Abstract
Free full text

MicroRNAs: key regulators of stem cells
Abstract
The hallmark of a stem cell is its ability to self-renew and to produce numerous differentiated cells. This unique property is controlled by dynamic interplays between extrinsic signalling, epigenetic, transcriptional and post-transcriptional regulations. Recent research indicates that microRNAs (miRNAs) have an important role in regulating stem cell self-renewal and differentiation by repressing the translation of selected mRNAs in stem cells and differentiating daughter cells. Such a role has been shown in embryonic stem cells, germline stem cells and various somatic tissue stem cells. These findings reveal a new dimension of gene regulation in controlling stem cell fate and behaviour.
Tissue development and homeostasis are mediated by embryonic stem (ES) cells and tissue stem cells (also known as adult stem cells)1. ES cells are derived from the inner cell mass of the blastocyst-stage embryo and give rise to the fetus; in the process they generate tissue stem cells, progenitor cells and eventually every cell type in an organism. Tissue stem cells include somatic and germline stem cells, which develop, maintain and repair their resident tissues in developing and adult organisms. Both ES cells and tissue stem cells are capable of producing various types of differentiated cells and replicating themselves. Self-replicating ability is a hallmark property of stem cells and is generally termed self-renewal. Self-renewal in tissue stem cells is often achieved by a unique pattern of asymmetric divisions. This generates one daughter cell that retains stem cell properties (self-renewal) and another daughter cell that is committed to specialized functions (differentiation)2. The self-renewing divisions of stem cells are controlled by both intercellular and intracellular mechanisms3. Whereas the intercellular mechanisms include signalling from the neighbouring niche cells, intracellular mechanisms include differential gene expression that is controlled at epigenetic, transcriptional, translational and post-translational levels. Recently, microRNAs (miRNAs) have emerged as important players in translational regulation and have implications in controlling stem cell fate and behaviour4.
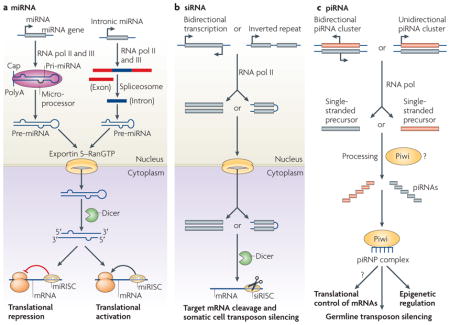
miRNAs are 20–25 nucleotide (nt) non-coding RNAs that bind to the 3′ untranslated region (UTR) of target mRNAs through an imperfect match to repress their translation and stability5 (BOX 1). This is achieved by forming a ribonucleoprotein complex, called the RNA-induced silencing complex (RISC), that contains an Argonaute family member. Occasionally, miRNAs have also been observed to activate target mRNA translation and to regulate their stability6. miRNAs are derived from precursor transcripts — called primary miRNAs (pri-miRNAs) — that are first processed in the nucleus into an intermediate form (pre-miRNAs) by the Microprocessor protein complex; this contains the Drosha and DiGeorge syndrome critical region gene 8 (DGCR8; also known as Pasha) proteins. The pre-miRNAs are then translocated by the exportin 5–RanGTP shuttle system into the cytoplasm, in which they are further processed by Dicer, an RNase III-like enzyme, into mature miRNAs. Recently, miRNAs have also been shown to be produced by two non-canonical, Drosha- or Dicer-dependent pathways. In the first pathway, the early processing step is done by spliceosome and a debranching enzyme that yields a short hairpin that is ready for further processing by Dicer. These non-canonical miRNAs have been termed mirtrons7,8. In the second pathway, short hairpin RNAs (shRNAs) are processed by unknown nucleases into pre-miRNAs and are further processed into miRNAs by Dicer. miRNAs that are derived in this way have been termed endogenous shRNA-derived miRNAs9.
Two new classes of small non-coding RNAs have recently also been identified — the endogenous small interfering RNAs (endo-siRNAs) and the Piwi-interacting RNAs (piRNAs) (BOX 1). The biogenesis of endo-siRNAs initiates with either bidirectional transcription or the transcription of an inverted repeat. The resulting double- stranded RNA or the hairpin RNA precursors is then transferred into the cytoplasm, processed by Dicer and loaded into a RISC complex in a similar manner to the miRNA pathway. The small interfering RISC complexes then target mRNAs with complete complementarity to the loaded siRNAs and mediate their degradation10.
Whereas miRNA and siRNA production is generally Dicer dependent, piRNAs are generated from a long single-stranded precursor — probably by Piwi proteins but not by Dicer. piRNAs also differ from miRNAs and siRNAs in that they are usually 26–31 nt long, they associate with Piwi-subfamily proteins and map mostly to the repetitive regions of the genome11. It is thought that piRNAs target transposons, and evidence is also available for epigenetic regulation in Drosophila melanogaster12. piRNAs might be important for stem cell self-renewal because their partner Piwi proteins are required for stem cell maintenance. Furthermore, a piRNA that was derived from a sub-telomeric region in D. melanogaster has been directly connected to germline stem cell self-renewal12. A brief description and comparison of these pathways is provided in BOX 1.
Recently, the stem cell and miRNA fields have converged with the identification of stem-cell-specific miRNAs13. In addition to canonical miRNAs, mirtrons and shRNA-derived miRNAs have also been identified in mouse ES cells9. It is now clear that miRNAs provide a new dimension to the regulation of stem cell functions. Based on their function in translational attenuation, miRNAs seem to regulate stem cell fate and behaviour by fine-tuning the protein levels of various factors that are required for stem cell or niche cell functions. In this article, we focus on current progress in understanding the biological function of miRNAs in ES cells and various tissue stem cells, with emphasis on their role in self-renewal and differentiation of stem cells.
miRNA function in ES cells
The ES cell system has been the main model for studying the function of miRNAs in stem cells, and, consequently, provides the most comprehensive knowledge on the subject.
Overall role of the miRNA pathway
The overall function of the miRNA pathway in ES cells has been evaluated in humans and mice by analysing the phenotypes of DGCR8 and Dicer mutants, respectively, as these two proteins have crucial roles in the production of mature miRNAs. In humans, DGCR8 (a double-stranded RNA-binding protein) partners with Drosha (an RNAse III enzyme) in the nucleus, and cleaves pri-miRNAs into ~70 nt pre-miRNA structures. These contain a 2 nt overhang at the 3′ end. Pre-miRNAs are then translocated into the cytoplasm by exportin 5–RanGTP and are further processed by Dicer to generate functional miRNAs14 (BOX 1). DGCR8 is exclusively involved in the miRNA pathway, whereas Dicer is involved in both miRNA and siRNA pathways. Therefore, the phenotype of Dicer mutants can be due to the absence of either miRNAs and/or siRNAs.
Dicer is essential for early mouse development, as its absence leads to embryonic lethality15. However, embryonic lethality does not seem to be due to the absence of ES cells, as Dicer-null mouse ES cells are viable, although they exhibit severe growth and differentiation defects and prolonged G1 and G0 phases of the cell cycle. It is not clear whether cell cycle regulation is required for stem cell differentiation, or whether the delayed kinetics of cell cycle progression is a cellular response to other defects that are caused by the absence of Dicer and/or miRNAs. In vitro, Dicer-null ES cells fail to express differentiation markers, such as hepatocyte nuclear factor 4A (HNF4A; which is endodermal) and brachyury, bone morphogenetic protein 4 (BMP4) and GATA1 (which are mesodermal), even after the induction of differentiation. This shows that miRNAs and/or endo-siRNAs might function in ES cell differentiation16. In reality, this differentiation defect can probably be attributed solely to the absence of miRNAs, because a related study revealed that only the profiles of miRNAs in ES cells, but not of other small RNAs, change in the absence of Dicer17. This study also revealed that one important function of miRNAs in ES cells is to regulate cell cycle progression, because almost one-half of the 110,000 miRNA molecules that have been detected are produced by 4 miRNA loci (miR-21, the miR-17–92 cluster, the miR-15b–16 cluster and the miR-290–295 cluster) that regulate cell cycle progression and oncogenesis.
Similar to the Dicer-null embryonic stem cells, DGCR8-deficient ES cells exhibit either delayed or reduced expression of differentiation markers, as well as delayed kinetics of cell cycle progression18. Most DGCR8-deficient ES cells are arrested in the G1 phase, which indicates that the main function of the miRNA pathway is to promote the ES cell cycle at the G1–S-phase transition. In addition, DGCR8-null ES cells exhibit differentiation defects as they fail to stably silence the expression of self-renewal markers, such as OCT4 (also known as OCT3 and POU5F1), REX1 (also known as ZFP42), nanog and SOX2 (REF. 18). In support of this, injected DGCR8-mutant ES cells into host mice fail to differentiate into different germ layers, in contrast with injected wild-type ES cells that can give rise to teratomas — tumours that can give rise to cell types of all three germ layers18. The similar phenotype of DGCR8 and Dicer mutants further confirms that Dicer in ES cells functions mainly in the miRNA pathway and that the predominant function of miRNAs is to regulate cell cycle progression during stem cell differentiation.
ES cells have distinct miRNA signatures
Whereas the Dicer and DGCR8 ES cell mutants provide an overall assessment of the function of the miRNA pathway in ES cells, cloning and deep sequencing of miRNAs from stem cells have revealed the identity of the specific miRNAs that are expressed in ES cells and might function in self-renewal and differentiation of ES cells. For example, in mice, the cloning and sequencing of small RNAs using conventional methods (BOX 2) revealed that the miR-290–295 cluster and miR-296 are specific to ES cells and that their levels decrease as the stem cells differentiate. In a simplified way, this indicates that the miR-290–295 cluster has specific roles in maintaining pluripotency13. However, more recent studies indicate that the real role of miR-290–295 is to induce differentiation (see below and FIG. 1). By contrast, levels of miR-21 and miR-22 increase substantially following the induction of differentiation, which indicates that these miRNAs might have important roles in stem cell differentiation (see TABLE 1 for a list of miRNAs that have been identified in various types of stem cell lineages). A similar study in human ES cells also shows that pluripotent stem cells have unique miRNAs, the levels of which decrease following differentiation19, even though no miRNA correlated to human ES cell differentiation has been identified. The functions of the ES cell-related miRNAs are being explored.

Embryonic stem (ES) cells have the ability to self-renew or differentiate into progenitor cells, which finally give rise to all of the adult tissue stem cells in the body. MicroRNAs (miRNAs) regulate both self-renewal and differentiation pathways of ES cells by regulating various factors that mediate these processes. miRNAs form an integral network with transcription factors in regulating stem cell processes. For example, RE1-silencing transcription factor (REST), a transcriptional repressor, downregulates miR-21, which targets Nanog, SOX2 and OCT4 (also known as OCT3 and POU5F1). These are essential for stem cell self-renewal. Therefore, REST promotes self-renewal26. Self-renewal is also promoted by the miR-290–295 cluster, which targets retinoblastoma like 2 (RBL2) — a repressor of DNA methyl transferases (DNMTs) — which methylate CpG islands and epigenetically silence OCT4 (REFS 29,30). Another mechanism of promoting self-renewal is by the regulation of let-7 miRNA processing by LIN28, which directly binds to a consensus sequence in the pre-let-7 miRNA loop region and inhibits processing by Drosha and Dicer32,33. miR-296 promotes ES cell differentiation and miR-22 inhibits ES cell differentiation. However, their mRNA targets have not been identified. Positive and negative regulations are shown by an arrow and a T line respectively; a dotted T line suggests a possible pathway that needs to be confirmed.
Table 1
RNAs identified in various kinds of stem cells
| Stem cell process | Cell type or lineage | miRNAs involved‡ | Targets§ |
|---|---|---|---|
| ES cells | |||
| Self-renewal | Mouse ES cells | miR-290–295 cluster, miR-296, miR-302 (REF. 13), miR-17–92 cluster and miR-15b–16 cluster17 | No targets identified |
| Human ES cells | miR-371, miR-372, miR-373*–373, miR-200c, miR-368 and miR-154* (REF. 19) | No targets identified | |
| Differentiation | Mouse ES cells | miR-21 and miR-22 (REF. 13) | miR-21 targets Nanog and Sox2 (REF. 26) |
| Human ES cells | miR-301, miR-374, miR-21, miR-29b and miR-29 (REF. 19) | No targets identified | |
| Somatic stem cells | |||
| Haematopoiesis | Progenitor cells | miR-128, miR-181, miR-16, miR-103 and miR-107 (REF. 35) | No targets identified |
| Pro-T lymphoid cells | miR-150 (REF. 41) | No targets identified | |
| Pro-B lymphoid cells | miR-181, miR-155, miR-24, miR-17, miR-16, miR-103 and miR-107 (REF. 35) | No targets identified | |
| Erythroid myeloid cells | miR-150, miR-155, miR-221, miR-222, miR-451, miR-16 (REFS 35,37) and miR-24 (REFS 35,38) | miR-24 targets human ALK4; and miR-221 and miR-222 target human KIT (REF. 39) | |
| Monocytes | miR-17-5p, miR-20a, miR-106a, miR-16, miR-103 and miR-107 (REF. 40) | miR-17-5p, miR-20a and miR-106a target AML1 | |
| Granulocytes | miRNA-155, miR-24, miR-17, miR-223, miR-16, miR-103 and miR-107 (REF. 35) | No targets identified | |
| Megakaryocytes | miR-155, miR-24, and miR-17 (REF. 35) | No targets identified | |
| Myogenesis | Skeletal cells | miR-1, miR-133 (REF. 45), miR-206 (REF. 46) and miR-26a (REF. 50) | miR-1 targets mouse Hdac4; miR-133 targets mouse Srf; miR-1 and miR-206 target mouse connexin 43 (also known as Gja1); and miR-26a targets mouse Ezh2 |
| Cardiac cells | Unknown | Unknown | |
| Neurogenesis | Neuronal | miR-124 and miR-128 (REFS 55,56) | miR-124 targets chicken SCP1 (REF. 57) |
| Astrocytes | miR-26, miR-29, miR-23 (REF. 55) | No targets identified | |
| Osteogenesis | Osteoblasts | miR-125b60 and miR-26a | miR-26a targets human SMAD |
| Skin development | Keratinocytes | miR-203 (REF. 64) | miR-203 targets mouse p63 |
ALK4, type I activin receptor; AML1, acute myeloid leukaemia 1; ES cell, embryonic stem cell; Ezh2, enhancer of zeste homologue 2; Hdac4, histone deacetylase 4; miRNA, microRNA; SCP1, small carboxy-terminal domain phosphatase 1; Srf, serum response factor.
Whereas a few of the identified miRNAs are conserved between human and mouse ES cells (miR-296, miR-301 and miR-302), a large proportion of the identified miRNAs are unique to human ES cells. The small overlap between the two data sets might reflect the fact that human and mouse ES cells are not found at the equivalent developmental stage. In addition, the small overlap seems to be partly due to the fact that the complete repertoire of miRNAs in stem cells has not been revealed in either species, and evolutionary differences are unlikely to be a major contributing factor. In support of this, a recent miRNA sequencing effort using the Solexa sequencing technology (BOX 2) discovered several miRNAs in human ES cells that overlapped with those detected in mouse ES cells, and also with those that are differentially expressed between self-renewing and differentiating daughter cells20. The development of this technology has significantly increased our ability to discover new miRNAs and other small RNAs, especially those with low expression levels or with modest differences in expression levels between various stages of stem cell differentiation21. It has to be noted that in conventional cloning and sequencing, miRNAs that are abundantly expressed in a cell are preferentially cloned and exclude those with low expression levels but possible significant functions in stem cells. Other deep-sequencing technologies, such as massively parallel signature sequencing22 and the Roche/454 platform12,23,24 (BOX 2), are also ready to use to identify a plethora of small RNAs in ES cells and various other tissues.
There are two issues that need to be addressed regarding the profiling of miRNA expression in relation to ES cell self-renewal and differentiation. First, individual miRNAs can show dynamic changes during stem cell differentiation, and therefore cloning and sequencing of miRNAs before and after differentiation would only allow us to assess ultimate changes, thereby precluding the assessment of dynamic changes of miRNAs during differentiation. This could be resolved by examining the expression of miRNAs at multiple time points during the course of stem cell differentiation. Second, the varying origin of ES cell lines and the heterogeneity of differentiated cells can skew and/or mask the true expression profile of miRNAs. This might have contributed to the varying results from different searches for ES cell-specific miRNAs. The recent application of single-cell real-time PCR to profile miRNA expression in primordial germ cells25 can be applied to evaluate the severity of this problem, as well as to help bypass it. In addition, homogeneous starting material and improved differentiation and cell selection protocols should also overcome the problem.
Interplay between miRNAs and other regulatory pathways
Research on ES cell-specific miRNAs has extended beyond their identification and phenotypic analysis, and is focusing on the regulation of their expression and their regulatory targets. These studies have in turn provided an opportunity to link miRNA regulation to other mechanisms of gene regulation in ES cells (FIG. 1). Although presently sporadic, this area of research should have a major role in integrating miRNA regulation to the biology of ES cells.
A recent study shows that miR-21 (REF. 26), the levels of which increase following the induction of mouse ES cell differentiation, has potential binding sites in the 3′ UTRs of the mRNAs that encode for nanog, SOX2 and possibly OCT4. These are key proteins that are involved in ES cell self-renewal13. Other differentiation-related miRNAs — miR-134, miR-296 and miR-470 — were recently found to target coding regions of nanog, SOX2 and OCT4 to promote differentiation27. Hence, differentiation-related ES cell-specific miRNAs seem to promote differentiation by acting inside these cells to reduce the cellular concentrations of key proteins involved in self-renewal. Thus, the role of these miRNAs in ES cells can be compared to a Trojan Horse. Interestingly, the transcription of miR-21 itself is regulated in ES cells by a transcriptional repressor called the RE1-silencing transcription factor (REST), which directly interacts with cis elements upstream of the miR-21 gene26. Furthermore, a recent genome-wide chrom atin immunoprecipitation coupled with DNA sequencing (ChIP-seq) study has shown, at high resolution, that key ES cell-specific transcription factors bind to promoters of ES cell-specific miRNAs and directly promote their expression28. These studies demonstrate the existence of a regulatory paradigm from a transcription factor to miRNA and to another transcription factor in promoting ES cell differentiation.
In addition to regulatory interactions with transcription factors, the miRNA pathway also interfaces with epigenetic mechanisms. A known example of this is the ES cell-specific miR-290–295 cluster, which also acts like a Trojan Horse. This cluster seems to promote ES cell differentiation by acting inside ES cells to downregulate key proteins that are involved in ES cell self-renewal. The miR-290–295 cluster achieves this role by stably targeting repressors of DNA methyl transferases (DNMTs), such as retinoblastoma like-2 (RBL2), which methylate CpG islands, leading to stable epigenetic repression of Oct4 transcription29,30 (FIG. 1). In fact, the miR-290–295 cluster seems to be responsible for the requirement of Dicer in ES cell differentiation. However, it remains to be addressed whether and how this epigenetic repression of Oct4 transcription is sufficient to promote differentiation, given that DNMTs are global regulators of CpG island methylation status in the entire genome.
Another important level of regulation of miRNA function resides at the level of miRNA biogenesis. This is shown by the regulation of the processing of the let-7 family of miRNAs by LIN28, a developmentally regulated RNA-binding protein that is involved in miRNA processing and promotes the reprogramming of human somatic cells into induced pluripotent stem (iPS) cells31. LIN28 selectively blocks the processing of pri-let-7 miRNAs in ES cells by specifically binding to conserved nucleotides in the loop region of the let-7 precursor and inhibiting the activity of Drosha and Dicer, thus having a central role in blocking miRNA-mediated differentiation in ES cells32,33. These findings also establish a novel role for the miRNA-precursor loop region in the regulated production of mature let-7 (REF. 34). These discoveries have started to reveal the intricate and complex web of regulatory networks through which miRNAs become part of the integrated mechanism of gene regulation that defines ES cell fate and behaviour.
miRNA function in adult stem cells
The functions of miRNAs in somatic tissue stem cells have also been increasingly explored. This section reviews the miRNAs that have been identified in various somatic tissues and discusses how they regulate adult stem cell proliferation and differentiation.
miRNAs regulate multiple steps of haematopoiesis
Haematopoiesis is initiated by self-renewing divisions of haematopoietic stem cells, which give rise to complex cell types of myeloid and lymphoid lineages (FIG. 2). A combined approach that involves assessment of miRNA expression using microarray and bioinformatic prediction of mRNA targets revealed that distinct miRNA signatures fine-tune each step of haematopoiesis. For example, miR-128 and miR-181 are expressed only in early progenitor cells and prevent the differentiation of all haematopoietic lineages35. Likewise, miR-16, miR-103 and miR-107 prevent proliferation of later progenitor cells, whereas miR-221, miR-222 and miR-223 control the terminal differentiation pathways35 (FIG. 2).
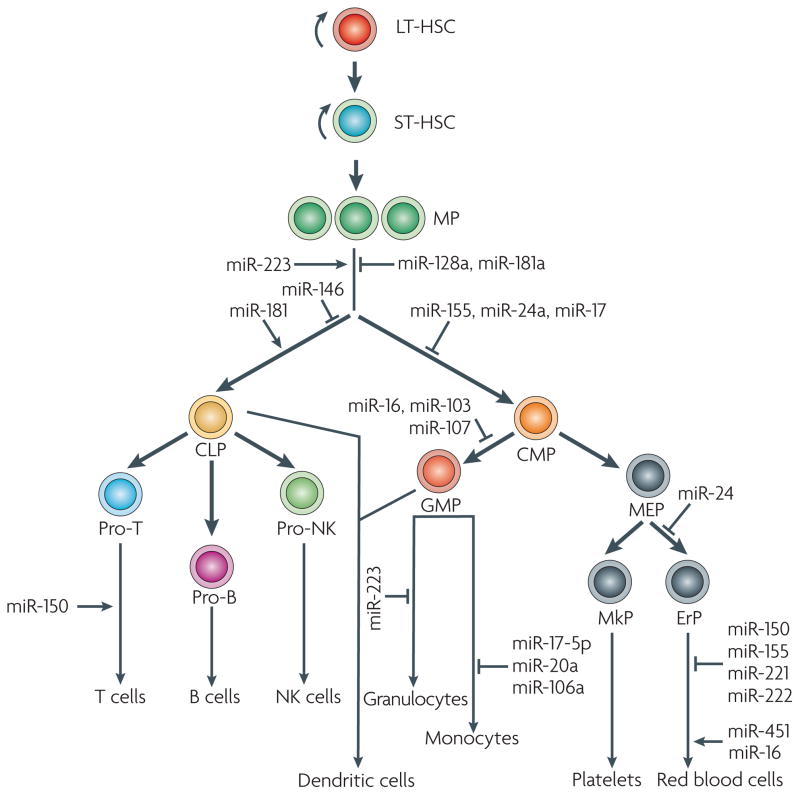
Haematopoiesis involves a cascade of cell proliferation and differentiation in which the long-term reconstituting haematopoietic stem cells (LT-HSCs) first give rise to short-term reconstituting HSCs (ST-HSCs), followed by multipotent progenitors (MPs). MPs differentiate into either common lymphoid progenitors (CLPs) or common myeloid progenitors (CMPs). CLPs and CMPs further differentiate to produce various cells of the bloodstream. A number of microRNAs (miRNAs) have been identified that fine-tune each step of the haematopoietic differentiation. miRNAs that function in specific steps are marked, and positive and negative regulations are illustrated by an arrow and a T line, respectively. ErP, erythroid progenitor; GMP, granulocyte–macrophage progenitor; MEP, megakaryocytic–erythroid progenitor; MkP, megakaryocyte progenitor; NK, natural killer.
The first lineage-specific decision during haematopoiesis is at the differentiation of common haematopoietic progenitor cells into common lymphoid or common myeloid progenitors. miR-223 and miR-181 are expressed less abundantly in the progenitor cells but are upregulated following differentiation into the myeloid and lymphoid lineages, respectively36. Largely consistent with this finding, ectopic expression of miR-181 in undifferentiated progenitor cells increases the population of B lymphoid lineage cells but not of myeloid lineage cells.
Within the myeloid lineage, several key mechanisms drive erythropoiesis. Bioinformatics analysis has suggested that miR-150, miR-155, miR-221 and miR-222 are progressively downregulated during erythropoiesis, whereas miR-451 and miR-16 are upregulated at late stages of erythropoiesis37. This implicates their function in these stages of erythropoiesis. Erythroid lineage differentiation is prevented by the repression of type I activin receptor (ALK4; also known as ACVR1B) by miR-24 (REF. 38). The growth factor activin, which, along with erythropoietin, promotes erythroid differentiation, binds to ALK4, resulting in a cascade of downstream signalling that culminates in erythroid differentiation. Another mechanism that prevents erythropoiesis involves miR-221 and miR-222, which block erythropoiesis by targeting KIT (also known as CD117), a cytokine receptor that is expressed on the surface of haematopoietic stem cells39.
Monocytopoiesis is regulated by miRNA-17-5p, miR-20a and miR-106a, which target the monocytic differential transcription factor acute myeloid leukaemia 1 (AML1; also known as RUNX1)40. AML1 promotes monocytic differentiation by activating transcription of macrophage colony-stimulating factor (M-CSF), which is essential for monocytic differentiation.
Within the lymphoid lineage, the differential expression of miR-150 regulates lineage decision between T cells and B cells41. Ectopic expression of miR-150 in the lymphoid progenitor cells predisposes them to form more T cells than B cells. This might be achieved by targeting mRNAs that are responsible for B cell maturation.
As in ES cells, lineage-specific transcription factors can indirectly determine the fate of adult stem cells by modulating the levels of lineage-specific miRNAs. For example, the haematopoietic transcription factor GATA1 is required for the expression of miR-144 and miR-451, which are specifically induced during erythroid differentiation42. GATA1 interacts with its cofactor FOG1 (also known as ZFPM1), and brings about transcriptional activation by binding to sequence-specific elements that are distal to the miRNA gene that produces both miR-144 and miR-451 (REF. 42). In parental erythroblast cells that lack GATA1, GATA2 occupies the site but does not activate transcription, and thereby prevents erythroid differentiation. In another study, PU.1, a transcription factor that is essential for monocytic differentiation, activates miR-424. In turn, this targets the transcription factor nuclear factor I/A (NFI-A) and induces monocytic differentiation43. These observations reveal specific transcription factors as upstream regulators of the miRNA pathway in cell fate regulation.
miRNAs in myogenesis and cardiogenesis
Myogenesis is also a highly regulated proliferation and differentiation process, in which the mesodermal progenitor cells give rise to myoblasts that later withdraw from the cell cycle and fuse to form multinucleated myotubes44. Recent reports have revealed several mechanisms that are mediated by miRNAs during myogenesis. miR-1 and miR-133 are co-transcribed and processed specifically in adult cardiac and skeletal muscle tissues, but promote myogenesis in distinct fashions45. miR-1 indirectly upregulates myocyte enhancer factor 2C (MEF2C), an essential muscle-related transcription factor, by targeting the transcriptional co-repressor histone deacetylase 4 (HDAC4) and thereby promoting myogenesis. However, miR-133 targets serum response factor (SRF), a transcription factor that is essential for muscle proliferation and differentiation45. In another mechanism, miR-1 and miR-206 promote myogenesis by targeting connexin 43 (Cx43; also known as Gja1) mRNA and tightly regulating its protein levels46,47. CX43, which forms the gap junctions, is essential for normal skeletal muscle differentiation, and it is thought that gap junctions promote better intercellular transfer of the signals that are involved in myogenesis. In mouse and human ES cells, miR-1 and miR-133 both promote mesoderm formation and are potent repressors of non-muscle gene expression. However, they have opposing functions during the further differentiation of the mesodermal precursors into cardiac muscle progenitors: miR-1 promotes cardiac progenitor formation, whereas miR-133 represses the process48. The effect of miR-1 in promoting the mesodermal differentiation of ES cells seems to be partially achieved by translational repression of the notch ligand delta-like 1 (DLL1), which in turn promotes the expression of cardiac mesoderm genes and suppresses the expression of non-mesoderm genes48. These findings indicate that muscle-specific miRNAs reinforce the silencing of non-muscle genes during cell lineage commitment from pluripotent ES cells.
Another miRNA, miR-26a, indirectly promotes skeletal muscle differentiation by targeting the histone methyl-transferase enhancer of zeste homologue 2 (EZH2)49. In undifferentiated mouse myoblasts, EZH2, along with the transcriptional regulators yin yang 1 (YY1) and HDAC1, epigenetically silences muscle-specific genes, and, following the induction of differentiation, EZH2 dissociates from muscle-specific genes. This paves the way for transcriptional activation of these genes50.
Similar to ES cells and haematopoiesis, muscle-specific transcription factors and miRNAs form a network to regulate myogenesis. For example, during cardiogenesis, SRF and the co-activator myocardin bind to cis-acting elements upstream of the genes that encode miR-1-1 and miR-1-2, and increase their cellular levels in cardiac progenitor cells51. These miRNAs in turn target HAND2, a transcription factor that is involved in ventricular cardiomyocyte expansion (proliferation), and help progenitor cells to choose between proliferation and differentiation. In another case, the myogenic factors myogenin and MYOD bind to cis elements upstream of loci that encode miR-1, miR-1-1, miR-1-2, miR-133, miR-133a-1 and miR-133a-2, and regulate their expression52.
Regulation of myogenesis also involves transforming growth factor-β (TGFβ)-mediated downregulation of miR-24. This involves the direct binding of SMAD3 and SMAD4 to the promoter region of the miR-24 gene and regulating its expression53. Reducing miR-24 expression leads to the downregulation of myogenic differentiation markers in the C2C12 myoblast cell line, whereas ectopic expression of miR-24-enhanced differentiation and partially rescued TGFβ-inhibited myogenesis53. These results demonstrate a crucial role for miRNAs in modulating TGFβ-dependent inhibition of myogenesis.
miRNAs specify astrocyte versus neuronal fates
Neuronal stem cells (NSCs) have to make crucial lineage-specific decisions as to whether to self-renew or differentiate into neurons or astrocytes54. As in other tissues, the differential expression of distinct miRNAs following the induction of neuronal differentiation dictates lineage-specific decisions. For example, miR-124 and miR-128 are specific to the neuronal lineage, whereas miR-26, miR-29 and miR-23 are preferentially and specifically expressed in astrocyte lineages respectively55. Consistent with these findings, overexpression of miR-124 and miR-128 in the NSCs prevents astrocyte differentiation and predisposes them towards neuronal differentiation56. miR-124 targets the 3′ UTR of small carboxy-terminal domain phosphatase 1 (SCP1; also known as CTDSP1) and induces neurogenesis57. SCP1 is an anti-neural factor that binds to a conserved response element (RE1) and prevents the expression of neural genes in non-neural tissues58. These studies show that miRNAs regulate the fate of neuronal stem cells by targeting key regulatory factors.
miRNAs prevent osteogenic differentiation
The function of miRNA in osteogenesis (bone formation) is now emerging. Osteoblasts (bone-forming cells) originate from multipotent mesenchymal stem cells, which differentiate into osteoblasts following induction by BMPs59. Two recent reports have revealed that miR-125b and miR-26a prevent the differentiation of mesenchymal stem cells into osteoblasts in distinct ways. miR-125b functions by inhibiting cell proliferation through an unknown mechanism60, whereas miR-26a prevents human adipose tissue-derived stem cells (ADSCs) from osteogenic differentiation by downregulating the translation of SMAD, a transcription factor that is required for late osteoblast differentiation61. These two studies support the role of miRNAs in maintaining the fate of osteogenic stem cells and progenitor cells.
miRNAs promote differentiation in skin
In contrast to osteogenesis, miRNAs in the skin have only been found to promote skin stem cell differentiation. Multipotent skin stem cells reside in the basal layer of the epidermis. They commit to the terminal differentiation pathway as they become suprabasal. In mouse epidermis, DICER1 is required for the maintenance of morphogenesis and for the maintenance of hair follicles. However, it was found not to be essential for skin stem cell fate determination and differentiation62,63. Furthermore, miR-203 was found to promote skin differentiation by limiting the proliferative capacity of the skin stem cells and induce cell cycle exit64. miR-203 is differentially expressed, with no expression in proliferative stem and progenitor cells but high expression in differentiated cells. A possible mechanism of miR-203 action is to target p63, which is an essential regulator of stem cell maintenance, although the mechanisms of action of p63 in stem cells are unclear.
miRNAs and piRNAs in the germ line
Germline stem cells are derived from primordial germ cells (PGCs) after they migrate into the embryonic gonad. In mammalian systems, only PGCs that migrate to the testicular cords on the male gonad become germline stem cells. During postnatal development, male germline stem cells undergo self-renewing divisions to produce many spermatogonia, which differentiate into spermatocytes, undergo meiotic divisions and eventually become mature sperm. By contrast, PGCs in the female gonad enter meiotic arrest before resuming meiosis during puberty.
Recent evidence suggests that, as in ES cells and somatic stem cells, the miRNA pathway has a vital role in both PGC specification and germline stem cell maintenance in invertebrates, and germline stem cell maintenance in vertebrates. In D. melanogaster, mature miRNAs are generated by a protein complex containing Dicer1 and Loquacious, a double-stranded RNA-binding protein. Depletion of maternal Dicer1 severely affects PGC formation in the early embryos65. In addition, knocking out either of these proteins66,67 affects female germline stem cell maintenance. Dicer1-null germline stem cells display delayed G1–S-phase transition68, which is similar to the phenotype displayed by Dicer-null ES cells. Mosaic analysis shows that the miRNA pathway might function intrinsically within the germline stem cells66,67. A similar role of Dicer1 for germline maintenance has also been shown in Caenorhabditis elegans69, which indicates that the function of the miRNA pathway in the germ line might be evolutionarily conserved.
In addition to miRNAs, the germ line has recently been found to be highly enriched for piRNAs that bind to Piwi-subfamily proteins11. Since Piwi proteins are required for germline development, germline stem cell self-renewal and gametogenesis70, piRNAs as Piwi partners might also be involved in germline stem cell functions. In support of this, a sub-telomeric piRNA in D. melanogaster has been correlated to germline stem cell maintenance12. In addition, piRNA-like (~32 nt) small RNAs have been found in neoblasts (stem cells) of the planarian Schmidtea mediterranea71, which hints at a possibility that piRNAs might also be found in the stem cells of higher organisms. It will be interesting to see if mammalian piRNAs are also involved in these processes. Since Piwi and its associated piRNAs in D. melanogaster have also been shown to be involved in epigenetic regulation12, piRNAs might provide a new mechanism of regulation of the stem cell fate in the germ line through epigenetic programming.
Conclusion and future perspectives
It is becoming increasingly evident that miRNAs have crucial roles in the self-renewal and differentiation of all three kinds of stem cells — ES cells, somatic tissue stem cells and germline stem cells. Although research on the role of miRNAs in stem cells is still in its infancy, several general conclusions can already be drawn from current studies for miRNA function in stem cells. First, the miRNA pathway has an important role in somatic stem cells in diverse tissues, although it is unclear whether this role is achieved by fine-tuning the expression of mRNA targets, as expected from the mode of miRNA action, or by a more drastic impact on the expression of the target mRNAs. Second, stem cell self-renewal and differentiation are mediated by distinct miRNAs that are differentially expressed in a cell-type- and stage-specific manner, although individual miRNAs can act at different stages of the stem cell lineage with multiple functions in promoting or repressing differentiation. Third, distinct profiles of miRNA expression in specific types of cells or during specific stages of differentiation are generated by conserved clusters of miRNAs that are coordinately transcribed, which in turn might function as master regulators of stem cell functions. Fourth, the regulatory function of the miRNA is often executed through, and controlled by, various transcription factors and other regulatory mechanisms of gene expression. These complex interactions between miRNAs and other regulators of gene expression at the epigenetic, transcriptional and post-transcriptional levels integrate miRNAs into the cellular network of regulation of gene expression that defines the stem cell fate and behaviour. Last but not least, the above modes of miRNA-mediated regulation seem to be well conserved among ES cells, various types of somatic tissue stem cells and germline stem cells. In fact, these might be the general modes of miRNA-mediated gene regulation during development.
miRNA functions in stem cells are being revealed at an extremely rapid rate. Further miRNAs will soon be discovered for their function in diverse stem cell systems by powerful deep-sequencing methods, combined with more effective methods for the isolation of stem cells and their differentiated progeny from various tissues. The mRNA targets of these miRNAs will be systematically identified by integrated experimental and bioinformatics approaches, and the effect of miRNAs towards their target mRNAs will be comprehensively assessed. The cellular effect of proteins translated from these target miRNAs will also be effectively measured. Equally importantly, the interplay between miRNAs and other cellular gene regulatory mechanisms, including other small RNA pathways (of which little is known thus far), will be systematically unravelled. These advances will not only lead to our in-depth understanding of how miRNA-mediated gene regulation contributes to the fate and behaviour of stem cells, but should also shed light on the mechanism through which miRNAs regulate developmental processes in general.
Acknowledgments
We thank members of the Lin laboratory for their valuable comments on the manuscript. We apologize to those whose works are not cited here owing to space limitations. The stem cell work done in the Lin laboratory is supported by National Institutes of Health Grants HD33760, HD37760S1 and HD42042, the Connecticut Stem Cell Research Fund, the G. Harold and Leila Mathers Foundation and the Stem Cell Research Foundation.
Glossary
| Blastocyst | An early stage of embryonic development at which cells begin to commit to two developmental lineages: the inner cell mass, which gives rise to the fetus, and the trophoblast, which gives rise to fetal support tissues, such as the placenta and the umbilical cord |
| Niche | The natural anatomical microenvironment that supports stem cell behaviour |
| Spliceosome | A ribonucleoprotein (RNP) complex that is involved in splicing of nuclear pre-mRNA. It is composed of five small nuclear (sn) RNPs and more than 50 non-snRNPs, which recognize and assemble on exon–intron boundaries to catalyse intron processing of the pre-mRNA |
| Piwi | An Argonaute or Piwi protein family member in Drosophila melanogaster that is required for germline stem cell self-renewal and also binds to ~25 nucleotide small RNAs. Piwi is the founding member that was used to define the protein family |
| Transposon | A mobile genetic element that can relocate within the genome of its host. An autonomous transposon encodes a transposase protein that catalyses its excision and reintegration in the genome, and can therefore direct its own transposition |
| Gap junction | An intercellular connection that directly connects cytoplasm of two cells so that exchange of molecules and ions can occur freely |
| Neoblast | An undifferentiated cell in annelids that proliferates to produce differentiated cells at the sites of repair |
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
KIT
The miRNA registry: http://microrna.sanger.ac.uk/sequences let-7 | miR-1 | miR-15b | miR-16 | miR-17 | miR-20a | miR-21 | miR-22 | miR-23 | miR-24 | miR-26a | miR-29 | miR-106a | miR-124 | miR-125b | miR-128 | miR-133 | miR-134 | miR-144 | miR-150 | miR-155 | miR-181 | miR-203 | miR-206 | miR-221 | miR-222 | miR-223 | miR-290 | miR-296 | miR-301 | miR-302 | miR-424 | miR-451 | miR-470
UniProtKB: http://www.uniprot.org ALK4 | AML1 | BMP4 | DGCR8 | Dicer | Drosha | exportin 5 | EZH2 | FOG1 | GATA1 | HNF4A | LIN28 | nanog | OCT4 | REST | REX1 | SOX2 | SRF
FURTHER INFORMATION
Haifan Lin’s homepage: http://www.cellbiology.yale.edu/faculty/lin_h/lin_h.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrm2621
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4118578?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nrm2621
Article citations
The potential of miRNA-based approaches in glioblastoma: An update in current advances and future perspectives.
Curr Res Pharmacol Drug Discov, 7:100193, 29 Jun 2024
Cited by: 0 articles | PMID: 39055532 | PMCID: PMC11268206
Review Free full text in Europe PMC
MiRNA-210 is involved in cigarette smoke extract-induced apoptosis of MLE-12 via the Shh signaling pathway.
Tob Induc Dis, 22, 29 May 2024
Cited by: 0 articles | PMID: 38813585 | PMCID: PMC11135024
Effects of miR-29c on proliferation and adipogenic differentiation of porcine bone marrow mesenchymal stromal cells.
Adipocyte, 13(1):2365211, 10 Jun 2024
Cited by: 1 article | PMID: 38858810
Stem Cell-Derived Exosomal MicroRNAs as Novel Potential Approach for Multiple Sclerosis Treatment.
Cell Mol Neurobiol, 44(1):44, 07 May 2024
Cited by: 0 articles | PMID: 38713302 | PMCID: PMC11076329
Review Free full text in Europe PMC
NANOG controls testicular germ cell tumour stemness through regulation of MIR9-2.
Stem Cell Res Ther, 15(1):128, 01 May 2024
Cited by: 0 articles | PMID: 38693576 | PMCID: PMC11062916
Go to all (452) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The Expression and Functional Roles of miRNAs in Embryonic and Lineage-Specific Stem Cells.
Curr Stem Cell Res Ther, 14(3):278-289, 01 Jan 2019
Cited by: 5 articles | PMID: 30674265
Review
Roles of MicroRNAs in Establishing and Modulating Stem Cell Potential.
Int J Mol Sci, 20(15):E3643, 25 Jul 2019
Cited by: 15 articles | PMID: 31349654 | PMCID: PMC6696000
Review Free full text in Europe PMC
Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl.
EMBO J, 36(21):3194-3211, 13 Oct 2017
Cited by: 31 articles | PMID: 29030484 | PMCID: PMC5666619
miRNAs in stem cell aging and age-related disease.
Mech Ageing Dev, 168:20-29, 25 Aug 2017
Cited by: 22 articles | PMID: 28847486
Review
Funding
Funders who supported this work.
NICHD NIH HHS (5)
Grant ID: HD33760
Grant ID: R03 HD042042
Grant ID: HD37760S1
Grant ID: HD42042
Grant ID: R01 HD033760