Abstract
Free full text

Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins
Abstract
Accumulation of misfolded oxidant-damaged proteins is characteristic of many diseases and aging. To understand how cells handle postsynthetically damaged proteins, we studied in Saccharomyces cerevisiae the effects on overall protein degradation of shifting from 30 to 38°C, exposure to reactive oxygen species generators (paraquat or cadmium), or lack of superoxide dismutases. Degradation rates of long-lived proteins (i.e., most cell proteins) were not affected by these insults, even when there was widespread oxidative damage to proteins. However, exposure to 38°C, paraquat, cadmium, or deletion of SOD1 enhanced two- to threefold the degradation of newly synthesized proteins. By 1 h after synthesis, their degradation was not affected by these treatments. Degradation of these damaged cytosolic proteins requires the ubiquitin–proteasome pathway, including the E2s UBC4/UBC5, proteasomal subunit RPN10, and the CDC48–UfD1–NPL4 complex. In yeast lacking these components, the nondegraded polypeptides accumulate as aggregates. Thus, many cytosolic proteins proceed through a prolonged “fragile period” during which they are sensitive to degradation induced by superoxide radicals or increased temperatures.
Introduction
A critical function of intracellular protein degradation is to serve as a quality control system that destroys misfolded proteins, which may arise through mutations, errors in gene expression, postsynthetic damage, and failures to fold correctly or to form multimeric complexes (Goldberg, 2003). The efficient elimination of such potentially toxic proteins is of clear selective advantage, and failure to do so can lead to the pathogenesis of various neurodegenerative diseases (Sherman and Goldberg, 2001). It has been proposed that in mammals, a large fraction of newly synthesized proteins are defective (Yewdell et al., 1996; Qian et al., 2006). However, it is unclear how often errors in gene expression occur or proteins fail to fold correctly or to form proper multimeric complexes. Protein conformations are labile structures, especially at 37°C, and are easily damaged by various agents found in cells (Goldberg, 2003), especially oxygen radicals, which are continually generated by intermediary metabolism (Davies, 1987). In addition, increases in environmental temperature can damage protein structures. In response to the accumulation of unfolded proteins, which occurs upon exposure to increased temperatures or various damaging agents (Lindquist, 1986), cells induce the heat-shock response that enhances survival under such stressful conditions (Goff and Goldberg, 1985; Ananthan et al., 1986). As part of this adaptive response, cells express molecular chaperones (Hartl and Hayer-Hartl, 2002), ubiquitin, and certain ubiquitination enzymes (Finley et al., 1987; Seufert and Jentsch, 1990), which collaborate to prevent the buildup of the misfolded aggregated proteins (Sherman and Goldberg, 2001).
Most studies of the degradation of misfolded proteins have focused on mutated proteins or ones that incorporated amino acid analogues in place of normal residues (Goldberg, 2003). Such proteins fail to fold correctly and, in eukaryotes, are degraded by the ubiquitin–proteasome pathway. This process requires Hsp70 and Hsp40 for the recognition of proteins for ubiquitin conjugation (Lee et al., 1996; Murata et al., 2001). The present studies investigated the breakdown of proteins in yeast that are damaged postsynthetically by heat shock (i.e., thermal damage) or reactive oxygen species (ROS) generated by exposure to paraquat or cadmium ions or endogenously by intermediary metabolism. Several studies have demonstrated that exposure of mammalian cells to oxygen radicals or hydrogen peroxide enhances overall proteolysis (Fagan et al., 1986; Davies and Goldberg, 1987; Matthews et al., 1989), although the nature of the proteins affected was not investigated. Also, most of these studies concluded that the oxidant-damaged proteins are digested independently of ATP or ubiquitin (Fagan et al., 1986; Davies and Goldberg, 1987; Matthews et al., 1989), probably by the 20S proteasome (Grune et al., 1996; Shringarpure et al., 2003), as has also been suggested to occur for many misfolded proteins (Qian et al., 2006). Such a mechanism is in sharp contrast to the ubiquitin-dependent degradation of unfolded proteins containing amino acid analogues (Seufert and Jentsch, 1990). Also, certain studies argued that ubiquitination is critical for the degradation of oxidant-damaged proteins (Dudek et al., 2005).
In this study, we investigated the involvement of the ubiquitin–proteasome pathway in breakdown of thermally or ROS-damaged proteins and tested whether any classes of cell proteins are particularly sensitive to this process. We show in this paper that the degradation of most cell proteins is surprisingly resistant to such insults but that a fraction of recently synthesized proteins is particularly sensitive to oxidative and thermal damage, which leads to their rapid ubiquitin-dependent degradation by a process distinct from the degradation of misfolded proteins in the secretory pathway (ER-associated degradation [ERAD]; Kostova and Wolf, 2003). Surprisingly, newly synthesized proteins are sensitive to thermally and ROS-induced degradation for 30–60 min after synthesis, presumably until they have assumed their final conformations in stable multimeric complexes.
Results
Increased temperature stimulates the degradation of recently synthesized proteins but not the bulk of cell proteins
Although it is well established that heat shock by causing an accumulation of misfolded proteins trigger the production of components of the ubiquitin–proteasome pathway and “molecular chaperones” (Ananthan et al., 1986; Sherman and Goldberg, 2001), the effects of increased temperatures on the degradation of cell proteins have not been systematically studied. To learn how increased temperatures influence the degradation of different classes of cell proteins, we studied the effects of shifting Saccharomyces cerevisiae from the standard growth temperature, 30°C, to 38°C, a temperature shift that induces the heat-shock response (Lee et al., 1996). Yeast growing exponentially were labeled for 5 min with 35S-methionine (Met) at 30°C. After washing twice in “chase” medium, which contained cycloheximide and high concentrations of nonradioactive Met, to prevent reincorporation of the 35S-Met and the synthesis of heat-shock proteins, cells were resuspended in chase media preadjusted to 20, 30, 38, or 42°C. Rates of proteolysis were then measured by following the conversion of radiolabeled cell proteins to trichloroacetic acid–soluble form (Fig. 1 A). The shift to 38°C initially caused a two- to threefold faster rate of degradation of the labeled proteins than in cells maintained at 30°C.
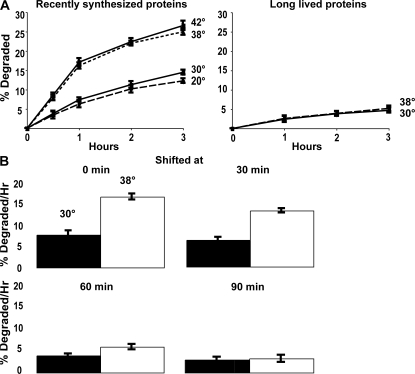
Shifting cells from 30 to 38°C stimulates the degradation of recently synthesized proteins, but not of long-lived proteins, and by 60 min after synthesis, proteins no longer show their response to temperature shift. (A) Proteins were labeled with 35S-Met for 5 min at 30°C to label recently synthesized proteins or 90 min to label long-lived proteins. After washing twice with chase media containing cyclohexamide, the cells were shifted to the indicated temperatures and rates of degradation were measured. The data shown in this and the subsequent experiments represent the means ± SD obtained from three experiments. (B) Cell proteins were labeled with 35S-Met for 5 min, washed, resuspended in chase media, and shifted to 38°C immediately or 30, 60, or 90 min later. The amounts of protein degradation after 1 h were measured. Degradation rates after 2 or 3 h showed similar differences as after 1 h (not depicted).
However, this rate of proteolysis was enhanced only briefly (Fig. 1 A). By an hour after synthesis, when the overall rate of proteolysis had slowed considerably (because of the loss of most rapidly degraded proteins), the rates of degradation at the two temperatures became indistinguishable (Fig. 1 A). These findings imply that some newly synthesized proteins are specifically sensitive to thermal damage by the same temperature shift (30 to 38°C) that is commonly used to induce the heat-shock response in yeast. Surprisingly, shifting to 42°C caused no further increase in proteolysis over that seen at 38°C, and in the cells at 20°C, overall rates of degradation were indistinguishable from those at 30°C, both in the first hour and at later times (Fig. 1 A). Shift to 38 or 42°C accelerated the degradation of ~13% of cell proteins that would otherwise be long lived so that they behaved like short-lived components.
These similar slopes after 1 h imply that the increased temperature did not significantly affect the degradation of the bulk of cell proteins, which are long-lived components that are degraded at a rate of ~1–2%/h (Fig. 1 A). In fact, their degradation appears quite independent of temperature, being very similar between 20 and 42°C, unlike the rates of the great majority of biochemical reactions.
To confirm this conclusion, yeast were grown at 30°C and exposed to 35S-Met for 90 min to label primarily long-lived cell components. After washing, the cells were resuspended at 30 or 38°C. As expected, overall rates of degradation of these proteins, which comprise the bulk of cell proteins, were much slower and did not increase upon shift to 38°C (Fig. 1 A). Because the shift to 38°C (or 42°C) does not alter the breakdown of long-lived components, the increased temperature specifically accelerates the degradation of ~10–13% of newly synthesized proteins that at 30°C would have become long-lived components.
Proteins are susceptible to heat-induced degradation for 30–60 min after synthesis
To determine how long after synthesis cell proteins are sensitive to thermally induced degradation, we compared the effects on proteolysis of shifting yeast from 30 to 38°C immediately after the 5-min pulse of 35S-Met or 30, 60, or 90 min later (Fig. 1 B). The cells shifted at 30 min showed a smaller increase in proteolysis than those shifted immediately after synthesis, and 60 min after synthesis the effect of temperature upshift was much smaller and not statistically significant. By 90 min, the shift to 38°C failed to increase degradation. Thus, during the hour after synthesis proteins appeared to become insensitive to thermal damage, as reflected in their resistance to heat-induced degradation. Because it is unlikely that the folding of many proteins would require 30–60 min, this “fragile period” presumably represents the time necessary for postsynthetic modifications, proper multimer formation, and localization, which together contribute to thermal resistance. Presumably, as proteins achieve stable multimeric structures and their final folded conformations, they become more resistant to thermal denaturation.
ROS stimulate the degradation of recently synthesized proteins
A variety of experiments using free radical generators or hydrogen peroxide (ROS) have indicated that oxidant damage to cell proteins can enhance their degradation (Fagan et al., 1986; Davies, 1987; Davies and Goldberg, 1987). To study the effects of oxygen radicals on the breakdown of recently synthesized and long-lived yeast proteins, we used paraquat, a superoxide generator (Lee et al., 2004), as well as cadmium, which can replace the Fe2+ in proteins and seems to generate oxygen radicals as a consequence of the released Fe2+ (Thevenod and Friedmann, 1999; Vido et al., 2001). Exposure of yeast to Cd++ causes induction of enzymes that defend against ROS, molecular chaperones, ubiquitination enzymes, and Cdc48p (Vido et al., 2001). During exponential growth, wild-type (WT) yeast were exposed to 80 μg/ml paraquat or 50 μg/ml cadmium. After 90 min, these cells and untreated controls were labeled with 35S-Met for 5 min, and then we compared the rates of protein degradation. At these low concentrations, neither cadmium nor paraquat reduced cell growth or protein synthesis during these experiments, but the proteins synthesized in their presence were initially degraded two- to threefold faster than controls (Fig. 2 A). As found after a shift to 38°C, the oxidant-induced acceleration of degradation was only clearly demonstrated during the first 60 min after synthesis. After this time, the proteins synthesized in the presence of cadmium or paraquat were degraded at similar rates to those in control cells (Fig. 2 A). These findings suggest that the degradation of the long-lived cell proteins is not affected by oxidative damage.
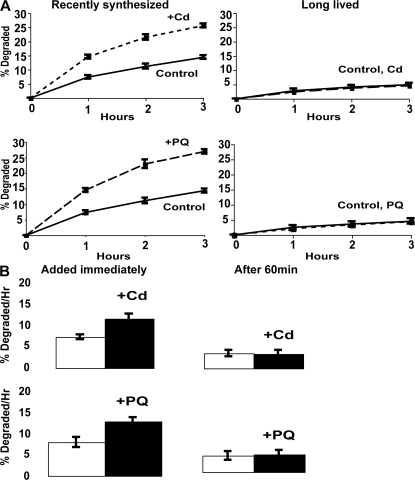
Exposure to paraquat or cadmium stimulates the degradation of recently synthesized but not long-lived proteins, and 60 min after synthesis, proteins no longer show this response to paraquat and cadmium. (A) During exponential growth at 30°C, yeast were exposed to 80 μg/ml paraquat or 50 μg/ml cadmium for 90 min, and then 35S-Met was added for 5 min at 30°C to label recently synthesized proteins or for 90 min to label long-lived proteins. After washing and resuspension in chase medium, rates of protein degradation were measured as in Fig. 1 A. (B) Proteins were labeled with 35S-Met for 5 min, washed, and then paraquat and cadmium were added immediately or 60 min after synthesis. The amounts of degradation after 1 h were measured. Degradation rates after 2 and 3 h showed similar differences as after 1 h (not depicted).
To verify this conclusion, growing yeast were exposed to paraquat or cadmium for 90 min. These cells and untreated controls were incubated with 35S-Met for 90 min to label primarily long-lived cell components, and after washing, the rates of protein degradation were compared. As expected, overall rates of degradation of these proteins were much slower (1–2%/h) and did not increase upon exposure to paraquat or cadmium (Fig. 2 A). Thus, the degradation of most cell proteins is quite resistant to these types of oxidative damage. It is also noteworthy that the magnitudes of the increases in degradation after exposure to paraquat, cadmium, 38°C, or 42°C (Figs. 1 A and 2 A) were similar. These findings suggest that after these insults, the maximal capacity of the degradative system had been reached. Accordingly, exposure of cells to these oxidants at 38°C did not further enhance proteolysis (unpublished data).
To confirm that the oxygen radicals stimulate only the degradation of newly synthesized proteins, we labeled cell proteins for 5 min during growth at 30°C, exposed a portion of the culture to paraquat or cadmium immediately or 60 min later, and then measured rates of degradation (Fig. 2 B). Addition of these ROS generators stimulated the breakdown of recently synthesized proteins. However, when the cadmium or paraquat was added 60 min later, the rates of proteolysis did not change (Fig. 2 B). The stimulation of proteolysis by this protocol was smaller than in Fig. 2 A, where the exposure to the oxidants was for 90 min, which should allow greater uptake of the oxidants. Because cycloheximide was present after the proteins were labeled, this resistance of proteolysis to the stimulation by ROS (Fig. 2) or heat shock (Fig. 1) after 1 h cannot be caused by the induction of proteins that protect against the oxidant or thermal damage. Instead, it probably reflects some maturational change in the newly synthesized proteins during this hour. Most likely, these insults can enhance degradation of newly synthesized proteins for only 1 h because, with time, cell proteins succeed in forming more stable multimeric complexes that are more resistant to ROS or thermal damage.
These findings raise two possibilities: either long-lived proteins are not chemically damaged by ROS or chemical damage occurs but does not trigger their unfolding and degradation, as it does to the recently synthesized proteins. To examine these alternatives, we assayed whether the long-lived proteins undergo oxidative damage using OxyBlot, which detects the presence of carbonyl groups in proteins (Sturtz et al., 2001). Exposure to paraquat for 90 min dramatically increased the presence of carbonyls in cell proteins and did so similarly, or even to a great extent, after treatment of the yeast with cycloheximide for 2 h to prevent the synthesis of new proteins and allow the degradation of short-lived polypeptides. Thus, the remaining long-lived cell proteins are certainly damaged by the oxygen-free radicals, although this modification fails to trigger rapid proteolysis (Fig. 3 A). The apparently greater appearance of protein carbonyls after cycloheximide treatment in Fig. 3 A is probably because some antioxidant enzymes are short-lived proteins (e.g., Sod1p).
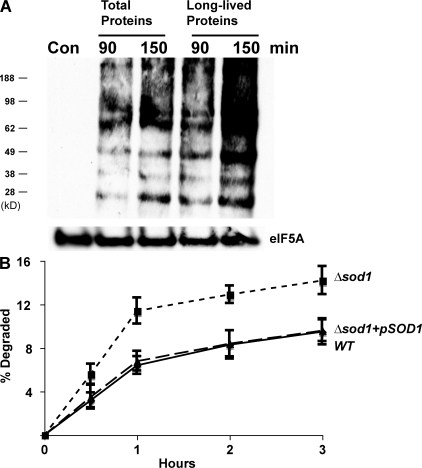
Although long-lived proteins are not degraded more rapidly after exposure to paraquat, they are oxidatively damaged, and a lack of SOD1 also enhances the degradation of recently synthesized proteins. (A) Yeast were grown exponentially, and a portion of cells were treated with cycloheximide for 2 h to allow the degradation of short-lived proteins (the times studied in Fig. 2 A). Both the control and treated cells were then exposed to paraquat for 90 or 150 min. The presence of carbonylated proteins in equal amounts of cell proteins was assayed after derivitization with DNP-hydrazine (DNPH) and then Western blotting with an anti–DNP-hydrazone antibody. A control lane without the treatments with DNPH and paraquat is included to show the specificity of the antibody. Equal loading of lanes was shown with an eIF5A antibody. (B) WT, Δsod1 mutant, and the Δsod1 mutant expressing SOD1 from a plasmid were labeled with 35S-Met for 5 min at 30°C. Rates of protein degradation were measured at 30°C as in Fig. 1 A.
Superoxide dismutase protects newly synthesized proteins against degradation
Prior studies on the effects of ROS on protein degradation used exogenous agents that cause free radical generation. However, oxygen radicals are also continually generated by cell metabolism. Cells have multiple enzyme systems to inactivate such radicals including the cytosolic Cu-Zn–containing superoxide dismutase 1 (Sod1p) and the mitochondrial Mn-containing superoxide dismutase 2 (Sod2p; Sturtz et al., 2001). Mutant strains lacking these enzymes, Δsod1 and Δsod2, accumulate oxidant-damaged proteins (Sturtz et al., 2001). To evaluate whether these enzymes help protect cell proteins against ROS-induced degradation and whether superoxide radicals can stimulate proteolysis, we compared rates of degradation in WT yeast and strains lacking either of these dismutases. After a 5-min pulse of 35S-Met, newly synthesized proteins were degraded at least twice as rapidly in the Δsod1 strain as in the WT (Fig. 3 B), but by 1 h after synthesis, the rates of proteolysis resembled those in the WT. Thus, Sod1p appears to be important in protecting newly synthesized proteins from degradation induced by endogenously generated superoxide radicals.
Unlike the Δsod1 strain, the Δsod2 mutant showed very similar rates of degradation to those in isogenic WT cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200803022/DC1). Thus, the mitochondrial Sod2p is not important in the protection of newly synthesized proteins, presumably because these proteins are restricted to the cytosolic compartment. Upon addition of paraquat or cadmium to the Δsod1 strain, there was a further increase in proteolysis, and its capacity to degrade damaged proteins exceeded those in the paraquat- or cadmium-treated WT (Fig. S1 B).
Thermal and ROS-induced degradation involves the ubiquitin–proteasome system
Although early studies indicated a requirement for ATP for the degradation of oxidant-damaged proteins (Goldberg and Boches, 1982), subsequent studies, primarily in mature red cells, indicated that oxidant-damaged proteins could be degraded by an energy- and ubiquitin-independent process (Fagan et al., 1986; Davies and Goldberg, 1987; Matthews et al., 1989). This process was reported to involve degradation of the damaged proteins by 20S proteasomes (Grune et al., 1996; Shringarpure et al., 2003). To learn if the rapid elimination of these oxidant- and heat-damaged proteins requires the ubiquitin–proteasome pathway (like the degradation of misfolded proteins containing amino acid analogues), we tested whether degradation of these postsynthetically damaged proteins requires the ubiquitin-conjugating enzymes (E2s) UBC4 and UBC5 (Seufert and Jentsch, 1990) and also RPN10, a cytosolic ubiquitin binding protein that also functions as a subunit of the 26S proteasome's 19S regulatory subcomplex (Glickman et al., 1998).
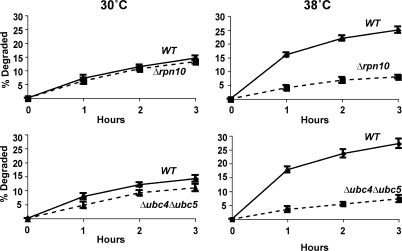
Involvement of these components seemed likely because both Δrpn10 and Δubc4Δubc5 strains have been reported to show growth defects at 38°C and, in the presence of amino acid analogues (Seufert and Jentsch, 1990; Cagney et al., 2001; Elsasser et al., 2004), Δubc4Δubc5 mutant is also sensitive to cadmium (Jungmann et al., 1993). However, at the concentrations of paraquat and cadmium used here or after exposure to 38°C, these mutants grew at similar rates to the WT for several hours. Nevertheless, these mutants were defective in their capacity to degrade the oxidant- and heat-damaged proteins. After a 5-min pulse of 35S-Met, degradation of short-lived and long-lived components at 30°C were not different in the WT and Δrpn10 strains, although degradation in the first hour was consistently slightly lower in the Δubc4Δubc5 double mutant, as was noted previously (Seufert and Jentsch, 1990). However, upon shift to 38°C (Fig. 4) or upon treatment with paraquat or cadmium (Fig. 5 A), both mutants failed to increase degradation of recently synthesized proteins (Figs. 4 and 5 A), which is in sharp contrast to WT cells.
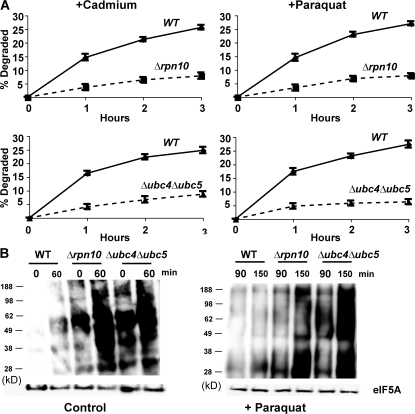
Paraquat and cadmium fail to stimulate the degradation of recently synthesized proteins in Δrpn10 and Δubc4Δubc5 strains and these strains contain higher amounts of oxidant-damaged proteins than WT during growth and after exposure to paraquat. (A) WT and mutants were grown exponentially at 30°C and treated with paraquat or cadmium for 90 min. Then cell proteins were labeled with 35S-Met for 5 min, and rates of degradation measured as in Fig. 2 A. (B) WT and mutants were grown exponentially, and a portion was exposed to paraquat for 90 min. The presence of carbonylated proteins in equal amounts of cell extracts was assayed by OxyBlot as in Fig. 3 A. To visualize oxidant damage in the control cells, approximately three times more protein was loaded than in cells treated with paraquat.
Thus, the selective degradation of oxidant- and thermally damaged proteins appears to involve the same components of the ubiquitin–proteasome pathway required for breakdown of proteins misfolded because of the incorporation of amino acid analogues (Seufert and Jentsch, 1990). Additional experiments using the autophagy-deficient mutant Δatg8 showed that autophagy is not involved in the degradation of heat damaged proteins (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200803022/DC1). This mutant showed a small reduction in breakdown of long-lived proteins which is catalyzed by vacuolar proteases (Lee et al., 1996). Further strong evidence that RPN10, UBC4, and UBC5 are important in the elimination of oxidant-damaged proteins came from analyzing extracts of these mutant cells by OxyBlot (Fig. 5 B). As predicted, the Δrpn10 and Δubc4Δubc5 cells during normal growth showed a higher content of protein carbonyl groups than WT cells, which increased markedly after exposure to paraquat for 1.5 or 2.5 h. These findings confirm that the ubiquitin–proteasome system is important in degradation of proteins damaged by endogenously generated as well as chemically induced ROS (Fig. 5 B).
The CDC48 ATPase and its partners ubiquitin fusion degradation (UFD) 1 and NPL4, but not UBC7 or UFD4, are essential for degradation of damaged proteins
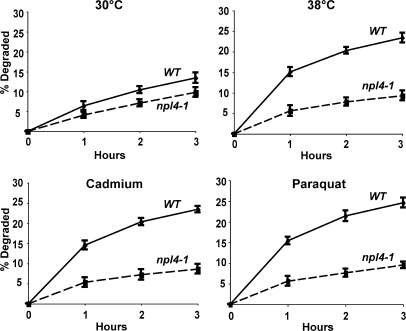
CDC48, a member of the AAA family of hexameric ATPases (the homologue of mammalian p97), and its associated partners UFD1 and NPL4, have been previously implicated in the degradation of misfolded secretory and resident proteins by the ERAD pathway (Ye et al., 2001), as well as certain cytosolic ubiquitin fusion proteins by the UFD pathway (Ghislain et al., 1996). Interestingly, the expression of CDC48 is enhanced by cadmium (Vido et al., 2001). To test if it also functions in this breakdown of oxidant- and thermally damaged cytosolic proteins, we studied their degradation in WT and cdc48-3, ufd1-1, and npl4-1 strains upon shift to 38°C and exposure to paraquat or cadmium. At 30°C, rates of degradation of pulse-labeled proteins were slower in the cdc48-3 (Fig. 6 A), ufd1-1 (Fig. 7), and npl4-1 mutants (Fig. 8) and, upon shift to 38°C or exposure to cadmium and paraquat, a very large defect in the degradation of recently synthesized proteins was evident (Fig. 6 A, Fig. 7, and Fig. 8). However, the degradation of long-lived proteins in these mutants occurred at approximately similar rates as in the WT. Further evidence for an important role of CDC48 and its cofactors in elimination of oxidant-damaged proteins was obtained by OxyBlot analysis on the cdc48-3 mutant. During normal growth, this mutant contained a greater content of oxidant-damaged proteins than did the WT and, after exposure to paraquat for 90 min, the content of protein carbonyls was also much greater than in WT yeast (Fig. 6 B). The mutants of CDC48 or its associated partners UFD1 and NPL4 all showed similar defects in proteolysis, which further supports the conclusions that together they play an important role in the clearance of heat- and ROS-damaged proteins in a process also involving RPN10, UBC4, and UBC5.
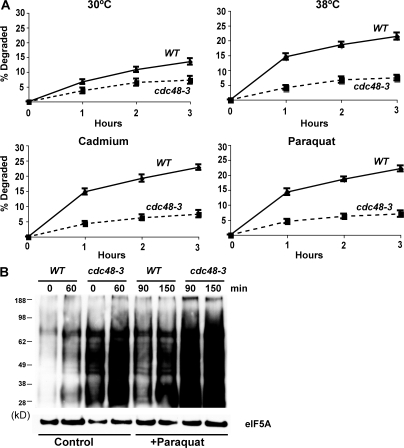
Shifting cells to 38°C and exposure to paraquat or cadmium fail to stimulate the degradation of recently synthesized proteins in the cdc48-3 mutant, and the cdc48-3 strain contains higher amounts of oxidant-damaged proteins than WT during growth and after exposure to paraquat. (A) WT and cdc48-3 strains were labeled with 35S-Met for 5 min at 30°C, and rates of degradation at 30 or 38°C were measured as in Fig. 1 A. To measure the degradation of oxidant-damaged proteins, WT and cdc48-3 strains were exposed to paraquat or cadmium for 90 min, followed by labeling with 35S-Met for 5 min, and rates of degradation were measured as in Fig. 2 A. (B) WT and the cdc48-3 mutant were grown exponentially, and a portion was exposed to paraquat for 90 min. The presence of carbonylated proteins in equal amounts of cell proteins was assayed by OxyBlot as in Fig. 3 A.
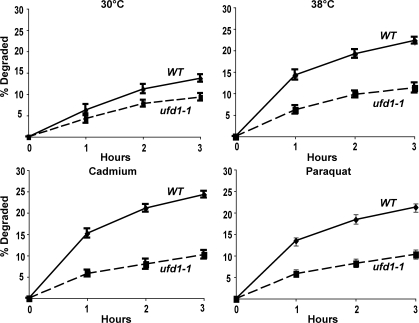
Shifting cells to 38°C and exposure to paraquat or cadmium fail to stimulate the degradation of recently synthesized proteins in the ufd1-1 mutant. WT and ufd1-1 strains were labeled with 35S-Met for 5 min at 30°C, and rates of degradation were measured at 30 or 38°C as in Fig. 1 A. To measure the degradation of oxidant-damaged proteins, WT and ufd1-1 strains were exposed to paraquat or cadmium for 90 min, followed by labeling with 35S-Met for 5 min, and rates of degradation were measured as in Fig. 2 A.
The E2 UBC7 plays an important role together with CDC48, UFD1, and NPL4 in the ubiquitination of misfolded ER resident proteins in the ERAD pathway (Hiller et al., 1996). To verify that the degradation of proteins damaged by heat and oxidative conditions involves proteins in the cytosol and is distinct from ERAD (as was suggested by its requirement for UBC4 and UBC5), we measured rates of degradation in the Δubc7 mutant upon shift to 38°C or exposure to paraquat or cadmium. A similar rapid increase in proteolysis was observed under these conditions in Δubc7 mutant as in WT cells (Fig. 9 A). Similar experiments tested whether the degradation of damaged cytosolic proteins is catalyzed by the UFD pathway. In Δufd4 mutants that lack the E3 ligase essential for the degradation of UFD substrates (Johnson et al., 1995), exposure to 38°C or to paraquat or cadmium caused a similar increase in breakdown of newly synthesized proteins to that in the WT (Fig. 9 B). Thus, this degradative process is distinct from the ERAD and UFD pathways, although all three require the same ATPase complex.
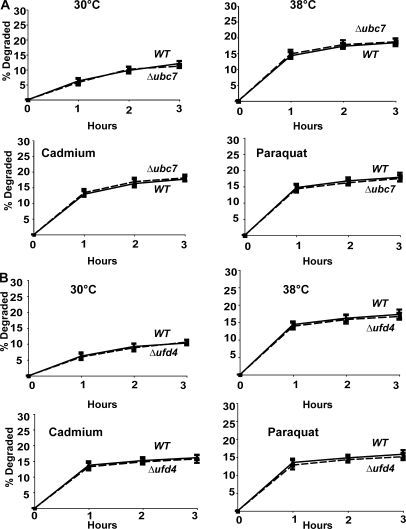
UBC7 and UFD4 are not required for increased degradation at 38°C and after exposure to paraquat or cadmium. (A) WT and Δubc7 strains were labeled with 35S-Met for 5 min at 30°C, and rates of degradation at 38°C and upon exposure to paraquat or cadmium were measured as in Fig. 6 A. (B) WT and Δufd4 strains were labeled with 35S-Met for 5 min at 30°C, and rates of degradation at 38°C and upon exposure to paraquat or cadmium were measured as in Fig. 6 A.
If degradation of damaged proteins is prevented, they accumulate as aggregates
A prominent feature of denatured or unfolded proteins is their tendency to aggregate and, in vivo, many mutated, recombinant, and analogue-containing proteins that fail to fold in the cytosol form intracellular inclusions (Prouty et al., 1975), a predominant feature of various inherited and neurodegenerative diseases (Kopito, 2000). We therefore examined whether the oxidant- or heat-damaged proteins, if they fail to be degraded in the Δrpn10, Δubc4Δubc5, or cdc48-3 strains, tend to aggregate and accumulate in particulate fractions. We analyzed the subcellular distribution of 35S-Met–labeled proteins in WT cells and these degradation-impaired mutants. After shift to 38°C or exposure to the ROS generators, yeast were collected at different times and lysed, and the rapidly sedimenting material was isolated by differential centrifugation. In the Δrpn10, Δubc4Δubc5, and cdc48-3 strains, after shift to 38°C there was a gradual accumulation of radiolabeled proteins in the ultracentrifugal pellets isolated at 275,000 g with time but not in those obtained at low speeds (9,300 g; Fig. 10 A and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200803022/DC1). After 2–3 h, the 35S-Met–labeled protein in the 275,000 g pellets accounted for approximately half of the radioactive protein that would have been degraded in the WT to acid-soluble material (Fig. 10 A).
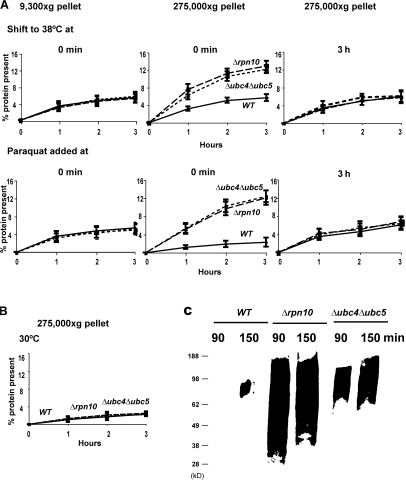
After shift to 38°C or exposure to paraquat to cause oxidant damage, recently synthesized proteins not degraded in Δrpn10 and Δubc4Δubc5 mutants selectively accumulate in the particulate fraction, unlike recently synthesized proteins in WT or mutant strains at 30°C without oxidant present. (A) WT, Δrpn10, and Δubc4Δubc5 strains were labeled with 35S-Met for 5 min, washed, and shifted to 38°C or treated with paraquat at 30°C. At the indicated times, lysates were prepared, and equal amounts of lysate proteins were subjected to differential centrifugation. The amounts of labeled protein present in pellets obtained by centrifuging for 20 min at 9,300 g and by ultracentrifugation at 275,000 g for 60 min were measured. To determine the amounts of labeled long-lived proteins that accumulate in these fractions, cells were shifted to 38°C or exposed to paraquat 3 h after labeling. (B) WT, Δrpn10, and Δubc4Δubc5 strains were labeled with 35S-Met for 5 min at 30°C. The amounts of radiolabeled protein present in 275,000 g pellet were measured as described in Fig. 10 A. (C) Yeast were grown exponentially and treated with paraquat as in Fig. 10 A. Cell lysates were subjected to centrifugation and the presence of carbonylated proteins in the 275,000 g pellet assayed by OxyBlot as in Fig. 3 A.
Very similar evidence for aggregation of the nondegraded oxidant-damaged proteins was obtained by differential centrifugation after exposing these strains to paraquat (Fig. 10 A). In contrast no such accumulation of recently synthesized proteins in their particulate fraction was seen at 30°C or without paraquat (Fig. 10 B). This tendency of the nondegraded proteins to form aggregates supports the conclusion that these proteins are in fact misfolded after thermal or oxidant damage. Further direct support for this conclusion was obtained by analyzing the proteins in these pellets for free-radical damage by OxyBlot. As expected, there were much higher amounts of protein carbonyls in the ultracentrifugal pellets from the Δrpn10 and Δubc4Δubc5 strains than from the WT (Fig. 10 C).
To confirm that the proteins accumulating in this fraction are in fact damaged components that would have been degraded in the WT, we analyzed whether these protein aggregates were formed upon damage of long-lived proteins. The WT and the Δrpn10 and Δubc4Δubc5 strains were labeled and, 3 h later, were shifted to 38°C or treated with paraquat when these treatments fail to enhance degradation. In contrast to the findings with recently synthesized components (Fig. 10 A), administrating these insults 3 h later did not cause an accumulation of long-lived proteins in the ultracentrifugal pellets. This lack of aggregation supports the earlier observations (Figs. 1 B and 2 B) that by this time these treatments did not trigger unfolding and accelerated degradation of long-lived proteins. Together, these observations indicate that for up to 1 h after synthesis, cell proteins are particularly susceptible to denaturation induced by heat-shock or ROS, which leads to their degradation by the ubiquitin pathway. Although ROS do react with long-lived proteins, the modified proteins do not appear to undergo widespread denaturation and aggregation.
Discussion
Postsynthetic damage mainly enhances degradation of newly synthesized proteins
Although it has long been appreciated that cells selectively degrade unfolded proteins (Goldberg and Dice, 1974), the effects of different types of postsynthetic damage on the breakdown of different classes of cell proteins have not been studied previously. The present study focused on the effects of increased or decreased temperatures and oxygen radicals because these insults are encountered in natural environments and in disease. Probably the most surprising of the present findings is that the degradation of the great majority of yeast proteins, the long-lived cell constituents, which are degraded at a rate of 1–2%/h, is not affected significantly by temperature shifts or ROS. Our various pulse-chase experiments demonstrated that only a fraction of newly synthesized proteins are degraded at increased rates upon shift from 30 to 38°C (Fig. 1 A). By 1 h, the degree of stimulation decreased and was no longer statistically significant, and this effect was completely lost by 90 min (Fig. 1 B). Thus, after heat shock or during exposure to free radicals, a larger fraction of the proteins labeled in a 5-min pulse behave like the inherently short-lived components. In yeast growing at 20 or 30°C, such rapidly degraded components comprise 3–4% of newly synthesized proteins, but the short-lived fraction reached 13% (Fig. 1 A) after shift to 38 or 42°C, and 10–13% of the proteins synthesized in the presence of cadmium or paraquat at 30°C (Fig. 2 A). These treatments accelerated the degradation of ~10% of proteins that would otherwise be long lived so that they behaved like short-lived components.
Various in vitro studies suggest that the times for model polypeptides to fold into their characteristic tertiary structures is seconds (or less) to minutes depending on its structural complexity. Many newly synthesized polypeptides undergo covalent modifications and assemble into larger structures including multimeric enzymes, multisubunit complexes, and organelles (Balch et al., 2008). However, there is no information available about the times required for such proteins to assume their mature quarternary structures. These times must vary widely and depend on whether the protein is a component of larger structures, contains cofactors, or is localized in a specific compartment. Because the formation of multimeric complexes may require many different interactions with other subunits and chaperones, these processes should take much more time than polypeptide folding. During this maturational period, these incomplete complexes are likely to be fragile unstable structures and, consequently, their assembly may easily go awry with increased temperature or ROS damage. Although it was initially surprising that some proteins proceed through a fragile period as long as 30–60 min, such a prolonged maturation seems quite reasonable for multimeric proteins undergoing complex multistep assembly processes. It is noteworthy that heat shock and ROS stimulate the degradation of only ~10% of newly synthesized proteins. This fraction includes many different polypeptides because SDS-PAGE of 35S-Met–labeled newly synthesized proteins shows many discrete bands that decreased after shift to 38°C (unpublished data). Identification of this subset of cell proteins will be important to determine whether in fact they tend to be components of large multimeric structures and whether those proteins most susceptible to thermal damage are also the ones most susceptible to ROS.
A corollary of the present findings is that heat shock and oxidative damage do not stimulate proteolysis by causing denaturation of cell proteins generally but, instead, these damaging agents prevent some newly synthesized proteins from assuming their final conformations, leading to their denaturation and rapid degradation. Recently synthesized polypeptides may exist as molten globules (Uversky, 2002) or partially formed complexes in transient associations with chaperones, but they should lack the stabilizing effects of complementary subunits and bound cofactors. These arguments also suggest that the temperature upshift and oxygen radicals are likely to selectively enhance the degradation of proteins that are components of highly complex multimeric structures.
Enhanced degradation of newly synthesized proteins was seen upon increasing the temperature from 30 to 38°C, which is a temperature range often used to elicit the heat-shock response in yeast. This finding fits with previous assertions that this stress response occurs in response to an accumulation of misfolded proteins in the cytosol or nucleus. It seems likely that the heat-shock response in fact evolved to adapt to those temperatures that most disrupt the maturation of newly synthesized proteins. It was also surprising both that a further increase in temperature to 42°C or decrease to 20°C caused no further change in the rates of degradation of the newly synthesized proteins and that rates of degradation of the bulk of cell proteins are remarkably independent of temperature. Unlike short-lived proteins, long-lived proteins in growing yeast are degraded not by the ubiquitin–proteasome pathway but by the vacuolar (autophagic) system (Lee et al., 1996), which showed no change between 20 and 42°C (Fig. 1 A). In contrast, increasing temperatures by 22°C would be expected to stimulate typical chemical reactions six- to ninefold.
Protein damage and degradation induced by superoxide radicals
To examine the effects of ROS, we used paraquat and cadmium, because prior studies in yeast or mammalian cells had shown that these free radical generators cause oxidative damage to polypeptide chains. In addition, cadmium ions are known to cause induction of heat-shock proteins, the ubiquitin-activating enzyme E1, and the proteasome subunit PRE1, as well as CDC48 and SOD1, which we show are essential for the degradation of the damaged newly synthesized proteins (Vido et al., 2001). Although paraquat or cadmium stimulated the degradation of newly synthesized proteins, by 1 h after synthesis, their degradation could no longer be stimulated by these ROS generators.
Endogenous production of superoxide radicals by metabolism also can damage recently synthesized proteins and, as shown in this paper, cause their rapid degradation. The mutant strains lacking SOD1 showed a very similar large increase in their degradation as was induced with paraquat or cadmium (Fig. 3 B). In other words, SOD1, by removing damaging superoxide radicals, seems to enhance the chances that newly synthesized proteins can assume their final conformations. Thus, SOD1, by eliminating this highly reactive species, functions together with the ubiquitin–proteasome pathway to prevent the accumulation of oxidant-damaged polypeptides and to help cells withstand the consequences of increased levels of ROS, as occurs in mammals with neurodegenerative disease and inflammation, as well as during normal aging.
Most likely, these modifications by themselves are not recognized by the degradative system but, instead, these modifications lead to unfolding or irreversible denaturation of the newly synthesized polypeptides. Accordingly, a large fraction of the thermally and oxidant-damaged proteins that were not degraded in the Δubc4Δubc5, Δrpn10, and cdc48-3 mutants accumulated as aggregates in rapidly sedimenting fractions (Fig. 10, A and C; and Fig. S3). These observations also suggest that the oxidant-damaged proteins are recognized by their ability to cause unfolding of newly synthesized proteins. Resistance of the long-lived proteins to ROS-induced degradation is not because they fail to react with the ROS. On the contrary, under conditions where no newly synthesized proteins were present (after exposure to cycloheximide for 2 h), exposure to paraquat caused as much or even more oxidant damage to cell proteins (appearance of carbonyl groups) as in control cells (Fig. 3 A). However, after such oxidant damage, the chemically modified long-lived proteins seem not to denature because in the degradation-deficient mutants, these proteins did not accumulate in the particulate fraction, which is in contrast to the oxidant-damaged newly synthesized proteins which tend to aggregate when not degraded.
Degradation of the damaged proteins requires the ubiquitin–proteasome pathway
A variety of studies on cultured cells, erythrocytes, and cell extracts have shown that oxidant-damaged proteins can be degraded under certain conditions independently of ATP and ubiquitin (Fagan et al., 1986; Davies and Goldberg, 1987; Matthews et al., 1989). This process, like the degradation of some misfolded newly synthesized proteins (Qian et al., 2006), has been attributed to 20S proteasomes (Grune et al., 1996; Shringarpure et al., 2003). Other works, however, have indicated a requirement for ATP (Goldberg and Boches, 1982) and ubiquitin conjugation (Dudek et al., 2005) in the degradation of oxidant-damaged proteins. However, the present findings on intact cells indicate that the degradation of oxidant- and heat-damaged proteins in yeast requires multiple components of the ubiquitin–proteasome pathway and does not occur via autophagy (Fig. S2). Specifically, the increased proteolysis upon exposure to 38°C or ROS requires the E2s UBC4 and UBC5, the ubiquitin binding protein RPN10 (Fig. 4; and Fig. 5, A and B), which exists in the cytosol and also functions in the 26S proteasome as a binding site for ubiquitin chains (Glickman et al., 1998), and a functional CDC48–UFD1–NPL4 complex (Fig. 6, A and B; Fig. 7; and Fig. 8). Each of these components is important in the degradation of other types of abnormal proteins. Mutants of Δubc4Δubc5 (Seufert and Jentsch, 1990) and Δrpn10 (unpublished data) show similar defects in the breakdown of misfolded proteins containing amino acid analogues. CDC48 and its cofactors UFD1-NPL4 are essential in the ERAD pathway for extracting misfolded proteins from the ER (Ye et al., 2001) and in the degradation of certain ubiquitin fusions by the UFD pathway. However, the process described in this paper is distinct from ERAD because it requires a different pair of E2s and not UBC7 (Fig. 9 A; Hiller et al., 1996), and it differs from the UFD pathway because it occurs in mutants lacking the ubiquitin ligase UFD4 (Fig. 9 B; Johnson et al., 1995).
Various in vitro experiments have shown that oxidant damage and unfolding enable polypeptides to enter and be degraded by 20S proteasome (Shringarpure et al., 2003) and also to be substrates for ATP-dependent degradation by 26S proteasomes without ubiquitination (Cascio et al., 2002). However, the efficient clearance of these damaged polypeptides in vivo requires ubiquitination. Though the responsible ubiquitin ligases in yeast are unknown, it seems likely that this process requires molecular chaperones for the recognition of partially folded polypeptides (like the mammalian E3 CHIP; Murata et al., 2001). In fact, we previously showed that the increased proteolysis at 37°C requires molecular chaperones of the Hsp40 family (YDJ1; Lee et al., 1996). It is also noteworthy that the magnitude of the increase in proteolysis was similar after these various insults. These findings suggest that the cell's maximal capacity for proteolysis was reached. In fact, no further increase in degradation was observed when cells were exposed to both paraquat and 38°C simultaneously. In these pulse-chase experiments, we routinely treated the cells with cycloheximide to prevent reutilization of amino acids released by proteolysis, but it also prevents cellular adaptations (e.g., the heat-shock response) that may enhance the cell's capacity to eliminate heat- or oxidant-damaged proteins.
Because molecular chaperones, the heat-shock response, and the ubiquitin–proteasome pathway are highly conserved across eukaryotes, it seems very likely that the present findings apply also to mammalian cells. In humans, the accumulation of carbonyl groups in proteins is commonly used as a criterion of oxidative stress. The failure of such modifications to trigger the degradation of most cell proteins may also explain the accumulation of these damaged proteins in aging or during oxidative stress. The tendency of the damaged short-lived proteins, if not degraded, to aggregate appears reminiscent of the aggregates of ubiquitinated proteins characteristic of many neurodegenerative diseases. Although such structures are generally assumed to result from damage to mature proteins, they may also result from damage and failure to degrade newly synthesized proteins.
Materials and methods
Yeast strains and plasmids
Molecular biological and genetic manipulations of yeast were performed using standard methods (Guthrie and Fink, 2001). S. cerevisiae strains SUB61 (MATα lys2-801, leu2-3, 2–112, ura3-52, his3-Δ200, trp1-1), SUB 453 (MATα, lys2-801, ura3-52, trp1-1, ubc4 HIS3, ubc5
HIS3, ubc5 LEU2), and SY417 (MATα, lys2-801, his3-Δ200, trp1-1, ura3-52, rpn10
LEU2), and SY417 (MATα, lys2-801, his3-Δ200, trp1-1, ura3-52, rpn10 LEU2) were generous gifts of D. Finley (Harvard Medical School, Boston, MA). Strains BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), BY4741Δsod1 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, sod1
LEU2) were generous gifts of D. Finley (Harvard Medical School, Boston, MA). Strains BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), BY4741Δsod1 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, sod1 kanMX4), BY4741Δsod2 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, sod2
kanMX4), BY4741Δsod2 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, sod2 kanMX4), BY4741Δufd4 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ufd4
kanMX4), BY4741Δufd4 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ufd4 kanMX4), BY4741Δubc7 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubc7
kanMX4), BY4741Δubc7 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubc7 kanMX4), and BY4741Δatg8 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, atg8
kanMX4), and BY4741Δatg8 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, atg8 kanMX4) were obtained from the yeast genome deletion library of Research Genetics. Strains YYH46 (MATa, ura3-52, leu2Δ1, trp1Δ63) and YYH1 (MATA, ura3-52, leu2Δ1, trp1Δ63, npl4-1) are described in Hitchcock et al. (2001). Strains YPC1505 (MATa, ura3Δ0, leu2Δ0, his3Δ1, trp1Δ63) and YPC1614 (MATa, ura3Δ0, leu2Δ0, his3Δ1, trp1Δ63, cdc48-3) are described in Ye et al. (2001). Strains YYH3 (MATa, his4-519, ura3-52, ade1-100, leu2-3, 112) and YYH4 (MATa, his4-519, ura3-52, ade1-100, leu2-3, 112, ufd1-1) are described in Johnson et al. (1995). Strains YYH46, YYH1, YPC1505, YPC 1614, YYH3, and YYH4 were provided by T. Rapoport (Harvard Medical School, Boston, MA). Centromeric plasmid expressing SOD1 (pLS101; Sturtz et al. 2001) was provided by V. Culotta (Johns Hopkins School of Public Health, Baltimore, MD).
kanMX4) were obtained from the yeast genome deletion library of Research Genetics. Strains YYH46 (MATa, ura3-52, leu2Δ1, trp1Δ63) and YYH1 (MATA, ura3-52, leu2Δ1, trp1Δ63, npl4-1) are described in Hitchcock et al. (2001). Strains YPC1505 (MATa, ura3Δ0, leu2Δ0, his3Δ1, trp1Δ63) and YPC1614 (MATa, ura3Δ0, leu2Δ0, his3Δ1, trp1Δ63, cdc48-3) are described in Ye et al. (2001). Strains YYH3 (MATa, his4-519, ura3-52, ade1-100, leu2-3, 112) and YYH4 (MATa, his4-519, ura3-52, ade1-100, leu2-3, 112, ufd1-1) are described in Johnson et al. (1995). Strains YYH46, YYH1, YPC1505, YPC 1614, YYH3, and YYH4 were provided by T. Rapoport (Harvard Medical School, Boston, MA). Centromeric plasmid expressing SOD1 (pLS101; Sturtz et al. 2001) was provided by V. Culotta (Johns Hopkins School of Public Health, Baltimore, MD).
Measurements of total protein degradation
Yeast cells were grown exponentially to OD600 0.8–1 at 30°C in SD medium supplemented with all amino acids (Sigma-Aldrich). 10 ml of cells were collected by centrifugation and resuspended in 2 ml of the same media, but without methionone, for 50 min. Then the cells were incubated with 50 μCi/ml 35S-Met for 5 min to label recently synthesized proteins or 90 min to label long-lived proteins. After two washes with ice-cold chase media at 4°C, cells were resuspended in 2 ml chase media with a high concentration of Met (6 mg/ml) and cycloheximide (0.5 mg/ml) to prevent reutilization of 35S-Met. At different time intervals, equal aliquots (450 μl) of cells were taken and mixed with 50 μl of 100% TCA to a final concentration of 10%. After incubation at 4°C overnight, the samples were centrifuged at 14,000 rpm for 10 min at 4°C, and the radioactivity in both the TCA-soluble and -insoluble fractions was measured by suspending the pellets in 4 ml of scintillation fluid (PerkinElmer). The rate of protein degradation is expressed as the percentage of initially incorporated radioactivity converted into acid-soluble fragments each time. The TCA-soluble cpm at time zero was substrated from each subsequent measurement. To measure the effects of temperature shift, cells were shifted to 38, 42, or 20°C during the chase period with chase media preadjusted to the respective temperatures. To measure the effect of 80 μg/ml paraquat or 50 μg/ml cadmium, cells were exposed to these agents for 90 min before the labeling with 35S-Met, except in Fig. 2 B where these agents were added after the pulse of 35S-Met. TCA-soluble and -precipitable cpm for typical experiments (Figs. 1 and and2)2) are shown in Tables S1 and S2 (available at http://www.jcb.org/cgi/content/full/jcb.200803022/DC1).
Detection of carbonyl groups in proteins
To detect the presence of carbonyl groups in proteins, yeast cells were grown exponentially at 30°C in YPD media. 20 ml of cells were then exposed to 80 μg/ml paraquat for 90 min. Cells were collected by centrifugation and lysed with glass beads in the presence of ice-cold sorbitol buffer (700 mM sorbitol, 50 mM Tris-HCl, pH 7.5, and 1 mM PMSF) at 4°C. Cell debris was removed by centrifugation at 2,000 g for 5 min at 4°C. Equal amounts of cell proteins were derivatized to yield 2,4-DNP-hydrazones by incubation with 2,4-DNPH for 15 min at room temperature. By using the OxyBlot detection kit (Millipore), proteins were separated by SDS-polyacrylamide gel electrophoresis, and the derivatized carbonyl groups were detected with an anti–DNP-hydrazone–specific antibody (Millipore).
Measurement of radiolabeled proteins in the ultracentrifugal pellets
Yeast cells were grown exponentially at 30°C, and 20 ml of cells were labeled with 35S-Met for 5 min, washed, and resuspended in chase media containing cycloheximide and high concentrations of Met (as described in Measurements of total protein degradation). Cells were shifted to 38°C or exposed to paraquat, and aliquots were collected at the indicated time points. Lysates were prepared by breaking the cells with glass beads in the presence of ice-cold sorbitol buffer at 4°C. Cell debris was removed by centrifugation at 2,000 g for 5 min. Lysates containing equal amounts of proteins were subjected to differential centrifugation. The amount of radiolabeled proteins present in the pellets was obtained by centrifuging for 20 min at 9,300 g, and after ultracentrifugation at 275,000 g for 60 min, they were measured by suspending the pellets in scintillation fluid. The accumulation of the 35S-Met protein in these fractions was expressed relative to the total amount of the radiolabeled protein at time zero. The presence of carbonyl groups in the 275,000 g pellet was detected by OxyBlot analysis.
Online supplemental material
Fig. S1 A shows that SOD2 is not involved in the degradation of recently synthesized proteins. Fig. S1 B shows that exposure of Δsod1 mutant to cadmium or paraquat further enhances the degradation of recently synthesized proteins. Fig. S2 shows that ATG8, which is essential for autophagy is not required for the increased degradation of short-lived proteins at 38°C. Fig. S3 shows that in cdc48-3 mutants, upon a shift to 38°C or exposure to paraquat or cadmium, recently synthesized proteins that are not degraded accumulate in the high-speed ultracentrifugal pellet. Table S1 shows the TCA-soluble and -insoluble counts for a typical pulse-chase experiment at 30 or 38°C or upon exposure to paraquat or cadmium. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803022/DC1.
Acknowledgments
We thank D. Finley, T. Rapoport, and V. Culotta for providing yeast strains and plasmids, D. Finley, O. Kandror, and J. Brault for helpful suggestions, and M. Dethavong for valuable assistance.
This research was supported by grants from the National Institutes of Health (GM5123-12), the Fund for Innovation from Elan, Inc., and the Ellison Medical Foundation.
Notes
Abbreviations used in this paper: DNPH, DNP-hydrazine; ERAD, ER-associated degradation; Met, methionine; ROS, reactive oxygen species; UFD, ubiquitin fusion degradation; WT, wild type.
References
- Ananthan, J., A.L. Goldberg, and R. Voellmy. 1986. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 232:522–524. [Abstract] [Google Scholar]
- Balch, W.E., R.I. Morimoto, A. Dillin, and J.W. Kelly. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919. [Abstract] [Google Scholar]
- Cagney, G., P. Uetz, and S. Fields. 2001. Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol. Genomics. 7:27–34. [Abstract] [Google Scholar]
- Cascio, P., M. Call, B.M. Petre, T. Walz, and A.L. Goldberg. 2002. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 21:2636–2645. [Europe PMC free article] [Abstract] [Google Scholar]
- Davies, K.J. 1987. Protein damage and degradation by oxygen radicals. I. general aspects. J. Biol. Chem. 262:9895–9901. [Abstract] [Google Scholar]
- Davies, K.J., and A.L. Goldberg. 1987. Proteins damaged by oxygen radicals are rapidly degraded in extracts of red blood cells. J. Biol. Chem. 262:8227–8234. [Abstract] [Google Scholar]
- Dudek, E.J., F. Shang, P. Valverde, Q. Liu, M. Hobbs, and A. Taylor. 2005. Selectivity of the ubiquitin pathway for oxidatively modified proteins: relevance to protein precipitation diseases. FASEB J. 19:1707–1709. [Abstract] [Google Scholar]
- Elsasser, S., D. Chandler-Militello, B. Muller, J. Hanna, and D. Finley. 2004. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279:26817–26822. [Abstract] [Google Scholar]
- Fagan, J.M., L. Waxman, and A.L. Goldberg. 1986. Red blood cells contain a pathway for the degradation of oxidant-damaged hemoglobin that does not require ATP or ubiquitin. J. Biol. Chem. 261:5705–5713. [Abstract] [Google Scholar]
- Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 48:1035–1046. [Abstract] [Google Scholar]
- Ghislain, M., R.J. Dohmen, F. Levy, and A. Varshavsky. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884–4899. [Europe PMC free article] [Abstract] [Google Scholar]
- Glickman, M.H., D.M. Rubin, V.A. Fried, and D. Finley. 1998. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18:3149–3162. [Europe PMC free article] [Abstract] [Google Scholar]
- Goff, S.A., and A.L. Goldberg. 1985. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 41:587–595. [Abstract] [Google Scholar]
- Goldberg, A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. [Abstract] [Google Scholar]
- Goldberg, A.L., and J.F. Dice. 1974. Intracellular protein degradation in mammalian and bacterial cells. Annu. Rev. Biochem. 43:835–869. [Abstract] [Google Scholar]
- Goldberg, A.L., and F.S. Boches. 1982. Oxidized proteins in erythrocytes are rapidly degraded by the adenosine triphosphate-dependent proteolytic system. Science. 215:1107–1109. [Abstract] [Google Scholar]
- Grune, T., T. Reinheckel, and K.J. Davies. 1996. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J. Biol. Chem. 271:15504–15509. [Abstract] [Google Scholar]
- Guthrie, C., and G.R. Fink, editors. 2001. Guide to Yeast Genetics and Molecular Biology (Methods in Enzymology). Academic Press, San Diego, CA. 623 pp.
- Hartl, F.U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 295:1852–1858. [Abstract] [Google Scholar]
- Hiller, M.M., A. Finger, M. Schweiger, and D.H. Wolf. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 273:1725–1728. [Abstract] [Google Scholar]
- Hitchcock, A.L., H. Krebber, S. Frietze, A. Lin, M. Latterich, and P.A. Silver. 2001. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell. 12:3226–3241. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson, E.S., P.C. Ma, I.M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456. [Abstract] [Google Scholar]
- Jungmann, J., H.A. Reins, C. Schobert, and S. Jentsch. 1993. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 361:369–371. [Abstract] [Google Scholar]
- Kopito, R.R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530. [Abstract] [Google Scholar]
- Kostova, Z., and D.H. Wolf. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309–2317. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee, D.H., M.Y. Sherman, and A.L. Goldberg. 1996. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4773–4781. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee, J.H., W.S. Yeo, and J.H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745–1755. [Abstract] [Google Scholar]
- Lindquist, S. 1986. The heat-shock response. Annu. Rev. Biochem. 55:1151–1191. [Abstract] [Google Scholar]
- Matthews, W., J. Driscoll, K. Tanaka, A. Ichihara, and A.L. Goldberg. 1989. Involvement of the proteasome in various degradative processes in mammalian cells. Proc. Natl. Acad. Sci. USA. 86:2597–2601. [Europe PMC free article] [Abstract] [Google Scholar]
- Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133–1138. [Europe PMC free article] [Abstract] [Google Scholar]
- Prouty, W.F., M.J. Karnovsky, and A.L. Goldberg. 1975. Degradation of abnormal proteins in Escherichia coli. Formation of protein inclusions in cells exposed to amino acid analogs. J. Biol. Chem. 250:1112–1122. [Abstract] [Google Scholar]
- Qian, S.B., M.F. Princiotta, J.R. Bennink, and J.W. Yewdell. 2006. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 281:392–400. [Abstract] [Google Scholar]
- Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9:543–550. [Europe PMC free article] [Abstract] [Google Scholar]
- Sherman, M.Y., and A.L. Goldberg. 2001. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 29:15–32. [Abstract] [Google Scholar]
- Shringarpure, R., T. Grune, J. Mehlhase, and K.J. Davies. 2003. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 278:311–318. [Abstract] [Google Scholar]
- Sturtz, L.A., K. Diekert, L.T. Jensen, R. Lill, and V.C. Culotta. 2001. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276:38084–38089. [Abstract] [Google Scholar]
- Thevenod, F., and J.M. Friedmann. 1999. Cadmium-mediated oxidative stress in kidney proximal tubule cells induces degradation of Na+/K(+)-ATPase through proteasomal and endo-/lysosomal proteolytic pathways. FASEB J. 13:1751–1761. [Abstract] [Google Scholar]
- Uversky, V.N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739–756. [Europe PMC free article] [Abstract] [Google Scholar]
- Vido, K., D. Spector, G. Lagniel, S. Lopez, M.B. Toledano, and J. Labarre. 2001. A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276:8469–8474. [Abstract] [Google Scholar]
- Ye, Y., H.H. Meyer, and T.A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. [Abstract] [Google Scholar]
- Yewdell, J.W., L.C. Anton, and J.R. Bennink. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157:1823–1826. [Abstract] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.200803022
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jcb/article-pdf/182/4/663/1338166/jcb_200803022.pdf
Free to read at intl.jcb.org
http://intl.jcb.org/cgi/content/abstract/182/4/663
Free after 6 months at intl.jcb.org
http://intl.jcb.org/cgi/content/full/182/4/663
Free after 6 months at intl.jcb.org
http://intl.jcb.org/cgi/reprint/182/4/663.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102644846
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1083/jcb.200803022
Article citations
TDP43 aggregation at ER-exit sites impairs ER-to-Golgi transport.
Nat Commun, 15(1):9026, 19 Oct 2024
Cited by: 0 articles | PMID: 39424779 | PMCID: PMC11489672
Exploring the origins of neurodevelopmental proteasomopathies associated with cardiac malformations: are neural crest cells central to certain pathological mechanisms?
Front Cell Dev Biol, 12:1370905, 12 Jul 2024
Cited by: 0 articles | PMID: 39071803 | PMCID: PMC11272537
Tetrahydrobiopterin metabolism attenuates ROS generation and radiosensitivity through LDHA S-nitrosylation: novel insight into radiogenic lung injury.
Exp Mol Med, 56(5):1107-1122, 01 May 2024
Cited by: 1 article | PMID: 38689083 | PMCID: PMC11148139
The ubiquitin codes in cellular stress responses.
Protein Cell, 15(3):157-190, 01 Feb 2024
Cited by: 12 articles | PMID: 37470788 | PMCID: PMC10903993
Review Free full text in Europe PMC
Proteostasis is differentially modulated by inhibition of translation initiation or elongation.
Elife, 12:e76465, 05 Oct 2023
Cited by: 0 articles | PMID: 37795690 | PMCID: PMC10581687
Go to all (118) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins.
Genetics, 171(4):1477-1484, 22 Aug 2005
Cited by: 25 articles | PMID: 16118187 | PMCID: PMC1456077
The Cdc48-Ufd1-Npl4 complex is central in ubiquitin-proteasome triggered catabolite degradation of fructose-1,6-bisphosphatase.
Biochem Biophys Res Commun, 394(2):335-341, 04 Mar 2010
Cited by: 10 articles | PMID: 20206597
A protein quality control pathway at the mitochondrial outer membrane.
Elife, 9:e51065, 02 Mar 2020
Cited by: 26 articles | PMID: 32118579 | PMCID: PMC7136024
Cdc48-Ufd1-Npl4: stuck in the middle with Ub.
Curr Biol, 12(10):R366-71, 01 May 2002
Cited by: 100 articles | PMID: 12015140
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: R01 GM051923
Grant ID: GM5123-12
Grant ID: R01 GM051923-13