Abstract
Free full text

The Rat Genome Database 2009: variation, ontologies and pathways
Abstract
The Rat Genome Database (RGD, http://rgd.mcw.edu) was developed to provide a core resource for rat researchers combining genetic, genomic, pathway, phenotype and strain information with a focus on disease. RGD users are provided with access to structured and curated data from the molecular level through to the level of the whole organism, including the variations associated with disease phenotypes. To fully support use of the rat as a translational model for biological systems and human disease, RGD continues to curate these datasets while enhancing and developing tools to allow efficient and effective access to the data in a variety of formats including linear genome viewers, pathway diagrams and biological ontologies. To support pathophysiological analysis of data, RGD Disease Portals provide an entryway to integrated gene, QTL and strain data specific to a particular disease. In addition to tool and content development and maintenance, RGD promotes rat research and provides user education by creating and disseminating tutorials on the curated datasets, submission processes, and tools available at RGD. By curating, storing, integrating, visualizing and promoting rat data, RGD ensures that the investment made into rat genomics and genetics can be leveraged by all interested investigators.
INTRODUCTION
The Rat Genome Database (RGD, http://rgd.mcw.edu) is the model organism database for the laboratory rat, Rattus norvegicus. The rat is a widely studied animal model for physiology, pharmacology, toxicology, nutrition, behavior, immunology and neoplasia (1). The biological relevance and wealth of phenotypic data that exists for the rat, when combined with genomic resources, provide the opportunity to accelerate the development of new diagnostic, preventative and treatment approaches in each of these fields. The primary goal of RGD is to provide support for researchers using the rat as a model organism in which to understand human health and disease through disease-oriented translational research.
In order to fulfill this goal, RGD provides a comprehensive, curated and integrated system for exploring genomic, genetic and phenotypic data across the many strains and substrains of rats. Core genomic and genetic datasets, including sequences, genes, mapping information, strains and quantitative trait loci, are acquired from a variety of sources (literature, other scientific databases and via local bioinformatic analyses).
Access to these datasets is provided using a variety of internally developed or externally developed and modified tools. These tools provide access to data within a genomic context, in a genome viewer or browser, as well as through report pages, which provide comprehensive, curated and updated reports on the specific feature queried, with links to a wealth of supplementary information including orthologs, references and physical interactions. In addition, RGD has recently begun curating and adding interactive pathway diagrams, providing links to detailed, expert curated information on the proteins involved in the pathways, the relationships between them, small molecules interactions, and links to related pathways.
RGD places significant focus on integration between core genetic and genomic datasets and phenotypic or pathogenic disease data. In order to simplify access to disease-associated datasets, RGD created a number of disease portals; entry points for researchers to access data related to a disease of interest, and links to tools displaying the genomic and genetic context of the data. To complement these disease portals, a Phenotypes and Models Portal was created at RGD to house strain and phenotype data along with tools to integrate the genomic and phenotypic datasets. Integration of large phenotype datasets from many rat strains is based on implementation of a variety of biological ontologies; more complex data structures are used to support integration of multiple ontology annotations, as well as qualifiers and actual values.
DATA ACCESS AND DATABASE STRUCTURE
The public website provides convenient access to this data through recently updated search tools, web page reports on genes, strains, QTLs and ontologies, and via specialized analysis and visualization tools. RGD data available via the website is updated daily. In addition, data curated at RGD is made available through other global genomics resources such as NCBI Ensembl and UCSC. Selected larger datasets are made available to high-throughput users via FTP.
RGD is built and deployed on a Sun Solaris platform. The website is powered by a content management system called SimpleCMS. Dynamic content is delivered through Java and Perl applications deployed on an Apache Web Server and Tomcat Servlet Container with an Oracle 10g database. Data pipelines were built as Java applications using the Spring framework.
RGD CORE GENOMIC DATASETS
RGD continues to acquire and integrate core genomic datasets (e.g. gene models, QTL, sequence and map data). The last year has seen significant growth in all the core curated datasets (Table 1).
Table 1.
RGD Core Genomic Data
| Category | Count |
|---|---|
| Total Rat Genes | 36317 |
    Known genes Known genes | 20428 |
    Pseudogenes Pseudogenes | 7963 |
    Splice variants Splice variants | 117 |
| QTLs | 1365 |
| Strains | 1441 |
| SSLPs | 12875 |
| References | 24813 |
Automated pipelines, manual procedures and submissions from the research community provide RGD users with the most complete catalogue of rat genomic, genetic and protein information available. The rat gene catalogue and human and mouse homolog data is updated via weekly automated EntrezGene pipelines. An automated pipeline updated ortholog relationships, and a nomenclature pipeline, which compares symbols and names among the rat, human and mouse, alerts curators to naming discrepancies. Various other automated pipelines, for example the Microarray probe set identifiers for Affymetrix, and Agilent run daily or weekly to ensure presentation of the latest data.
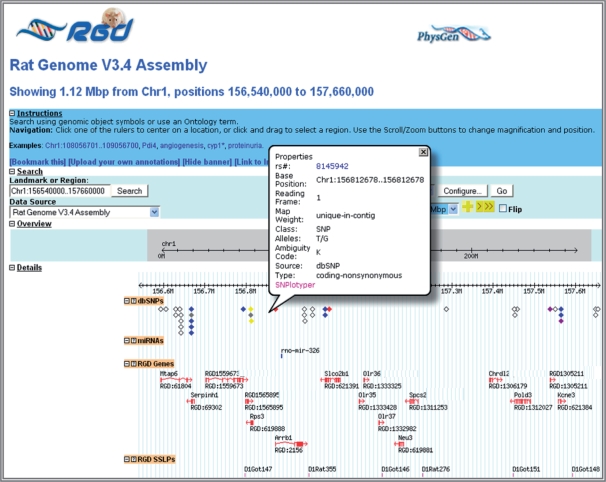
Within the last year, additional core genomic datasets have been incorporated into RGD. As in human and other model organisms, SNPs are becoming the dominant genetic marker in the rat. SNPs have been incorporated into RGD using two tools. The RGD Genome Browser (GBrowse), implemented using the highly successful Generic Model Organism System Database Project (GMOD) generic genome browser (2), offers SNP tracks for SNPs catalogued at NCBI's dbSNP and Ensemble (Figure 1), and SNPs from the STAR project (3) have been incorporated into the RGD SNPlotyper tool for haplotype analysis (Figure 1). Advances in the methodology for miRNA gene prediction have led to an expansion in the number of miRNA gene annotations in many organisms including the rat. Rather than curating these genes ourselves, RGD obtains the core annotations from miRBase, the Sanger Institute's repository for published miRNA sequences and annotation (4), and provide this data as a track in Gbrowse with links back to miRBase. In addition to miRNAs, RGD has incorporated a GBrowse track depicting and providing annotation on a variety of additional classes of rat ncRNAs, including small nucleolar (sno)RNAs, and small nuclear (sn)RNAs, sourced from the non-coding database NONCODE (5).
TOOL DEVELOPMENT
The volume and complexity of genomic data currently available for the rat poses a major challenge for exploration and visualization by biomedical researchers. In the last year, RGD has made enhancements to a number of tools to support and simplify researcher's exploration of these datasets. Improved search and display tools, extended ontologies and increased interoperability between existing tools are some of the features that will help scientists streamline their research. These changes have, in large part, been guided by the desire to identify and use rat strain data in the study of disease traits, with subsequent translation of this information to humans both for disease prevention and drug targeting.
SEARCH
RGD released a new site search in August 2008, allowing users to find objects in RGD based on keyword or position information. Over 8.5 million terms within RGD have been ranked and indexed to return genes, QTLs, strains, markers and references housed at RGD. Search results are presented in an easy-to-use report format that includes position information along with additional fields specific to the genomic object type. Genomic objects have been indexed for the rat, mouse and human to allow for comparison between species. In addition, ontology term matches in the Gene (GO), Disease, Pathway and Mammalian Phenotype ontologies are returned by each search. This new site search provides the ability to search on multiple terms, as well as exclusion of common words, stemming, phrase searches and negative terms. All data in RGD is indexed and ranked to ensure correct positioning of objects in the search result list. Customization to a specific assembly is available by selecting one of the assembly maps available for each species. Search results can be exported by downloading results as a CSV or tab delimited file and viewed on the RGD GViewer using a species-specific map.
GENOME BROWSER
The RGD GBrowse has undergone a variety of enhancements in the last year, achieved in part through implementation of GBrowse version 1.69 (2). Features in this enhanced version can be found at http://gmod.org/wiki/GMOD_News. The RGD GBrowse now displays popup balloons when the user hovers over a genomic object; these are used to provide object and track specific summarized annotation and links to other internal and external applications (Figure 1). Within a number of tracks, we have provided color coding based on the genic/genomic location of the object; this allows users, for example, to scan the genome and differentiate between coding and intergenic SNPs (Figure 1).
Additional SNP visualizations are available from GBrowse by following links to the RGD SNPlotyper tool (Figure 1). Having identified a SNP or region of interest through examination of annotation and genomic context in GBrowse, RGD users can investigate the genotype of specific rat strains of interest in this region by selecting and sending the region in question to the SNPlotyper tool which returns a visualization of the genotype across selected commonly used strains.
PATHWAYS
As part of the continuous effort to add new dimensions and depth to the pathway ontologies and associated data, RGD has begun publishing interactive diagrams for selected pathways using the Pathway Studio software developed by Ariadne Genomics (6). The diagrams offer an instant ‘snapshot’ of a pathway's components and the relationships between them (Figure 2) and include direct links to RGD gene report pages. In addition to the links to the RGD gene reports, a brief overview of each pathway is provided as well as a link to the pathways ontology report including the complete list of annotated genes involved in the pathway and the ability to download this information. As applicable, the diagrams also point to downstream activated pathways providing the ability to ‘walk’ from one pathway to another. Direct links to pathways activated during disease states are available from the disease portals.
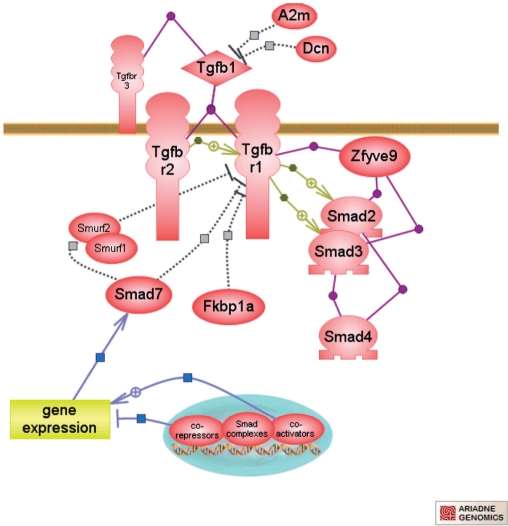
New interactive pathways available at RGD include the transforming growth factor-beta superfamily mediated pathway.
The interactive pathway diagrams have been built using the pathway ontology developed at RGD. This ontology was created to provide: (a) a means to standardized annotations of genes to pathway terms, including altered and disease pathways and (b) as a platform to illustrate the relationships and dependencies between pathways. Pathway data from multiple sources (KEGG, Reactome, MetaCyc, GenMAPP) and data as described in the literature have been incorporated in the ontology and integrated into its hierarchical structure. Pathways of relevance for a particular Disease Portal are selected for full curation. Review articles for given pathways are used to identify the key components and annotate the respective genes.
DISEASE PORTALS
To facilitate and guide efficient access for researchers focusing on complex diseases, a number of disease portals for a variety of classes of disease have been created: Neurophysiologic, Cardiovascular, Obesity and Metabolic Syndrome, and Cancer. Most recently released is the Cancer Disease Portal, which initially focuses on Breast and Urogenital Cancers. Here, the user is able to view gene, QTL and strain data for all Breast and/or Urogenital Cancers or refine their selection further to a specific disease or class of tumor. The Cancer Portal provides access to data on phenotypes, biological processes and pathways, as well as to Strain Report pages detailing rat strains associated with the disease.
INTEGRATION OF PHYSIOLOGY DATA
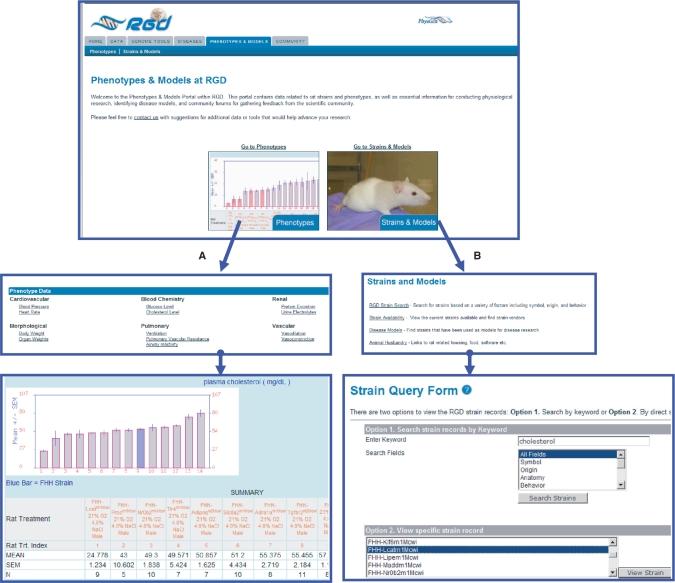
The Phenotype and Models portal at RGD, released in April 2008, was designed to provide an entryway to physiological data and disease model information for researchers using rat as a model to study physiological function and human disease (Figure 3). To provide researchers with the data needed in selection of appropriate rat disease models, the portal includes both phenotype data for cardiovascular, pulmonary, blood biochemistry, renal, vascular and general morphological results, as well as strain availability, disease models and strain details. In order to integrate physiological data for a wide variety of inbred rat strains under different experimental and environmental conditions, an ontology-based integration tool is under development. RGD captures biological information for genes, strains and QTL through curation using ontologies including the Gene Ontology (7), Mammalian Phenotype ontology (8), a disease ontology and the Pathway ontology created at RGD (9). Ontology development to describe measurement methods, experimental conditions, clinical measurements and rat strains is ongoing, initially focusing on cardiovascular phenotypes (10,11). Researchers can use these multiple ontologies to explore and filter the data to address specific questions; the system aims to allow the simultaneous querying and exploration of multiple ontologies to enable researchers unfamiliar with the data to rapidly focus on areas of interest or expand their view to encompass related experiments. In addition to providing access to these integrated datasets, the portal includes information on animal husbandry, breeding schemes for developing rat models, detailed phenotyping protocols and links to a wide variety of external physiological resources.
OUTREACH/EDUCATION
RGD has ongoing educational and community activities promoting the use of the analysis tools and data curated and stored at RGD. Recent efforts have focused on developing video tutorials designed to instruct rat researchers in the tools and data available. The videos and audio narration walk the user through the dataset, tool or disease portal, and can be accessed from the Rat Community page on SciVee (http://www.scivee.tv/node/5812). RGD continues to host the Rat Community Forum (http://rgd.mcw.edu/RCF/) as a source for subscribers to ask and answer questions pertaining to rat research.
DISCUSSIONS AND FUTURE DIRECTIONS
RGD has made a continuous effort to provide disease-oriented data, tools and resources for the rat research community. This effort will continue as future disease portals are developed with input from the scientific community. The use of biomedical ontologies is growing within RGD in the curation process for genetic, genomic, strain, QTL and phenotype data. Tool development will include an emphasis on integrating data across multiple strain sequences as new genomes are sequenced and linking biological function to provide a platform for translational research. These approaches will allow RGD to continue to build on the existing disease-oriented views to assist users in identifying comprehensive datasets to bridge genomics to human health.
FUNDING
This work was supported by the National Institutes of Health through the National Heart, Lung, and Blood Institute (HL64541 to H.J.J.) and the National Human Genome Research Institute (HG002273 to S.N.T.). Funding for open access charge: Rat Genome Database funding from the National Heart Lung and Blood Institute grant (HL64541) to Howard J. Jacob, Ph.D (PI for Rat Genome Database).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
In addition to the listed authors, we wish to thank the following group of committed people: Kent Brodie, Diane Munzenmaier, Andrew Patzer, and Stacy Zacher.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkn842
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/37/suppl_1/D744/3313500/gkn842.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/37/suppl_1/D744
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/37/suppl_1/D744.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/37/suppl_1/D744
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/118366952
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gkn842
Article citations
Genetic characterization of outbred Sprague Dawley rats and utility for genome-wide association studies.
PLoS Genet, 18(5):e1010234, 31 May 2022
Cited by: 25 articles | PMID: 35639796 | PMCID: PMC9187121
Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells.
Sci Rep, 10(1):15263, 17 Sep 2020
Cited by: 34 articles | PMID: 32943704 | PMCID: PMC7499307
A new approach to identifying hypertension-associated genes in the mesenteric artery of spontaneously hypertensive rats and stroke-prone spontaneously hypertensive rats.
J Hypertens, 37(8):1644-1656, 01 Aug 2019
Cited by: 6 articles | PMID: 30882592 | PMCID: PMC6615961
The Year of the Rat: The Rat Genome Database at 20: a multi-species knowledgebase and analysis platform.
Nucleic Acids Res, 48(d1):D731-D742, 01 Jan 2020
Cited by: 91 articles | PMID: 31713623 | PMCID: PMC7145519
HPO2Vec+: Leveraging heterogeneous knowledge resources to enrich node embeddings for the Human Phenotype Ontology.
J Biomed Inform, 96:103246, 27 Jun 2019
Cited by: 14 articles | PMID: 31255713 | PMCID: PMC6710011
Go to all (54) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Rat Genome Database (RGD): mapping disease onto the genome.
Nucleic Acids Res, 30(1):125-128, 01 Jan 2002
Cited by: 58 articles | PMID: 11752273 | PMCID: PMC99132
Exploring human disease using the Rat Genome Database.
Dis Model Mech, 9(10):1089-1095, 01 Oct 2016
Cited by: 18 articles | PMID: 27736745 | PMCID: PMC5087824
Review Free full text in Europe PMC
The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease.
Nucleic Acids Res, 43(database issue):D743-50, 29 Oct 2014
Cited by: 142 articles | PMID: 25355511 | PMCID: PMC4383884
A Primer for the Rat Genome Database (RGD).
Methods Mol Biol, 1757:163-209, 01 Jan 2018
Cited by: 9 articles | PMID: 29761460 | PMCID: PMC6487669
Funding
Funders who supported this work.
NHGRI NIH HHS (1)
Grant ID: HG002273
NHLBI NIH HHS (1)
Grant ID: HL64541