Abstract
Free full text

Protein Synthesis by Platelets: Historical and New Perspectives
Abstract
In the late 1960’s numerous investigators independently demonstrated that platelets are capable of synthesizing proteins. Studies continued at a steady pace over the next 30 years and into the 21st century. Collectively, these investigations confirmed that platelets synthesize proteins and that the pattern of protein synthesis changes in response to cellular activation. More recent studies have characterized the mechanisms by which platelets synthesize proteins and have shown that protein synthesis alters the phenotype and functions of platelets. Here, we chronologically review our increased understanding of protein synthetic responses in platelets and discuss how the field may evolve over the next decade.
“Seek simplicity and distrust it”
Alfred North Whitehead“Is there any thing whereof it may be said, See, this is new?”
Ecclesiastes 1:10
I. Early Studies of Protein Synthesis by Platelets
The blood platelet was discovered by Bizzozero in the late 19th century [1, 2] and shortly thereafter Wright determined that megakaryocytes are platelet precursors [3]. The elegant studies of Bizzozero and Wright were critical for assigning hemostatic roles to platelets and understanding the roots of platelet production [1-5]. These early studies also described the essential anatomy of the blood platelet, which is unique and deceptively simple. One of the key features of platelets is that they circulate without a nucleus. Because they lack nuclei, platelets were considered incapable of regulated gene expression and protein synthesis.
In 1966, however, Andrew Warshaw and his colleagues [6] demonstrated that platelets incorporate 14C-leucine as they oxidize glucose. They also found that puromycin, a protein synthesis inhibitor that blocks mRNA translation [7], reduced incorporation of 14C-leucine providing the first suggestion that anucleate platelets synthesize protein. This initial study was not only compelling in regards to the synthetic potential of platelets, but the experiments were rigorously performed and thoughtfully interpreted. The investigators determined that the average erythrocyte and leukocyte contamination was less than 1 cell per 3,000 platelets, contributing no more than 0.1% of the observed radiochemical yield in their experiments [6]. Warshaw and his group were also among the first to study the functions of platelets for extended periods of time. The premise of their study was that clot retraction occurs over hours and requires energy derived from glucose. Using an in vitro model of clot retraction they determined that thrombin induces glucose oxidation in platelets that lasts for at least 8 hours and is lessened, but not obliterated, when protein synthesis is inhibited [7]. Just a little over 40 years later, we found that protein synthesis regulates platelet-dependent clot retraction, validating their initial results [8].
A year after their initial discovery Warshaw’s group conducted a more focused study that, in their view, constituted the first definitive demonstration that mammalian platelets synthesize protein [9]. At roughly the same time, Francois Booyse and Max Rafelson Jr. published two articles demonstrating that platelets incorporate amino acids into contractile proteins [10, 11]. These investigators also concluded that platelets use stable messenger RNA (mRNA) transcripts to synthesize protein and speculated that the stability of mRNAs directing protein synthesis may determine the lifespan of the platelet [11]. Numerous studies ensued over the next five years (1968-1973) addressing whether platelets synthesize protein [12-23]. Two independent groups developed cell-free systems to demonstrate that platelets contain ribosomes and other constituents necessary for protein synthesis [14, 21]. There was also considerable effort to separate platelets into different populations and to identify a relationship between age and synthetic potential. Data from these studies suggested that large platelets, which were presumably younger cells, have the greatest synthetic potential [15, 19] and ultrastructural analyses identified rough endoplasmic reticulum and ribosomes in platelets after the induction of thrombocytopenia [24].
II. The Next 30 Years Provided More Evidence that Platelets Synthesize Proteins
Although several studies independently concluded that platelets synthesize protein, the physiological significance of this process was not clear. There were questions regarding the magnitude of protein synthesis, the contributions of leukocyte contaminants, and whether protein production was confined to mitochondria [16, 25]. In addition, it was hard to envision how protein synthesis controlled the function of platelets especially since aggregation, which was avidly studied at that time, was considered a terminal event [25]. Thus, after the initial burst of studies in the late 1960’s published observations became less frequent, but nonetheless remained steady over the next 30 years.
A recurrent theme in the 1970’s and 1980’s was that investigators were able to reproducibly demonstrate that platelets incorporate amino acids into protein [26-40]. The source and specificity of protein synthesis was confirmed with several types of classic translation inhibitors [26, 27, 32, 33, 35] and one study demonstrated that an extract from oriental hornet venom blocked protein synthesis by platelets [34]. Protein synthesis by platelets was shown to be influenced when platelets were exposed to extracellular factors [30], cigarette smoke [40], or during phagocytosis of foreign particles [28]. Agam and colleagues [41] also observed that platelets transcribe RNA, confirming an earlier report of DNA-dependent synthesis of RNA in platelets [20], which presumably occurred in mitochondria. In addition, Agam’s group observed DNA synthesis in platelets [41], a finding that was subsequently confirmed by Gerald Soslau in 1983 [42].
For the majority of studies described above, the index for protein synthesis was incorporation of radiolabelled amino acids into trichloroacetic-acid-precipitable material. This allowed for global assessment of protein synthesis but did not identify the types of proteins synthesized by platelets. In 1987, however, Kieffer et al. [38] separated proteins by electrophoresis and demonstrated that several of the proteins stained by Coomassie blue in newly-formed platelets isolated from splenectomized patients with idiopathic thrombocytopenic purpura (ITP) also incorporated radiolabelled amino acids. The pattern of protein synthesis was nearly identical, but reduced, in circulating platelets isolated from normal subjects. Using crossed-immunoelectrophoresis Kieffer’s group concluded that platelets synthesize GPIb, αIIbβ3, fibrinogen, thrombospondin, albumin, von Willebrand factor, various contractile proteins, HLA and coagulation factor XIIIa. Shortly thereafter, Francis Belloc and colleagues [36] provided definitive evidence that platelets synthesize and assemble the different subunits of thrombospondin and fibrinogen. Their results also indicated that normal and Glanzmann’s Thrombastenia platelets retain the capacity to synthesize fibrinogen, but in thrombastenia there is a defect in storage of the newly synthesized protein [36]. Athough these studies demonstrated that platelets synthesize additional amounts of constitutively expressed proteins, the biologic significance of this response in regards to platelet function was not resolved.
In 1988 Peter Newman’s group used polymerase chain reaction (PCR) to demonstrate that messenger RNA (mRNA) resides in platelets [43]. Although presumed by other investigators, this pivotal result was the first definitive evidence that platelets expressed mRNA templates necessary for protein synthesis [43]. Santoso and associates [44] subsequently used this technology to demonstrate that platelets express mRNA for HLA class I. They also found that platelets translate this mRNA into protein [44], confirming the earlier predictions of Nelly Kieffer’s group that platelets synthesize HLA [38]. Two subsequent studies demonstrated that environmental signals facilitate translational responses in platelets. Singh and Kaul [45] found that exogenous cholesterol induced the synthesis of Receptor-Ck protein, which was associated with the messenger ribonucleoprotein (mRNP) pool of platelets [45]. In the same year, Lemaitre and colleagues [46] found that n-3 polyunsaturated fatty acids increased the expression of glutathione-dependent peroxidase (GPx) protein and activity in platelets, responses that were abolished by the translational inhibitor cycloheximide.
III. The Last Decade: What Has It Revealed Regarding Protein Synthesis by Platelets?
Surprisingly, nearly 40 peer-reviewed manuscripts were published between 1966 and 1997 that together demonstrated anucleate platelets synthesize protein. These studies provided the framework for investigations during the last decade, which identified new proteins under synthetic control and characterized previously-unrecognized mechanisms used by activated platelets to synthesize proteins.
In 1998, we found that activated platelets translate B-cell lymphoma 3 (Bcl-3) mRNA into protein [47]. After its synthesis, Bcl-3 binds the SH3 domain of Fyn and regulates platelet-dependent clot retraction [8, 47]. Bcl-3 synthesis is dramatically enhanced by engagement of αIIbβ3 integrins, which regulate the intracellular distribution of mRNAs and the mRNA-binding protein eukaryotic initiation factor 4E (eIF4E) within platelets [48, 49]. Bcl-3 synthesis also requires signaling through the mammalian Target of Rapamycin (mTOR) [8, 47, 49]. Platelets express high levels of mTOR and each of its downstream targets, S6K1 and 4E-BP1 (eIF4E-binding protein-1) [8, 48]. The mTOR inhibitor, rapamycin, specifically blocks activation-dependent phosphorylation of S6K1 and 4E-BP1 and as a result controls the translation of a subset of mRNAs in platelets that include Bcl-3 [8, 48]. Studies from Evangelista and colleagues [50] also imply that mTOR controls de novo synthesis of cyclooxygenase-1 (COX-1) in platelets.
Earlier studies from independent laboratories demonstrated that activated, but not resting, platelets release and/or express interleukin-1β (IL-1β) [51-54]. This led us to the discovery that platelets synthesize IL-1β protein [55]. Anucleate platelets regulate IL-1β synthesis by splicing pre-mRNA [56], a novel mechanism of post-transcriptional signaling that has subsequently been observed in the cytoplasm of dendrites and proliferating fibroblasts [57, 58]. Spliceosome factors and IL-1β pre-mRNA are transferred from megakaryocytes to platelets (figure 1). Thrombin, lipopolysaccharide or clustering of FcαR1 induces IL-1β pre-mRNA splicing in platelets [56, 59, 60]. The mature transcript is subsequently translated into IL-1β protein [55, 56, 59, 60], which increases the adhesiveness of endothelial cells for polymorphonuclear leukocytes [55]. The majority of newly-synthesized IL-1β is retained in the platelet [55], a finding that was recently confirmed by Shashkin and colleagues [60] in platelet preparations that were filtered through a 5-μm mesh and then passed twice through columns to deplete CD14, CD15 and CD45 positive leukocytes. The tendency of platelets to accumulate intracellular IL-1β, rather than actively secrete it, may partially explain why a recent report did not detect mature IL-1β protein in platelet releasates [61].
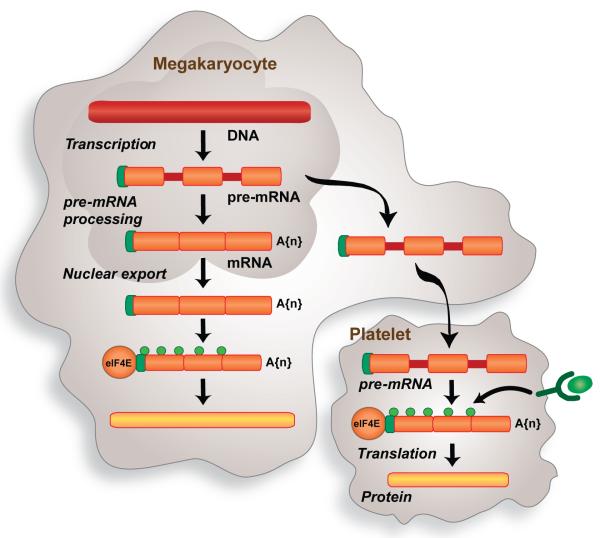
During the final stages of differentiation, splicing factors accumulate in the cytoplasm of megakaryocytes. The splicing components, along with specific pre-mRNAs, are transferred to anucleate platelets. Stimulated platelets activate their splicing machinery and generate mature mRNAs that are subsequently translated into protein. This novel pathway of control provides platelets with a mechanism to alter their transcriptome and proteome profile in response to cellular activation.
Activation-dependent splicing of IL-1β pre-mRNA sparked the search for other candidates, yielding tissue factor (TF) as a gene under a similar type of regulatory control [62]. TF is a critical procoagulant protein that is involved in the initiation and propagation stages of clot formation [63]. TF pre-mRNA splicing is controlled by Cdc2-like kinase 1 (Clk1), which induces phosphorylation of splicing factor 2 (SF2) in platelets [62]. Inhibition of Clk1 in platelets prevents TF pre-mRNA from being spliced and as a consequence, delays the onset of plasma clot formation [62]. A recent independent report demonstrates that ligation of the human IgA receptor FcαR1 on platelets induces TF pre-mRNA splicing and protein production [59]. Panes and colleagues [64] also observed varying expression of mature TF mRNA in platelets and confirmed that activated human platelets synthesize TF protein. The contribution of platelet-derived TF to in vivo clot formation and resolution, however, is not yet known [65].
Several independent investigations have also reported that platelets synthesize other proteins. Brogren and colleagues [66] recently demonstrated that platelets constitutively synthesize plasminogen activator inhibitor-1 (PAI-1), a response that is markedly increased in the presence of thrombin. A follow-up study from the same group suggests that PAI-1 synthesis by platelets is the primary source of plasma PAI because platelet-derived and plasma PAI-1 possess identical glycosylation patterns [67]. In addition, the authors demonstrated that the amount of PAI synthesized in vitro by platelets far exceeds what would be required to maintain normal plasma levels of PAI-1 [66, 67]. Platelets also compensate for fluctuations in intracellular ascorbate levels by modulating the expression of the Na+-dependent transporter SVCT2 at the translational level [68]. Depletion of ascorbic acid levels enhances SVCT2 synthesis by platelets and markedly reduces thrombus rigidity [68]. Evangelista and colleagues [50] also found that aspirin-treated platelets recover their capacity to generate thromboxane A2 through mechanisms that involve de novo synthesis of COX-1. COX-2 pre-mRNA splicing and expression of its protein has likewise been reported in platelets, but the significance of COX-2 protein in regards to arachidonate metabolism has not been established [60]. Circumstantial evidence also suggests that translational control mechanisms regulate tetrahydrobiopterin biosynthesis in platelets [69] and metabolic labeling experiments indicate that activated platelets synthesize many additional proteins, most of which are yet to be identified [47, 55, 70].
IV. Moving Forward on the Protein Synthetic Front
The findings to date demonstrate that platelets have developed extranuclear mechanisms to process and efficiently translate mRNAs into protein. Specialized post-transcriptional and translational mechanisms are now considered part of the platelet functional repertoire. It is also likely that additional new pathways and mechanisms that regulate protein synthesis will be discovered in the near future. Among these are cytoplasmic polyadenylation elements (CPEs), nucleotide sequences embedded in the 3′-untranslated region (3′-UTR) that facilitate translation by extending the length of the poly(A)-tail [71]. Comprehensive SAGE (serial analysis of gene expression) indicates that the 3′-UTRs of platelet mRNAs tend to be longer and more complex when compared to mRNAs of nucleated cells [72]. Platelet-derived mRNAs are also enriched for CPEs and Dittrich and colleagues [72] have identified several highly expressed transcripts with CPE elements whose corresponding proteins have not been previously identified or characterized in platelets. CPE-like elements are present in the 3′-UTR of inducible poly(A)-binding protein (iPABP), a protein that was previously detected in activated platelets by Houng and colleagues [73].
Other control elements, such as AU-rich sequences and Brd boxes, are also commonplace in platelet mRNAs [72]. Given the complexity, stability and abundance (~4000-6000 transcripts) of their transcriptome [55, 72, 74-78], it is attractive to predict that platelets use a variety of translational and post-translational mechanisms to fine-tune their protein profile once they enter the circulation. Our group has focused on signal-dependent translation [7, 79], but there is also recent evidence that constitutive protein synthesis occurs in platelets. Rosenwald and colleagues [80] found that platelets constitutively express eIF4E and also eIF-2α, a rate-limiting protein responsible for the transfer of initiator methionyl-tRNA to the 40S subunit, and that cooperatively these two translation initiation factors facilitate continual protein synthesis in stored platelets. A recent study by Thon and Devine [81] clearly demonstrates that platelets use their translational apparatus to synthesize integrin αIIbβ3 during storage. Full-length mRNA for αIIbβ3 was present throughout a 10-day storage period and translation of the message into protein was demonstrated by incorporation of [35S]-methionine into sequence-confirmed immunoprecipitated αIIbβ3 protein. These studies [80, 81], along with previous ones [36, 38], demonstrate that platelets have the capacity to continually synthesize proteins during their short lifespan. Whether or not constitutive protein synthesis is required to maintain threshold concentrations of essential platelet proteins, however, is not known.
Protein synthesis by platelets was described over 40 years ago, a mechanism of control that has been advanced by our group and others. Platelets synthesize numerous proteins, and several lines of evidence indicate that others will be discovered in the future [79]. There is also much to be learned regarding signaling intermediates that control pre-mRNA splicing, mRNA translation and the biologic significance of protein synthesis in this anucleate cell. Questions of this nature will occupy investigators for years to come and will undoubtedly reveal fascinating new discoveries about platelets, an anucleate cell that continues to defy conventional logic and preconceived notions.
Acknowledgements
The authors thank Diana Lim for preparing the figures and the trainees and collaborators who contributed to work cited in this review. We are also indebted to the funding agencies that have supported our work over the years, especially the American Heart Association, Deutsche Forschungsgemeinschaft, and the National Institutes of Health.
References
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1538-7836.2008.03211.x
Read article for free, from open access legal sources, via Unpaywall:
http://www.jthjournal.org/article/S1538783622106306/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/j.1538-7836.2008.03211.x
Article citations
Metabolic landscape in venous thrombosis: insights into molecular biology and therapeutic implications.
Ann Med, 56(1):2401112, 19 Sep 2024
Cited by: 0 articles | PMID: 39297312 | PMCID: PMC11413966
Review Free full text in Europe PMC
Western Diet Modifies Platelet Activation Profiles in Male Mice.
Int J Mol Sci, 25(15):8019, 23 Jul 2024
Cited by: 0 articles | PMID: 39125586 | PMCID: PMC11311362
Platelets, plasma, and proteostasis: a translation tightrope.
Blood Adv, 8(6):1567-1569, 01 Mar 2024
Cited by: 1 article | PMID: 38530305 | PMCID: PMC10982960
Supernatant of platelet-Klebsiella pneumoniae coculture induces apoptosis-like death in Klebsiella pneumoniae.
Microbiol Spectr, 12(3):e0127923, 30 Jan 2024
Cited by: 0 articles | PMID: 38289116 | PMCID: PMC10913751
Genetically engineered transfusable platelets using mRNA lipid nanoparticles.
Sci Adv, 9(48):eadi0508, 01 Dec 2023
Cited by: 3 articles | PMID: 38039367 | PMCID: PMC10691771
Go to all (152) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[The synthesis of proteins in unnucleated blood platelets].
Postepy Hig Med Dosw (Online), 67:672-679, 23 Jul 2013
Cited by: 7 articles | PMID: 24018431
Review
Historical perspective and future directions in platelet research.
J Thromb Haemost, 9 Suppl 1:374-395, 01 Jul 2011
Cited by: 51 articles | PMID: 21781274 | PMCID: PMC3163479
Review Free full text in Europe PMC
Signal-dependent protein synthesis by activated platelets: new pathways to altered phenotype and function.
Arterioscler Thromb Vasc Biol, 28(3):s17-24, 01 Mar 2008
Cited by: 121 articles | PMID: 18296586 | PMCID: PMC2594008
Review Free full text in Europe PMC
Forty five years with membrane phospholipids, phospholipases and lipid mediators: A historical perspective.
Biochimie, 125:234-249, 05 Apr 2016
Cited by: 19 articles | PMID: 27059515
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (11)
Grant ID: R01 HL090870-01
Grant ID: R01 HL092746
Grant ID: R01 HL091754
Grant ID: R21 HL091283-01
Grant ID: R01 HL066277-09
Grant ID: R29 HL056713-04
Grant ID: R01 HL090870
Grant ID: R21 HL091283
Grant ID: R01 HL091754-02
Grant ID: R01 HL066277
Grant ID: R01 HL092746-03





