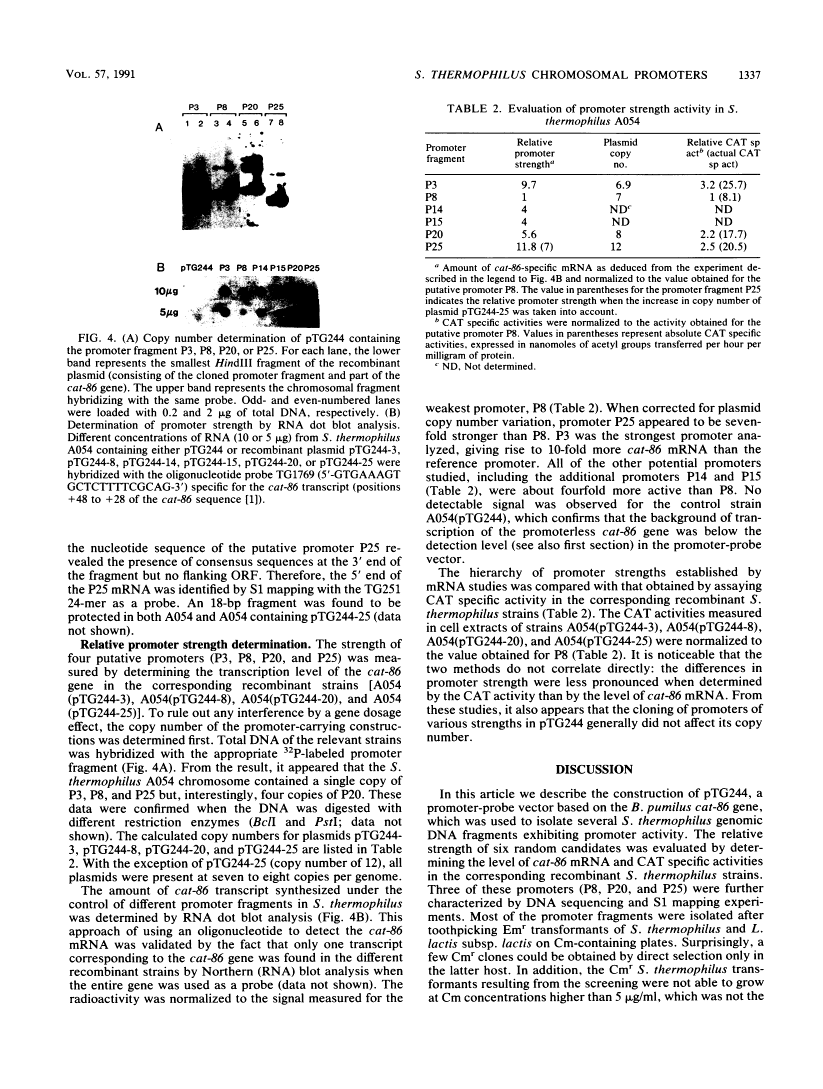
A promoter probe vector, pTG244, was constructed with the aim of isolating transcription initiation signals from Streptococcus thermophilus (Streptococcus salivarius subsp. thermophilus). pTG244 is based on the Escherichia coli-streptococcus shuttle vector pTG222, into which the promoterless chloramphenicol acetyltransferase gene of Bacillus pumilus (cat-86) was cloned. Random Sau3A fragments from the S. thermophilus A054 chromosomal DNA were cloned upstream of the cat-86 gene by using E. coli as the host. The pool of recombinant plasmids were introduced into S. thermophilus and Lactococcus lactis subsp. lactis in order to search for promoter activity in these hosts. For S. thermophilus, it was necessary to first select erythromycin-resistant transformants and then to screen for chloramphenicol resistance among these. Direct selection of chloramphenicol-resistant clones was, however, possible in L. lactis subsp. lactis. Six fragments exhibiting promoter activity were characterized in S. thermophilus by measuring the levels of cat-86 transcription and/or chloramphenicol acetyltransferase specific activity. Three of the promoter-carrying fragments were sequenced. The 5' ends of their corresponding mRNAs were determined by S1 mapping and shown to correspond to a purine residue in all cases. Upstream from these potential transcription start points, sequences homologous to the E. coli sigma 70 and the Bacillus subtilis vegetative sigma 43 (or sigma A) consensus promoters were identified.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.5M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos NP, Jr, Mongkolsuk S, Kaufman JD, Lovett PS. Chloramphenicol-induced translation of cat-86 mRNA requires two cis-acting regulatory regions. J Bacteriol. 1985 Nov;164(2):696–703. [Europe PMC free article] [Abstract] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. [Abstract] [Google Scholar]
- Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. [Abstract] [Google Scholar]
- Denhardt DT. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. [Abstract] [Google Scholar]
- Deuschle U, Kammerer W, Gentz R, Bujard H. Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J. 1986 Nov;5(11):2987–2994. [Europe PMC free article] [Abstract] [Google Scholar]
- Dhaese P, Hussey C, Van Montagu M. Thermo-inducible gene expression in Bacillus subtilis using transcriptional regulatory elements from temperate phage phi 105. Gene. 1984 Dec;32(1-2):181–194. [Abstract] [Google Scholar]
- Gasson MJ, Davies FL. Prophage-Cured Derivatives of Streptococcus lactis and Streptococcus cremoris. Appl Environ Microbiol. 1980 Nov;40(5):964–966. [Europe PMC free article] [Abstract] [Google Scholar]
- Graves MC, Rabinowitz JC. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for "extended" promoter elements in gram-positive organisms. J Biol Chem. 1986 Aug 25;261(24):11409–11415. [Abstract] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. [Abstract] [Google Scholar]
- Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. [Europe PMC free article] [Abstract] [Google Scholar]
- Hen R, Sassone-Corsi P, Corden J, Gaub MP, Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. [Europe PMC free article] [Abstract] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. [Europe PMC free article] [Abstract] [Google Scholar]
- Kieny MP, Lathe R, Lecocq JP. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. [Abstract] [Google Scholar]
- Kondo JK, McKay LL. Plasmid transformation of Streptococcus lactis protoplasts: optimization and use in molecular cloning. Appl Environ Microbiol. 1984 Aug;48(2):252–259. [Europe PMC free article] [Abstract] [Google Scholar]
- Lovett PS. Translational attenuation as the regulator of inducible cat genes. J Bacteriol. 1990 Jan;172(1):1–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Ludwig W, Seewaldt E, Kilpper-Bälz R, Schleifer KH, Magrum L, Woese CR, Fox GE, Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985 Mar;131(3):543–551. [Abstract] [Google Scholar]
- Mercenier A, Robert C, Romero DA, Castellino I, Slos P, Lemoine Y. Development of an efficient spheroplast transformation procedure for S. thermophilus: the use of transfection to define a regeneration medium. Biochimie. 1988 Apr;70(4):567–577. [Abstract] [Google Scholar]
- Mercenier A, Slos P, Faelen M, Lecocq JP. Plasmid transduction in Streptococcus thermophilus. Mol Gen Genet. 1988 May;212(2):386–389. [Abstract] [Google Scholar]
- Messing J, Crea R, Seeburg PH. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. [Europe PMC free article] [Abstract] [Google Scholar]
- Moran CP, Jr, Lang N, LeGrice SF, Lee G, Stephens M, Sonenshein AL, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. [Abstract] [Google Scholar]
- Murray NE, Brammar WJ, Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. [Abstract] [Google Scholar]
- Poolman B, Royer TJ, Mainzer SE, Schmidt BF. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989 Jan;171(1):244–253. [Europe PMC free article] [Abstract] [Google Scholar]
- Poolman B, Royer TJ, Mainzer SE, Schmidt BF. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990 Jul;172(7):4037–4047. [Europe PMC free article] [Abstract] [Google Scholar]
- Robbins JC, Spanier JG, Jones SJ, Simpson WJ, Cleary PP. Streptococcus pyogenes type 12 M protein gene regulation by upstream sequences. J Bacteriol. 1987 Dec;169(12):5633–5640. [Europe PMC free article] [Abstract] [Google Scholar]
- Romero DA, Slos P, Robert C, Castellino I, Mercenier A. Conjugative mobilization as an alternative vector delivery system for lactic streptococci. Appl Environ Microbiol. 1987 Oct;53(10):2405–2413. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Schroeder CJ, Robert C, Lenzen G, McKay LL, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus beta-galactosidase sequences. J Gen Microbiol. 1991 Feb;137(2):369–380. [Abstract] [Google Scholar]
- Sleigh MJ. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986 Jul;156(1):251–256. [Abstract] [Google Scholar]
- Somkuti GA, Steinberg DH. Genetic transformation of Streptococcus thermophilus by electroporation. Biochimie. 1988 Apr;70(4):579–585. [Abstract] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. [Abstract] [Google Scholar]
- Tinoco I, Jr, Borer PN, Dengler B, Levin MD, Uhlenbeck OC, Crothers DM, Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. [Abstract] [Google Scholar]
- van de Guchte M, van der Vossen JM, Kok J, Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989 Jan;55(1):224–228. [Europe PMC free article] [Abstract] [Google Scholar]
- van der Vossen JM, Kok J, Venema G. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985 Aug;50(2):540–542. [Europe PMC free article] [Abstract] [Google Scholar]
- van der Vossen JM, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. [Europe PMC free article] [Abstract] [Google Scholar]
- Williams DM, Duvall EJ, Lovett PS. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu CW, Goldthwait DA. Studies of nucleotide binding to the ribonucleic acid polymerase by a fluoresence technique. Biochemistry. 1969 Nov;8(11):4450–4458. [Abstract] [Google Scholar]
- Wu CW, Goldthwait DA. Studies of nucleotide binding to the ribonucleic acid polymerase by equilibrium dialysis. Biochemistry. 1969 Nov;8(11):4458–4464. [Abstract] [Google Scholar]
Associated Data
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.57.5.1333-1339.1991
Read article for free, from open access legal sources, via Unpaywall:
https://aem.asm.org/content/aem/57/5/1333.full.pdf
Free to read at aem.asm.org
http://aem.asm.org/cgi/content/abstract/57/5/1333
Free after 4 months at aem.asm.org
http://aem.asm.org/cgi/reprint/57/5/1333
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Advances in Genetic Tools and Their Application in Streptococcus thermophilus.
Foods, 12(16):3119, 19 Aug 2023
Cited by: 0 articles | PMID: 37628118 | PMCID: PMC10453384
Review Free full text in Europe PMC
Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275.
Sci Rep, 4:4974, 15 May 2014
Cited by: 46 articles | PMID: 24827399 | PMCID: PMC4021336
Effect of inactivation of stress response regulators on the growth and survival of Streptococcus thermophilus Sfi39.
Int J Food Microbiol, 129(3):211-220, 03 Dec 2008
Cited by: 16 articles | PMID: 19128851
Characterization of the cro-ori region of the Streptococcus thermophilus virulent bacteriophage DT1.
Appl Environ Microbiol, 71(3):1237-1246, 01 Mar 2005
Cited by: 12 articles | PMID: 15746324 | PMCID: PMC1065193
DNA-binding activity of the Streptococcus thermophilus phage Sfi21 repressor.
Virology, 303(1):100-109, 01 Nov 2002
Cited by: 12 articles | PMID: 12482661
Go to all (29) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Isolation and characterization of Streptococcus cremoris Wg2-specific promoters.
Appl Environ Microbiol, 53(10):2452-2457, 01 Oct 1987
Cited by: 179 articles | PMID: 2447829 | PMCID: PMC204128
Isolation and characterization of transcription signal sequences from Streptococcus thermophilus.
Curr Microbiol, 34(4):216-219, 01 Apr 1997
Cited by: 3 articles | PMID: 9058540
Isolation and characterization of Lactococcus lactis subsp. lactis promoters.
Appl Environ Microbiol, 57(2):333-340, 01 Feb 1991
Cited by: 28 articles | PMID: 1707605 | PMCID: PMC182715
Genomic organization of lactic acid bacteria.
Antonie Van Leeuwenhoek, 70(2-4):161-183, 01 Oct 1996
Cited by: 41 articles | PMID: 8879406
Review