Abstract
Free full text

Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1- phosphatidylinositol 3-kinase complex
Abstract
Beclin 1, a mammalian autophagy protein that is implicated in development, tumor suppression, neurodegeneration and cell death, exists in complex with Vps34, the class III phosphatidylinositol 3-kinase (PI-3K) that mediates multiple vesicle trafficking pathways including endocytosis and autophagy. However, the precise role of the Beclin 1-Vps34 complex in autophagy regulation remains to be elucidated. Combining mouse genetics and biochemistry, we uncover a large in vivo Beclin 1 complex containing the known proteins Vps34, p150/Vps15 and UVRAG, as well as novel proteins Atg14L (yeast Atg14-like) and Rubicon (RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein). Characterization of these novel proteins reveals that Atg14L enhances Vps34 lipid kinase activity and up-regulates autophagy, whereas Rubicon reduces Vps34 activity and down-regulates autophagy. We show that Beclin 1 and Atg14L synergistically promote formation of double-membraned organelles that are associated with Atg5 and Atg12, whereas forced expression of Rubicon results in aberrant late endosomal/lysosomal structures and impaired autophagosome maturation. We hypothesize that by forming distinct protein complexes, Beclin 1 and its binding proteins orchestrate the precise function of the class III PI-3K in regulating autophagy at multiple steps.
Macroautophagy (herein referred to as autophagy) is a regulated process that sequesters and delivers portion of the cytoplasm to the lysosomes for degradation. Currently the autophagic process in mammals is poorly understood. Identification and characterization of mammalian autophagy proteins are crucial to elucidate details of mammalian autophagy. Beclin 1 (encoded by Becn1, the orthologue of yeast ATG6/Vps30), is among the earliest characterized mammalian autophagy proteins1. Initially identified as a Bcl-2 binding protein2, Beclin 1 has been shown both in vitro and in vivo to participate in autophagy regulation and to play important roles in development3, tumorigenesis1, 3–5 and neurodegeneration6–8. Like yeast Atg6, Beclin1 also forms a complex with Vps34/class III phosphatidylinositol 3-kinase9–11. Yeast has at least two Atg6/Vps34 protein complexes: one containing Atg14 and participating in autophagy, and the other containing Vps38 and functioning in non-autophagic pathway10. However, no concrete evidence has indicated multiple Beclin 1-Vps34 complexes or multiple functions associated with the Beclin 1-Vps34 in mammals11.
To reveal the mechanism whereby the Beclin 1-Vps34 interaction regulates autophagy, we combined mouse genetics and biochemistry to identify Beclin 1-associated protein complexes in vivo. We genetically engineered mice in which endogenous Beclin 1 is functionally replaced by an enhanced green fluorescent protein-tagged Beclin 1 protein (Beclin 1-EGFP) (Figs. 1a & S1a–b). In these mice (Becn1−/−; Becn1-EGFP/+), only the Beclin 1-EGFP fusion protein, but not endogenous Beclin 1 was detected by anti-Beclin 1 antibody (Fig. 1a). These mice were born at the expected Mendelian ratio (Fig. S1c), survived postnatally and were phenotypically normal at adult stage, suggesting a full “rescue” of the embryonic lethality of Becn1−/− mice by functional Becn1-EGFP transgene.
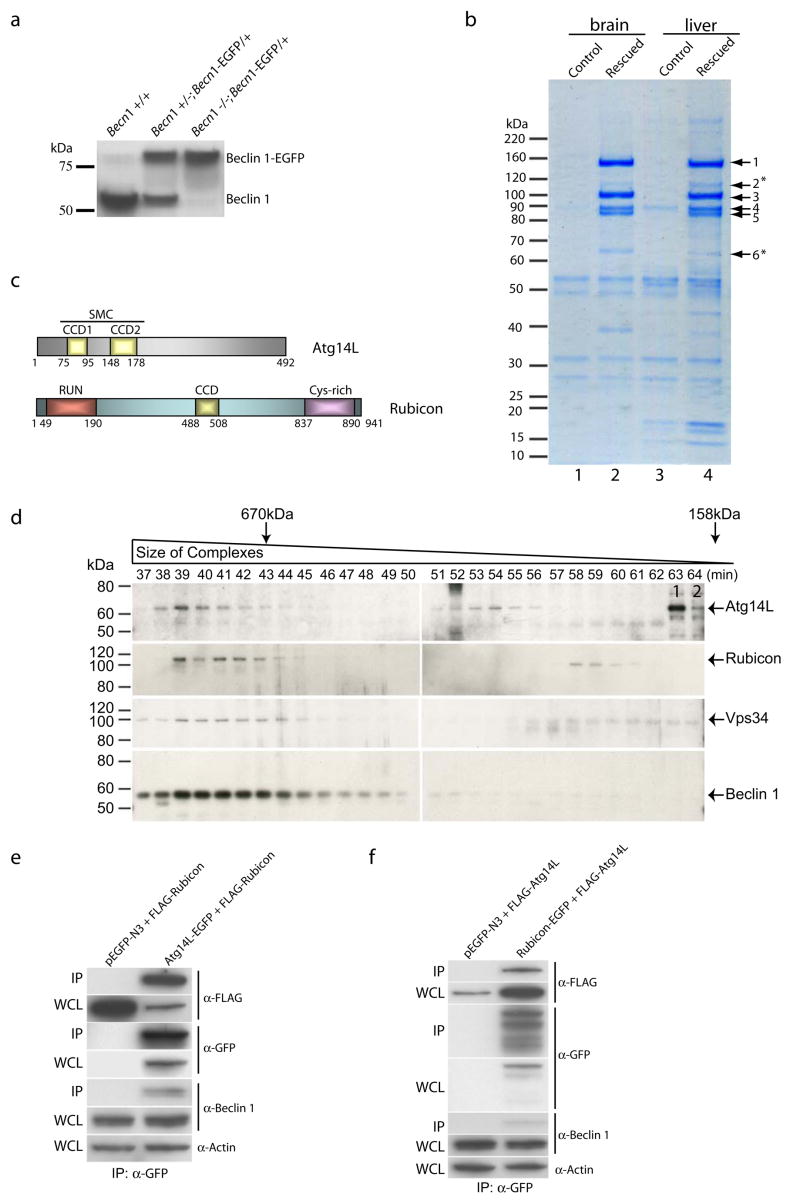
(a) Western blot analysis showed the replacement of endogenous Beclin 1 with Beclin 1-EGFP in Becn1−/−; Becn1-EGFP/+ mice, as detected by anti-Beclin 1 antibody.
(b) Coomassie-stained SDS-PAGE revealed the Beclin 1-interacting proteins immuno-isolated from the brain and liver tissues of the “rescued” mice (lanes 2 and 4) and of the control Becn1+/− littermates (lanes 1 and 3), using anti-GFP antibody. Proteins in the gel bands were extracted and identified by mass spectrometry as Vps15/p150 (band 1), Vps34/class III PI-3K (band 3), UVRAG (band 4), Beclin 1-EGFP (band 5) and two novel proteins Atg14L (band 6, asterisk, gi|27369860) and Rubicon (band 2, asterisk, gi|45708948). Notably, UVRAG levels varied with different affinity purification conditions, suggest relatively unstable association of UVRAG with the complex.
(c) Schematic diagrams of the domain structures of Atg14L and Rubicon. Atg14L contains two coiled-coil domains (CCD1 and CCD2), which are also homologous to the SMC domain (Structural Maintenance of Chromosomes). Rubicon contains an N-terminal RUN (for RPIP8, UNC-14 and NESCA) domain, a C-terminal cysteine-rich domain, and a central CCD domain.
(d) Western blot analysis of Atg14L, Rubicon, Vps34 and Beclin 1 in the gel filtration fractions from wild type mouse liver extract showed co-elution of these proteins in fractions 38–45. Atg14L was also eluted in later fractions 51–56. The fractions for the peak elution of thyroglobulin (670 kDa) and γ-globulin (158 kDa) are labeled by arrows. The control siRNA-transfected NIH 3T3 cell lysate was loaded as a positive control (labeled with “1”) for the migration position of the Atg14L protein on SDS-PAGE; the Atg14L siRNA-transfected NIH 3TC cell lysate was loaded as a negative control (labeled with “2”).
(e-f) Co-immunoprecipitation confirmed protein-protein interaction between Atg14L and Rubicon. HEK 293 cells were co-transfected with Atg14L-EGFP and FLAG-Rubicon (e), or Rubicon-EGFP and FLAG-Atg14L (f). Cell lysate was used for immunoprecipitation with anti-GFP antibody, and the resulting immunoprecipitates were blotted with anti-FLAG antibody. Our results showed immunoprecipiration of Rubicon by Atg14L (e) and vice versa (f). WCL: whole cell lysate; IP: immunoprecipirated.
Using these “rescued” mice, we isolated the Beclin 1-EGFP protein complexes by affinity purification from liver, brain (Fig. 1b) and thymus (data not shown), and identified their components using mass-spectrometry (Fig. S2). The “rescued” mice, not the control mice, are associated with at least six readily detectable protein bands common to both liver and brain (Fig. 1b). These bands include Beclin 1-EGFP (#5) (~90 kD), three previously reported Beclin 1-binding proteins p150/Vps15 (#1)12, Vps34 (#3)9–11 and UVRAG (#4)13, and two novel proteins (#2 and #6, asterisks). The first novel protein (#6, ~60 kD, gi|27369860) contains 492 amino acids and has a conserved SMC (Structural Maintenance of Chromosomes) motif or two coiled-coil domains (aa 75–95 and aa 148–178) near the N-terminus (Fig. 1c). Interestingly, the sequence of this protein shows modest similarity to yeast Atg14 (overall 15% identity) (Fig. S3a). Thus, we named this protein Atg14L for yeast Atg14-Like. The second novel protein (#2, ~124 kD, gi|45708948) contains 941 amino acids and has a conserved RUN domain (aa 49–190) near the N-terminus, a cysteine-rich domain (aa 837–890) near the C-terminus, and a coiled-coil domain (aa 488–508) in the central region (Fig. 1c). Thus, we named this protein Rubicon for RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein. No sequence homology was observed between Rubicon and Vps38 or Atg14 (data not shown). Noticeably, the protein levels of affinity purified Beclin 1-EGFP, p150/Vps15, Vps34 and UVRAG and are comparable and reproducibly higher than those of Atg14L and Rubicon (Fig. 1b), suggesting a stable “core” Beclin 1-Vps34 complex consisting of Beclin 1, Vps34, p150 and UVRAG. Additionally, we did not detect the previously identified Beclin 1-associated proteins, such as nPIST6, Bcl-22, Ambra-114 or Bif115, thus raising a possibility that their interactions with Beclin 1 may be relatively unstable, transient or occur only under specific conditions.
We next examined the specific binding of Atg14L or Rubicon to Beclin 1 in transfected mammalian cells. We showed that FLAG- or EGFP-tagged Atg14L or Rubicon co-immunoprecipitated with endogenous Beclin 1 (Fig. S3b–c) as well as Vps34 (Fig. S3d-e). We also constructed a series of deletion mutants to analyze the sequence domains required for Beclin 1-Atg14L/Rubicon associations (Fig. S3f). We found that while the coiled-coil domain (CCD) of Beclin 1 is sufficient for the binding of Atg14L, the CCD and evolutionarily conserved domain (ECD) of Beclin 1 are necessary for the binding of Rubicon (Fig. S3g-h). Furthermore, both CCD domains of Atg14L are required for the efficient binding of Atg14L to Beclin 1 and Vps34 (Fig. S3i), whereas the central region of Rubicon containing the CCD domain is important for the binding of Rubicon to Beclin 1 and Vps34 (Fig. S3j). Interestingly, the RUN or cysteine-rich domain of Rubicon appears to be inhibitory to the binding of Rubicon to Beclin 1 and Vps34 (Fig. S3j).
We then characterized the composition of the Beclin 1 complexes. Using anti-Atg14L and anti-Rubicon antibodies, we found that Atg14L is co-immunoprecipitated with Vps34 (Fig. S4a) and Beclin 1 (data not shown), but not with Rubicon (Fig. S4a); Rubicon is co-immunoprecipitated with Vps34 and UVRAG, but not with Atg14L (Fig. S4a). Therefore, Atg14L and Rubicon appear to exist in separate Beclin 1 complexes.
We performed gel filtration experiments with the tissue extract prepared from either wild-type (Fig. 1d) or “rescued” (Fig. S4b) mouse liver. For each sample, eighty fractions of the eluent were collected and analyzed by immunoblotting. We found that the endogenous Vps34, Beclin 1 (or Beclin 1-EGFP), Atg14L and Rubicon proteins were primarily co-eluted in the fractions 38–45 (Figs. 1d & S4b), suggesting that these fractions contain a major Beclin 1-Vps34 complex (size > 700 kD) that includes both Atg14L and Rubicon. We also performed gel filtration experiments with the cell lysate prepared from stable cell lines expressing either Atg14L-EGFP (Fig. S4c) or Rubicon-EGFP (Fig. S4d). Again, endogenous Beclin 1 was co-eluted with Atg14L-EGFP (Fig. S4c) and Rubicon-EGFP (Fig. S4d). Interestingly, starvation treatments of these stable cells did not affect the elution profiles of Atg14L-EGFP, Rubicon-EGFP and Beclin 1 (Fig. S4c-d).
To test the possibility that Atg14L and Rubicon are in separate protein complexes while co-eluted, we added anti-Rubicon antibody to the tissue extract before the gel filtration run and immunoblotted the resulting fractions with anti-Atg14L antibody. Our data show that Atg14L was co-eluted with the anti-Rubicon antibody (Fig. S4e, bands labeled by *, ** and ***), suggesting that the binding of anti-Rubicon antibody to, and therefore the presence of Rubicon in, the Atg14L-containing complex.
Moreover, we observed mutual co-immunoprecipitation (co-IP) of Atg14L and Rubicon from transfected cells, further supporting that Atg14L and Rubicon can be present in the same protein complex (Fig. 1e-f); UVRAG was also co-immunoprecipitated with Atg14L or Rubicon, and the interaction between UVRAG and Rubicon is significantly enhanced in the presence of Beclin 1 (Fig. S4f-h).
Taken together, we conclude that Atg14L, Rubicon, UVRAG, Beclin 1, p150/Vps15 and Vps34 can form a major Beclin 1-Vps34 complex in vivo. However, Atg14L was also eluted in the later fractions (51–56) containing Beclin 1 or Beclin 1-EGFP (but not Rubicon) (Figs. 1d & S4b), suggesting that Atg14L is also associated with a smaller Beclin 1 complex without Rubicon.
We performed several autophagy assays to dissect the role of Atg14L in autophagy. First, we knocked down Atg14L expression in cultured cells by RNA interference (RNAi) and analyzed the levels of LC3II, a lipid-conjugated form of LC3 that is normally localized on autophagosomes, by immunoblotting 16–18. Like Beclin 1 siRNA, Atg14L siRNA caused increased levels of LC3II, as compared to control siRNA (Fig. 2a, left). Second, we examined levels of p62/SQSTM1, a known autophagy substrate that is normally accumulated upon autophagy impairment19–21. Again, like Beclin 1 siRNA, Atg14L siRNA resulted in increased p62/SQSTM1 levels (Fig. 2a, left), and the increase in p62/SQSTM1 and LC3II levels upon the Beclin 1 or Atg14L siRNA treatment was also significant after starvation (Fig. 2a, right). Therefore, knocking-down of Atg14L or Beclin 1 impaired the autophagy-mediated clearance of p62/SQSTM1 and LC3II.
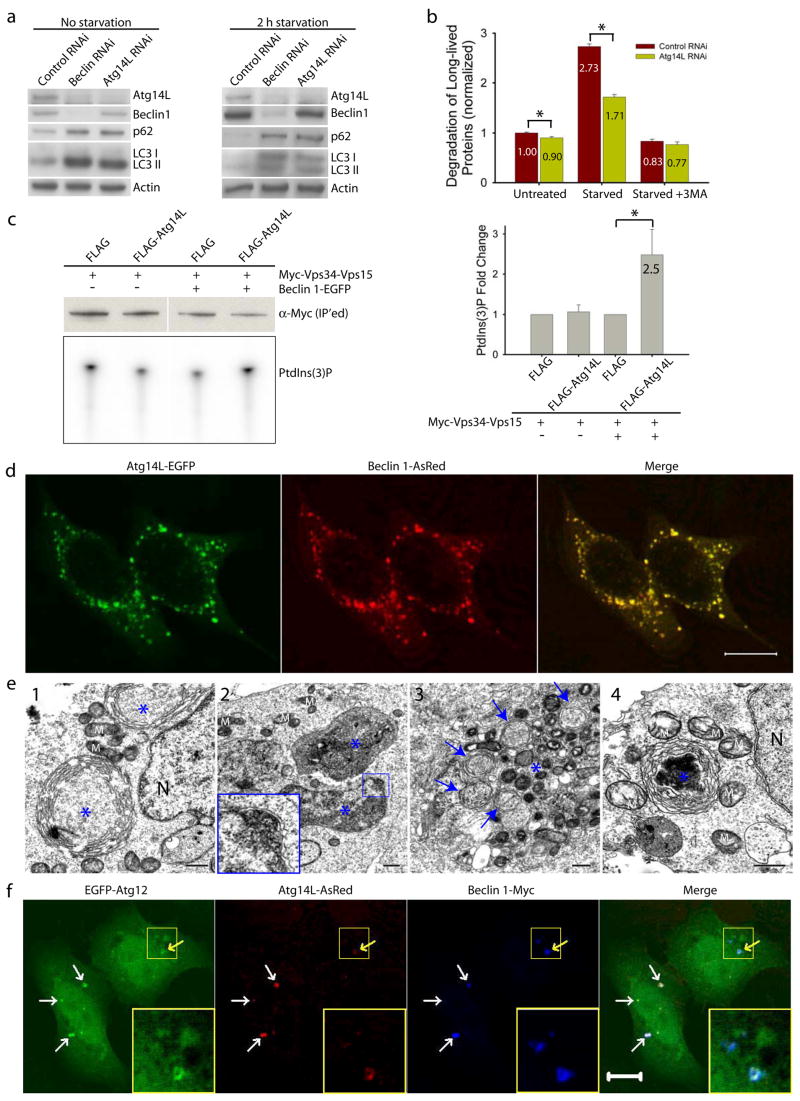
(a) Beclin 1 or Atg14L siRNA reduced Atg14L levels and increased p62/SQSTM1 and LC3 II levels under both normal and nutrient-starvation conditions in NIH 3T3 cells.
(b) Atg14L siRNA, as compared to control siRNA, decreased long-lived protein degradation in NIH 3T3 cells under both normal (p=0.007) and starvation (p=5E-6) conditions (asterisks, one-tailed Student’s t-test with equal variances, n = 4). This difference was diminished when the starved cells were treated with 3-methyladenine (3MA), a PI-3K inhibitor.
(c) Vps34 kinase assay. HEK 293T cells were co-transfected with myc-Vps34-Vps15 and FLAG-Atg14L or FLAG vector, either in the absence or in the presence of Beclin 1-EGFP. Myc-Vps34-Vps15 was immunoprecipitated by anti-myc antibody for the in vitro kinase assay. The resulting radioactive PtdIns(3)P was separated by thin layer chromatography (TLC) (lower left panel), quantified and normalized against the amount of immunoprecipitated myc-tagged Vps34 as measured by Western blot (upper left panel). The quantified results (right panel) showed that over-expressing Atg14L significantly up-regulated the Vps34 kinase activity by 2.5 folds, but only when Beclin 1 was also over-expressed (asterisk, p=0.04, one-tailed Student’s t-test with unequal variances, n=5).
(d) Co-localization of co-expressed Atg14L-EGFP (green) & Beclin 1-AsRed (red) in punctate structures in transiently transfected HeLa cells. Scale bar: 10 μm.
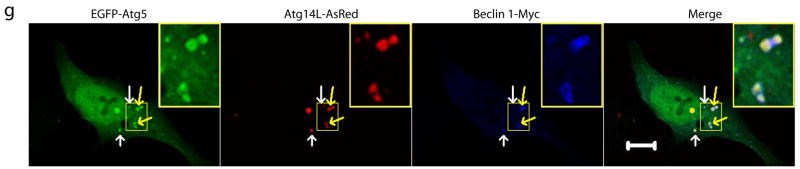
(e) Electron microscopic images show large structures (blue asterisks) that are often enwrapped with double membranes in the HEK 293T cells co-transfected with Atg14L-EGFP and Beclin 1-AsRed: (e1) concentric membrane “rings”; (e2) two large structures (3–5 μm in diameter) contain materials with high electron density; inset, enwrapping double membranes; (e3) numerous autophagosomes (blue arrows) in cytoplasm; (e4) immuno-electron microscopic image of a Atg14L-Beclin 1 structure (labeled with anti-GFP antibody and developed by DAB) enwrapped with concentric membrane “rings”. Abbreviations: M – mitochondria, N – nucleus. Scale bar: 500 nm.
(f-g) EGFP-Atg12 (f) or EGFP-Atg5 (g) (green) was colocalized with the large structures (arrows) that were labeled by Atg14L-AsRed (red) and Beclin 1-myc (blue) in transfected HeLa cells. Some of these structures appeared to be “ring”-shaped (yellow arrows & inset). Scale bar: 10 μm.
Third, we knocked down Atg14L expression in MLE12 cells that stably expressed GFP-LC3. In control siRNA-treated cells, many small GFP-LC3 puncta were observed, reflecting the presence of basal levels of autophagosomes (Fig. S5a, left). In contrast, Atg14L siRNA transfection resulted in accumulation of large-size GFP-LC3 puncta (Fig. S5a, right). These large GFP-LC3 puncta were co-colocalized with p62/SQSTM1 (Fig. S5b) and ubiquitin (Fig. S5c), indicating that these are ubiquitinated protein inclusions as previously shown in Atg5 or Atg7-deficient mouse-tissues22, 23. Ultrastructural analysis showed that Atg14L siRNA transfection results in reduced number of autophagosomes (data not shown). These above analyses suggest that reduced Atg14L expression abolishes autophagosome formation and increases ubiquitinated protein levels.
Fourth, as compared to control siRNA treatment, Atg14L siRNA treatment caused a slight decrease in the rate of long-lived protein degradation (~10%, p=0.007) under nutrient-rich conditions and a strong reduction in the rate of long-lived protein degradation (~37%, p=5E-6) after nutrient withdrawal; and this effect of Atg14L siRNA treatment was diminished in the presence of 3-methyladenine (3MA), an inhibitor of autophagy (Fig. 2b).
Fifth, we investigated whether Atg14L modulates Vps34 kinase activity using a novel kinase assay which included Vps15/p15024. Our result showed that co-expression of FLAG-Atg14L with myc-Vps34-Vps15 plasmids resulted in Vps34 activity that was 2.5-fold as high as resulted from co-expression of control FLAG with myc-Vps34-Vps15 plasmids (Fig. 2c). Interestingly, the Atg14L-mediated stimulation of Vps34 activity occurred only when co-expressing Beclin 1. This result suggests that over-expression of Atg14L enhances Vps34 activity in a Beclin 1-dependent manner.
We observed that Atg14L-EGFP or Beclin 1-EGFP stably expressed in cells was primarily diffuse in cytoplasm (Fig. S5d), which is consistent with the previous report in Beclin 1-EGFP transgenic tissues25. However, co-expression of Atg14L-EGFP and Beclin 1-AsRed resulted in their colocalization on punctate structures (Fig. 2d). Electron microscopy (EM) analysis of these Atg14L-EGFP and Beclin 1-AsRed co-transfected cells revealed many large size “organelles” (~3–5 μm) (Fig. 2e). Some of these structures display concentric “rings” with double membranes (Fig. 2e1, asterisks); many are large vacuole-like structures filled with materials of high electron density (Fig. 2e2, asterisks) and enwrapped with double-membranes (Fig. 2e2 inset), which are readily distinguishable from typical aggresomes or protein aggregates (usually not associated with limiting membranes). These structures are positive for Atg14L-EGFP, as shown by immuno-EM (Fig. 2e4). We also observed increased number of autophagosomes in these transfected cells (Fig. 2e3, blue arrows).
We next studied the nature of these Beclin 1-Atg14L-resided structures. We found that these structures were negative for Golgi (Fig. S5e) or ER (Fig. S5f) markers. In contrast, they were co-localized with GFP-LC3 (Fig. S5g), suggesting that these Beclin 1-Atg14L structures likely recruit LC3. Moreover, they were co-localized with co-expressed EGFP-Atg12 (Fig. 2f) or EGFP-Atg5 (Fig. 2g), suggesting that these Beclin 1-Atg14L structures may be involved in the early steps of autophagosome biosynthesis by recruiting Atg12 and Atg5.
To investigate the role of Rubicon in autophagy, we knocked down endogenous Rubicon protein levels. In contrast to Atg14L or Beclin 1 siRNA, Rubicon siRNA caused reduced steady-state levels of LC3II and p62/SQSTM under normal or nutrient starvation conditions (Fig. 3a), suggesting that knock-down of Rubicon promotes autophagic activity. Conversely, in cells stably or transiently transfected with Rubicon-EGFP, the p62/SQSTM1 protein levels were markedly enhanced as compared to those in the cells transfected with EGFP (Fig. 3b), suggesting that over-expression of Rubicon inhibits autophagy.
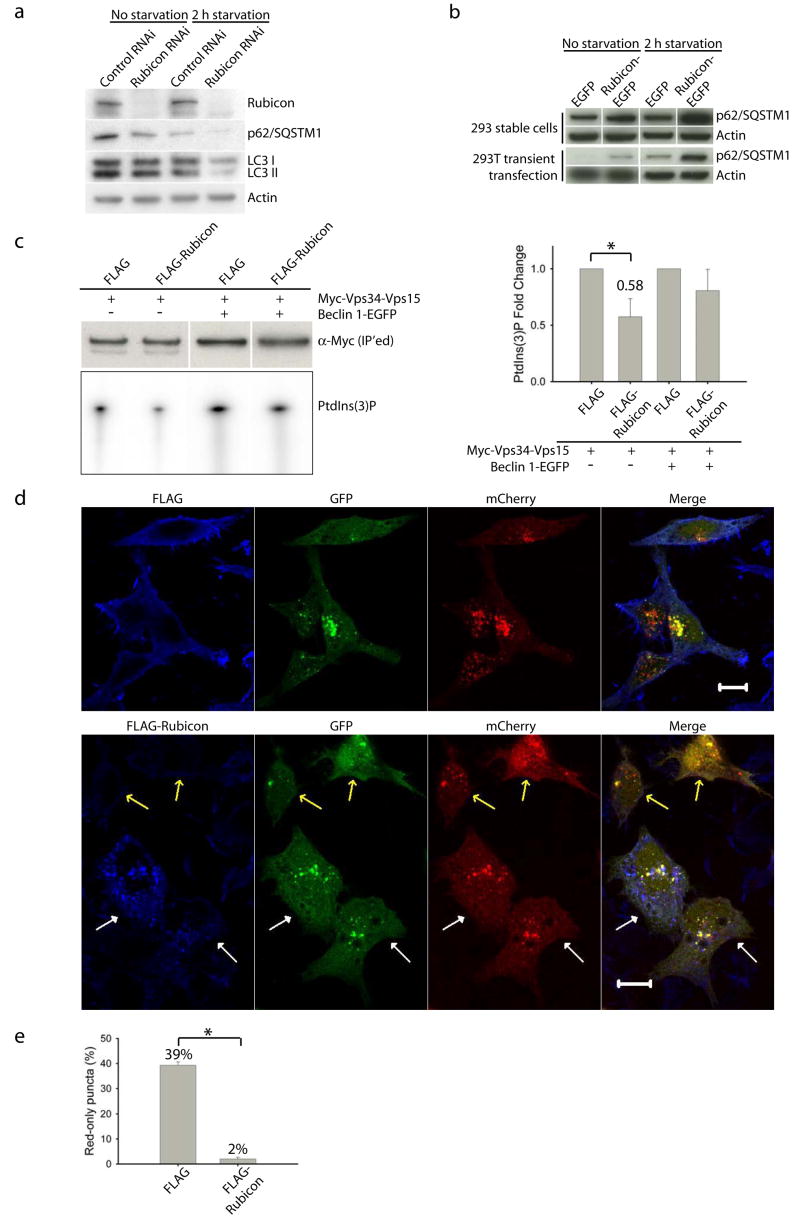
(a) Rubicon siRNA treatment of the NIH 3T3 cells led to decreased levels of p62 and LC3 II under both normal and nutrient-starvation conditions.
(b) Over-expression of Rubicon resulted in increased levels of p62 under both normal and nutrient-starved conditions in HEK 293 cells either stably expressing (upper panel) or transiently transfected with (lower panel) Rubicon-EGFP. The control cells were either stably expressing or transiently transfected with the EGFP-N3 vector.
(c) Vps34 kinase activity. HEK 293T cells were co-transfected with myc-Vps34-Vps15 and FLAG-Rubicon or FLAG vector, either in the absence or in the presence of Beclin 1-EGFP. Myc-Vps34-Vps15 was immunoprecipitated by anti-myc antibody and used for the in vitro kinase assay. The resulting radioactive PtdIns(3)P was separated by TLC, quantified and normalized against the amount of immunoprecipitated myc-tagged Vps34 as measured by Western blot (upper left panel). The quantified results (right panel) showed that over-expressing Rubicon significantly down-regulated the Vps34 kinase activity to 0.58 fold, but only when without Beclin 1 over-expression (asterisk, p=0.04 using one-tailed Student’s t-test with unequal variances, n=4).
(d) Effect of over-expressing FLAG-Rubicon on autophagosome acidification, as monitored by mCherry-GFP-LC3 fluorescence. HeLa cells were transiently co-transfected with mCherry-GFP-LC3 and FLAG-Rubicon (or control FLAG vector). Cells co-expressing mCherry-GFP-LC3 and control FLAG vector contained many red-only puncta along with yellow (indicating presence of both red and green) puncta, suggesting both autolysosomes and nascent autophagosomes (upper panel). In contrast, cells co-expressing mCherry-GFP-LC3 and FLAG-Rubicon contained primarily yellow puncta, suggesting only nascent autophagosomes (lower panel, white arrows). Notably, some cells, which were co-transfected with mCherry-GFP-LC3 and FLAG-Rubicon but expressed high levels of mCherry-GFP-LC3 and undetectable FLAG-Rubicon, contained many red-only puncta (low panel, yellow arrows).
(e) Quantitation of the results in (d) showed that over-expressing FLAG-Rubicon drastically reduced the percentage of red-only puncta (mCherry-LC3) from 39% in the control FLAG vector-transfected control cells to 2% in the FLAG-Rubicon-transfected cells (asterisk, p=2E-26, one-tailed Student’s t-test with unequal variances, n=30), indicating that over-expression of Rubicon blocks autophagosome acidification or maturation.
To examine whether Rubicon also modulates the Vps34 lipid kinase activity, we performed the above described lipid kinase assay. Our result showed that co-expression of FLAG-Rubicon with myc-Vps34-Vps15 resulted in remarkably reduced Vps34 activity, but only in the absence of Beclin 1-EGFP over-expression (Fig. 3c). This result suggests that over-expression of Rubicon inhibits the Vps34 kinase activity, and this effect does not require Beclin 1..
Previously mCherry-GFP-LC3 has been used to examine autophagosome maturation, e.g., autophagosome acidification following fusion with late endosomes/lysosomes26. We found that cells co-expressing mCherry-GFP-LC3 and FLAG-Rubicon contained primarily yellow fluorescent mCherry-GFP-LC3 puncta (immature autophagosomes) (Fig. 3d, lower panel, white arrows), whereas cells expressing only mCherry-GFP-LC3 (Fig. 3d, lower panel, yellow arrows) or co-expressing mCherry-GFP-LC3 and control vector FLAG (Fig. 3d, upper panel) contained considerable numbers of red fluorescent mCherry-GFP-LC3 puncta (mature autophagosomes) (Fig. 3e). This result suggests that over-expression of Rubicon may block autophagy through inhibiting autophagosome maturation.
Interestingly, Rubicon-EGFP (or FLAG-Rubicon, data not shown) expression exhibited punctate subcellular localization (Fig. 4a). While the Rubicon-EGFP puncta (some were “ring”-shaped), were occasionally labeled with the early endosomal marker EEA1 (Fig. S5h), they were primarily co-localized with the late endosomal/lysosomal marker Lamp1 (Fig. 4a). Moreover, some of the Rubicon-EGFP puncta were positively stained with an antibody against lysobisphosphatidic acid (LBPA) (Fig. 4b), an unusual eukaryotic lipid found only in multi-vesicular body (MVB)27, suggesting that some of the Rubicon-EGFP structures may be related to MVB28. EM analysis of Rubicon-EGFP-transfected cells showed many abnormal large vacuole-like structures (1–5 μm in diameter) (Fig. 4c). Some of these structures contained high electron density (Fig. 4c1–2, orange arrows), characteristic of late endosomes/lysosomes; some had relatively less content with overall low electron density, which may represent enlarged early stage endosomes (Fig. 4c3, black arrows); notably, some appeared to enclose numerous small vesicles of multiple-layers (Fig. 4c2&4, purple arrows), while others resembled MVB28 (Fig. 4c1–2, blue arrows). Through immuno-EM using anti-GFP gold particles, we observed that these vacuole-like structures in the Rubicon-EGFP-transfected cells were positive for Rubicon-EGFP; moreover, Rubicon-EGFP was associated with the limiting membranes of these particular structures (Fig. 4d3–4). Therefore, these structures corresponded to the fluorescent Rubicon-EGFP puncta (Fig. 4a). In addition, our immuno-EM result also confirmed the co-localization of Rubicon-EGFP and Lamp1 at ultrastructural level (Fig. 4d5–7).
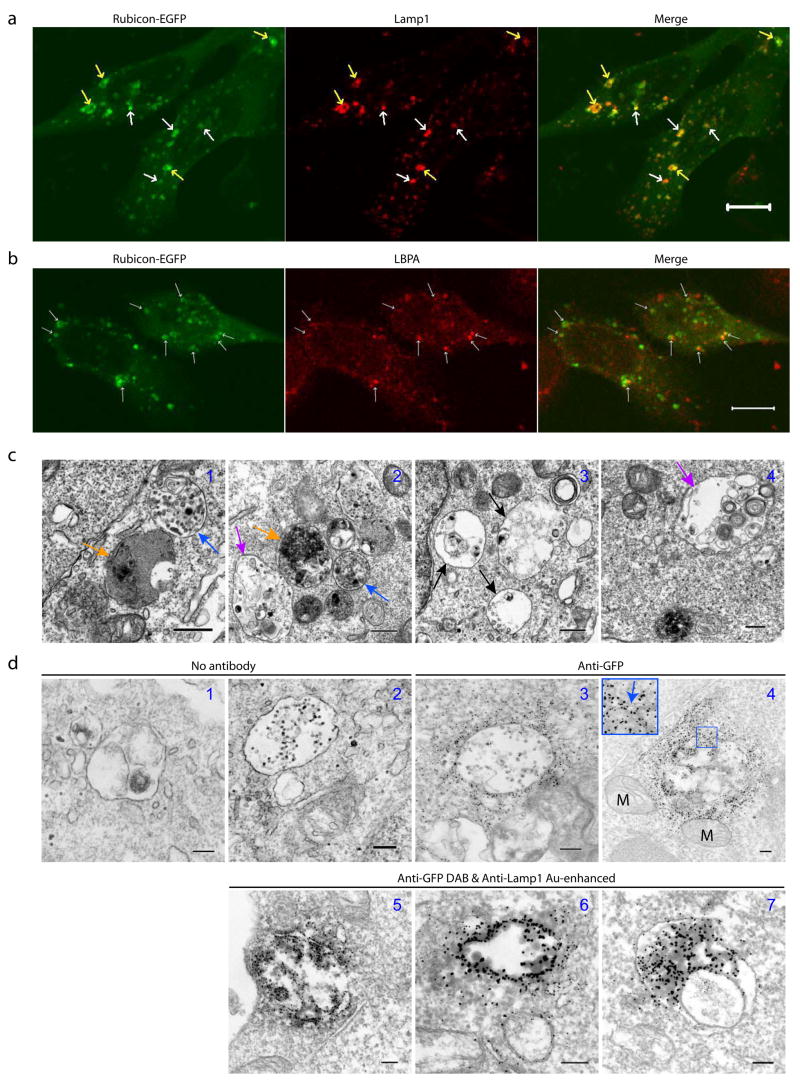
(a) Co-localization of Rubicon-EGFP-associated structures (green) with the late endosome/lysosome marker Lamp1 (red) (arrows) in the HeLa cells that were transfected with Rubicon-EGFP. Note that some of the Rubicon-EGFP-associated structures displayed “ring”-shape (yellow arrows). Scale bar: 10 μm.
(b) Partial co-localization of Rubicon-EGFP-associated structures (green) with the MVB marker LBPA (red) (arrows) in the HeLa cells that were transfected with Rubicon-EGFP. Scale bar: 10 μm.
(c) Representative ultrastructural images show aberrant expansion of late endosomes/lysosomes structures in the HEK 293T cells over-expressing Rubicon-EGFP. These abnormal organelles are large in size and contained high (orange arrows) or low (black arrows) electron density. Some enclose small vesicles (purple arrows) and some resemble MVB (blue arrows). Scale bars: 500 nm.
(d) Representative ultrastructural images showe late endosome/lysosome-like structures that are labeled by anti-GFP gold particles (panels 3–4) in the HEK 293T cells transiently transfected with Rubicon-EGFP. These structures are enwrapped by double membranes (panel 4 inset) and co-labeled by anti-GFP (developed by DAB) and anti-Lamp1 (gold enhanced) (panels 5–7). Note that mitochondria are mostly negative for Rubicon-EGFP (panel 4). The negative controls are without antibody (panels 1–2). M – mitochondria. Scale bars: 200 nm.
Bioinformatic analysis revealed that the cysteine-rich domain of Rubicon shares sequence homology to the FYVE domain (Fig. 5a), a well-characterized motif specific for PtdIns(3)P binding29. When examined experimentally, unlike the control PtdIns(3)P-binding protein 2×FYVE-EGFP, Rubicon-EGFP was not pulled down by PtdIns(3)P-conjugated sepharose beads (data not shown). However, co-expressed Rubicon-AsRed and p40 (phox)-PX-EGFP, another reporter for PtdIns(3)P binding, showed extensive co-localization (Fig. 5b, upper), suggesting that the Rubicon-associated structures are enriched in PtdIns(3)P. Moreover, wortmanin, an inhibitor of Vps34 kinase, effectively dispersed the p40 (phox)-PX-EGFP puncta but not the Rubicon-AsRed structures (Fig. 5b, lower), suggesting that the maintenance of these Rubicon-associated structures does not depend on PtdIns(3)P. Furthermore, through immunofluorescent imaging of Rubicon truncation mutants, we found that the cysteine-rich domain of Rubicon is required for the formation of the Rubicon-positive structures that are enriched in PtdIns(3)P and associated with the aberrant endosomes/lysosomes (Fig 5c). Finally, we found that the distribution of Beclin 1-AsRed or FLAG-Beclin 1-CE was exclusive from Rubicon-EGFP puncta (Fig. 5d-e); and Beclin 1 siRNA treatment did not affect the formation of the Rubicon-EGFP puncta (Fig. 5f). Therefore, the formation of these Rubicon-associated late endosomal-lysosomal structures are Beclin 1-independent.
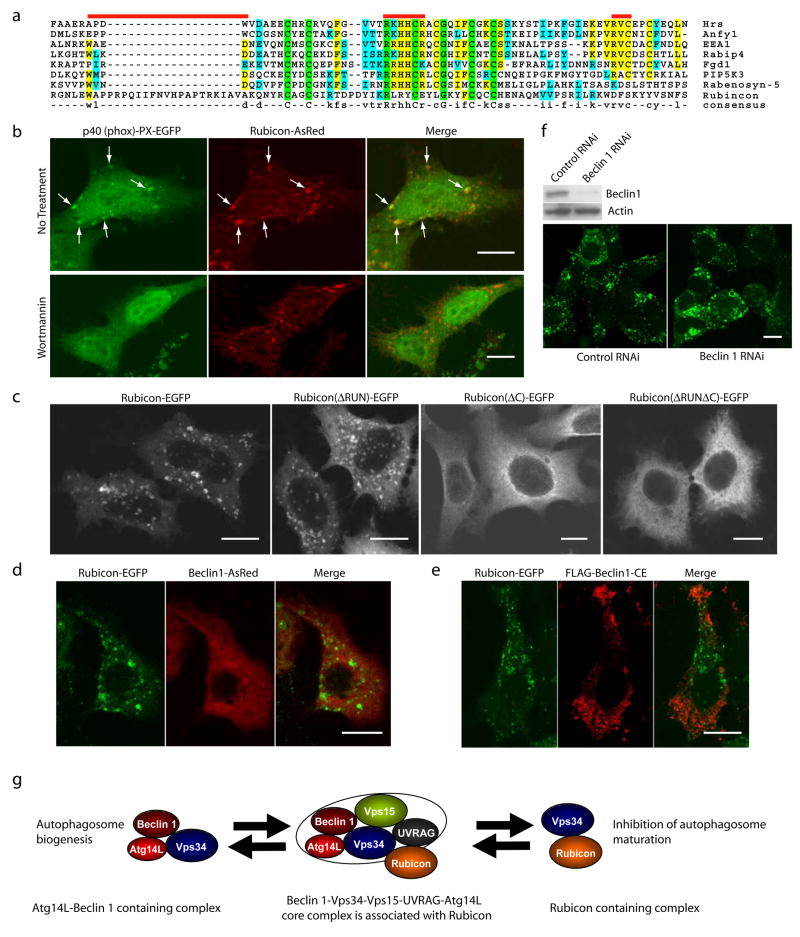
(a) Local sequence alignment between the C-terminal cystein-rich domain of Rubicon and FYVE domains of several known FYVE-containing proteins. Rubicon does not possess the key consensus sequences of a typical FYVE domain, i.e., N-terminal WxxD, central R[R/K]HHCR and C-terminal RVC (indicated by red bars).
(b) Co-localization of the PtdIns(3)P-enriched lipid domain marker p40 (phox)-PX-EGFP (green) and Rubicon-AsRed (red) on large punctate structures (arrows) in the co-transfected HeLa cells (upper panels). Upon short treatment with 75 nM wortmannin (another PI-3K inhibitor) for 1 h, while the PtdIns(3)P-enriched lipid domains disappeared, the Rubicon-AsRed-positive structures was maintained (lower panels). Scale bars: 10 μm.
(c) Subcellular localization of transiently transfected Rubicon-EGFP, Rubicon(ΔRUN)-EGFP, Rubicon(ΔC)-EGFP or Rubicon(ΔRUNΔC)-EGFP in HeLa cells. In contrast to punctate Rubicon-EGFP and Rubicon(ΔRUN)-EGFP, Rubicon(ΔC)-EGFP and Rubicon(ΔRUNΔC)-EGFP were dispersed in cytoplasm. Abbreviations: ΔRUN – RUN domain deletion, ΔC – cysteine-rich domain deletion. Scale bars: 10 μm.
(d-e) Absence of full-length Beclin 1 (d) or Beclin 1-CE mutant (e) (red) on the Rubicon-EGFP-positive structures (green) in the HEK 293 cells stably expressing Rubicon-EGFP. These cells were transiently transfected with either (d) Beclin 1-AsRed or (e) FLAG-Beclin 1-CE (i.e., the FLAG-tagged Beclin 1 mutant containing both CCD and ECD that mediate the Beclin 1-Rubicon interaction as shown in Fig. S3h). Scale bars, 10μm.
(f) The formation of the Rubicon-EGFP-positive structures was not affected by siRNA knocking-down of Beclin 1 in the HEK 293 cells stably expressing Rubicon-EGFP.
(g) A hypothetic model for the Beclin 1-Vps34protein complexes and their functions#. In this model, a core Beclin 1 complex is composed of Vps34/PI-3K, p150/Vps15, Beclin 1, UVRAG and likely substoichiometric Atg14L (indicated by the tight binding and functional connection between Atg14L and Beclin 1). Under physiological conditions, a large Beclin 1-Vps34 complex is formed, including the core complex and Rubicon. This large complex may be reduced to form smaller complexes such as a Atg14L-Beclin 1-containing complex and a Rubicon-containing complex. These smaller complexes are likely the functional units participating in autophagy regulation through modulating the Vps34 lipid kinase activity.
In summary, our study uncovers Atg14L and Rubicon, two novel components in the Beclin 1-Vps34 protein complexes, and reveals their distinct roles in regulating autophagy and Vps34 kinase activity. We show that Atg14L and Rubicon may regulate autophagy via modulating Vps34 activity. Our study also suggests the existence of multiple Beclin 1 protein complexes that are engaged in distinct functions in autophagy regulation (Fig. 5g). The significance of these distinct Beclin 1 complexes remains to be fully elucidated. The dynamic change in protein composition between different functional Beclin 1-Vps34 complexes may play a central role in mediating the Beclin 1-Vps34 activity that governs multiple cellular events including autophagy.
Materials and Methods
Reagents and antibodies, details of mouse genetics, affinity purification and mass spectrometric analysis, and microscopy are available in the Supplemental Information.
Mouse genetics
Becn1-EGFP/+ mice were generated using BAC mouse transgenics30. Becn1-EGFP/+ transgenic and Becn1+/− mice were genetically crossed to generate the “rescued” mice, in which both endogenous Becn1 alleles are deleted and only Becn1-EGFP transgene is expressed.
Affinity purification and mass spectrometric analysis
Affinity purification of Beclin 1 interacting proteins and mass spectrometric identification of these proteins were carried out as described in Wang et al.(2006)19 with slight modification.
Plasmid constructs and stable cell lines
Total RNA was extracted from postnatal day 12 mouse whole brain using RNeasy mini kit (Qiagen, USA). Full length cDNA was synthesized with Omniscript RT kit (Qiagen, USA) and used as templates for PCR amplifications with KOD HiFi DNA polymerase (Novagen, USA). Becn1 was cloned into EcoRI and BamHI sites of pEGFP-N3 and pAs-Red vectors (Clonetech, USA). Atg14L was cloned into EcoRI and BamHI sites of pEGFP-N3, pAsRed and pCMV-FLAG (Sigma, St Louis, MO) vectors. Rubicon was cloned into HindIII and BamHI sites of pEGFP-N3, pAs-Red and pCMV-FLAG2 vectors. UVRAG was cloned into XhoI and BamHI sites of pEGFP-N3 vector. Single or combinations of Becn1 domains were cloned into EcoRI and BamHI sites of pCMV-FLAG2 vector. Truncated Atg14L mutants were cloned into EcoRI and BamHI sites of pEGFP-N3 vector. Truncated Rubicon mutants were cloned into KpnI and BamHI sites of pEGFP-N3 vector. EGFP-Atg12 and EGFP-Atg5 constructs were generously provided by Dr. X. Jiang. Myc-hVps34-hVps15-V5-His/pVITRO2 plasmid was described previously24. HEK 293 stable cells stably transfected with pEGFP-N3 vector, Beclin 1-EGFP, Atg14L-EGFP or Rubicon-EGFP were generated as described in the online Supplemental Information.
Cell cultures
Human embryonic kidney (HEK) 293 and 293T, Hela and NIH 3T3 cells were maintained in standard medium, i.e., Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA). MLE12 cells were generously provided by Dr. C Münz and maintained in DMEM/F12 medium (ATCC, Manassas, VA) supplemented with 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM beta-estradiol, 10 mM HEPEs, 2 mM L-glutamine, 2% FBS and 1% penicillin-streptomycin. Transient DNA transfection was performed with standard calcium phosphate precipitation procedure, FuGene 6 or Lipofectamine 2000 kit, following protocol provided by the manufacturers. SiRNA transfection of NIH 3T3 cells and GFP-LC3 MLE12 stable cells was performed with Lipofectamine RNAi MAX kit following the reverse transfection protocol provided by the manufacturer. The sequences of siRNA are: Beclin 1, CAGUUUGGCACAAUCAAUA; Atg14L, UUUGCGUUCAGUUUCCUCACUGCGC; and Rubicon, GCCUUCAGUCUAUGCCACA.
In vitro protein immunoprecipitation
DNA plasmids were transfected into HEK 293T cells. For co-IP experiments, two or three plasmids were transfected simultaneously in equal amount. Cells were lysed in IP lysis buffer (20 mM HEPEs/pH7.4, 1 mM MgCl2, 0.25 mM CaCl2, 0.2% triton X-100, 150mM NaCl, EDTA-free protease inhibitor cocktail (PIC), 200 μg/mL phenylmethylsulfonyl fluoride (PMSF), pepstatin 4 μg/mL, and DNase I). For IP with GFP, Atg14L or Rubicon antibody, Dynabeads M-270 E-proxy (Invitrogen, Carlsbad, CA) were conjugated with each antibody, and incubated with cell lysate at 4°C for 2 h. After the beads were washed in IP lysis buffer for 5 times, proteins were eluted by incubating beads in elution buffer (0.5 mM EDTA/pH8 and 0.5 M NH3·H2O) at room temperature for 20 min, frozen in liquid nitrogen and dried with vacuum speed centrifuge. For FLAG-tagged protein IP, anti-FLAG M2 affinity resin (Sigma, St Louis, MO) was used and the manufacture’s protocol was followed.
Vps34 kinase assay
Vps34 kinase assay was performed as described31. Myc-hVps34-hVps15-V5-His/pVITRO2 plasmid was transfected into HEK 293T cells in combination with other FLAG- or EGFP-tagged plasmids. Cells were lysed in 1% Nonidet P-40 lysis buffer (20 mM Tris/pH 7.5, 137 mM NaCl, 1 mM MgC12, 1 mM CaC12, 100 mM NaF, 10 mM sodium pyrophosphate, 100 μM Na3VO4, 10% glycerol, 0.35 mg/ml PMSF, protease and phosphatase inhibitor cocktails). IP was performed with anti-myc affinity gel beads, following manufacture’s protocol. Beads (associated with purified proteins) were washed in lysis buffer three times, followed by three washes in washing buffer (100 mM Tris-HCl/pH7.4 and 500 mM LiCl) and two washes in reaction buffer (10 mM Tri-HCl/pH7.4, 100 mM NaCl and 1 mM EDTA). Beads were resuspended in 60 μl of reaction buffer, followed by addition of 10 μ1 of 100 mM MnCl2 and 10 μl of sonicated phosphatidylinositol (2 μg/μl). The reaction was started by the addition of 10 μl of 440 μM ATP containing 10 μCi of γ-32P-ATP, and beads were incubated for 10 min at room temperature. Reaction was terminated by adding 20 μl 8 M HCl, and organic phase was extracted with 160 μl chloroform:methanol (1:1). Extracted phospholipid products were resolved by TLC using a coated silica gel and a solvent composed of chloroform:methanol:H2O:ammonium hydroxide (v/v/v/v, 9:7:1.7:0.3), followed by visualization with Typhoon 9400 Variable Imager (GE Healthcare Biosciences, Piscataway, NJ).
Gel Filtration
Liver and brain extracts from both Becn1+/− and Becn1−/−; Becn1-GFP/+ mice (4 months of age) were prepared as described in the online Supplemental Information. Cell extracts were prepared as previously described19. Tissue and cell extracts were diluted with equal volumes of 2× pull-out buffer (1× containing 20 mM HEPEs/pH 7.4, 1 mM MgCl2, PIC, 100 μg/mL PMSF, 2 μg/mL pepstatin, 0.2% triton X-100 and 150 mM NaCl) and incubated for 15 min at 4°C. The samples were then subject to ultracentrifugation at 100,000 g and the resulting supernatants were used for gel filtration experiments. Superdex 200 HR10/30 column (Pharmacia) was equilibrated with 2 bed volumes of filtered running buffer (1× pull-out buffer less PIC and triton). The column was calibrated using Biorad gel filtration calibrant mixtures that are composed of thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1,350 Da). A spike of these calibrants (10 μl) was also added to each sample (240 μl) as internal calibrants. Both calibrants and samples were run at a flow rate of 0.2 ml/min. For each run, 2 bed volumes of running buffer were used to elute the sample and a total of 80 fractions were collected 25–29 min after starting the runs and at a rate of 1 fraction/min. Two bed volumes of running buffer were used to wash the column at the same flow rate in between two consecutive runs.
Long-lived protein degradation assay
Long-lived protein degradation was accessed as described32. In brief, NIH 3T3 cells were transfected with either control or Atg14L siRNA and plated in 12-well plates. After 48 h, standard medium was changed to leucine-free medium supplemented with 1 μCi/ml 3H-L-leucine. After being pulse-labeled for 24 h, cells were washed 3 times and further cultured in standard medium supplemented with excess unlabeled leucine (5 mM) for 16 h to chase out short-lived proteins. Cells were then washed 3 times and further cultured for 4 h in standard medium, Earle’s Balanced Salt Solution (EBSS), or EBSS supplemented with 10 mM 3-MA, all containing 5 mM unlabeled leucine. Both media and cell lysate were subject to trichloroacetic acid (TCA) precipitation. Long-lived protein degradation was calculated as the ratio of TCA-soluble medium radioactivity to TCA-precipitated cell lysate radioactivity.
Acknowledgments
This work was supported by National Institutes of Health Grants RNS055683A (Z.Y.), RR00862 and RR022220 (B.T.C.) and by the Howard Hughes Medical Institute (N.H.). We thank Dr. A. Tolkovsky for GFP-LC3 HeLa cells, Dr. C. Münz for GFP-LC3 MLE12 cells, Dr. T. Johansen for mCherry-EGFP-LC3 plasmid, Dr. X. Jiang for EGFP-Atg12 and EGFP-Atg5 constructs. We thank W. Yang for help with cell culture and generation of stable cells, and H. Shio, E. Sphicas and Dr. A. North in the Bio-Imaging Resource Center at The Rockefeller University for help with microscopy.
Footnotes
Author contributions
Z. Yue conceived the project and coordinated all efforts in the study; Z. Yue and Q. J. Wang, Y. Zhong, N. Heintz and B.T. Chait planned the project; Y. Zhong and Q. J. Wang performed most of the assays; X. Li assisted in p40 (phox)-PX-EGFP localization and Vps34 kinase analyses; Y. Yan and J. M. Backer developed the myc-Vps34-Vps15 constructs and Vps34 kinase assay protocol; Z. Yue, Q. J. Wang, Y. Zhong wrote the paper.
#Note that this hypothetical model does not mean to propose direct binary interactions.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb1854
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2664389
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ncb1854
Article citations
Promising and challenging phytochemicals targeting LC3 mediated autophagy signaling in cancer therapy.
Immun Inflamm Dis, 12(10):e70041, 01 Oct 2024
Cited by: 1 article | PMID: 39436197 | PMCID: PMC11494898
Review Free full text in Europe PMC
The Rubicon-WIPI axis regulates exosome biogenesis during ageing.
Nat Cell Biol, 26(9):1558-1570, 22 Aug 2024
Cited by: 2 articles | PMID: 39174742
Epigallocatechin-3-gallate protects bovine ruminal epithelial cells against lipopolysaccharide-induced inflammatory damage by activating autophagy.
J Anim Sci Biotechnol, 15(1):109, 09 Aug 2024
Cited by: 0 articles | PMID: 39118120 | PMCID: PMC11311925
The Role of LC3-Associated Phagocytosis Inhibits the Inflammatory Response in Aspergillus fumigatus Keratitis.
Invest Ophthalmol Vis Sci, 65(8):4, 01 Jul 2024
Cited by: 0 articles | PMID: 38953845 | PMCID: PMC11221612
Egg multivesicular bodies elicit an LC3-associated phagocytosis-like pathway to degrade paternal mitochondria after fertilization.
Nat Commun, 15(1):5715, 08 Jul 2024
Cited by: 0 articles | PMID: 38977659 | PMCID: PMC11231261
Go to all (643) article citations
Other citations
Wikipedia
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels.
J Biol Chem, 289(38):26021-26037, 01 Aug 2014
Cited by: 49 articles | PMID: 25086043 | PMCID: PMC4176228
Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.
Mol Biol Cell, 19(12):5360-5372, 08 Oct 2008
Cited by: 731 articles | PMID: 18843052 | PMCID: PMC2592660
Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex.
PLoS Genet, 10(10):e1004626, 02 Oct 2014
Cited by: 78 articles | PMID: 25275521 | PMCID: PMC4183436
The Beclin 1 network regulates autophagy and apoptosis.
Cell Death Differ, 18(4):571-580, 11 Feb 2011
Cited by: 1377 articles | PMID: 21311563 | PMCID: PMC3131912
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (5)
Grant ID: P41 RR000862
Grant ID: RR022220
Grant ID: P41 RR000862-36
Grant ID: RR00862
Grant ID: U54 RR022220
NINDS NIH HHS (3)
Grant ID: R01 NS060123-01A1
Grant ID: R01 NS060123
Grant ID: R21 NS055683
PHS HHS (1)
Grant ID: RNS055683A





