Abstract
Free full text
Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR® Yfiler® PCR amplification kit
Associated Data
Abstract
The Y-chromosomal short tandem repeat (Y-STR) polymorphisms included in the AmpFlSTR® Yfiler® polymerase chain reaction amplification kit have become widely used for forensic and evolutionary applications where a reliable knowledge on mutation properties is necessary for correct data interpretation. Therefore, we investigated the 17 Yfiler Y-STRs in 1,730–1,764 DNA-confirmed father–son pairs per locus and found 84 sequence-confirmed mutations among the 29,792 meiotic transfers covered. Of the 84 mutations, 83 (98.8%) were single-repeat changes and one (1.2%) was a double-repeat change (ratio, 1:0.01), as well as 43 (51.2%) were repeat gains and 41 (48.8%) repeat losses (ratio, 1:0.95). Medians from Bayesian estimation of locus-specific mutation rates ranged from 0.0003 for DYS448 to 0.0074 for DYS458, with a median rate across all 17 Y-STRs of 0.0025. The mean age (at the time of son’s birth) of fathers with mutations was with 34.40 (±11.63) years higher than that of fathers without ones at 30.32 (±10.22) years, a difference that is highly statistically significant (p <
< 0.001). A Poisson-based modeling revealed that the Y-STR mutation rate increased with increasing father’s age on a statistically significant level (α
0.001). A Poisson-based modeling revealed that the Y-STR mutation rate increased with increasing father’s age on a statistically significant level (α =
= 0.0294, 2.5% quantile
0.0294, 2.5% quantile =
= 0.0001). From combining our data with those previously published, considering all together 135,212 meiotic events and 331 mutations, we conclude for the Yfiler Y-STRs that (1) none had a mutation rate of >1%, 12 had mutation rates of >0.1% and four of <0.1%, (2) single-repeat changes were strongly favored over multiple-repeat ones for all loci but 1 and (3) considerable variation existed among loci in the ratio of repeat gains versus losses. Our finding of three Y-STR mutations in one father–son pair (and two pairs with two mutations each) has consequences for determining the threshold of allelic differences to conclude exclusion constellations in future applications of Y-STRs in paternity testing and pedigree analyses.
0.0001). From combining our data with those previously published, considering all together 135,212 meiotic events and 331 mutations, we conclude for the Yfiler Y-STRs that (1) none had a mutation rate of >1%, 12 had mutation rates of >0.1% and four of <0.1%, (2) single-repeat changes were strongly favored over multiple-repeat ones for all loci but 1 and (3) considerable variation existed among loci in the ratio of repeat gains versus losses. Our finding of three Y-STR mutations in one father–son pair (and two pairs with two mutations each) has consequences for determining the threshold of allelic differences to conclude exclusion constellations in future applications of Y-STRs in paternity testing and pedigree analyses.
Electronic supplementary material
The online version of this article (10.1007/s00414-009-0342-y) contains supplementary material, which is available to authorized users.
Introduction
A reliable knowledge of the particular Y-chromosomal short tandem repeat (Y-STR) polymorphisms used in the forensic context is essential for the correct interpretation of the resulting profiles. Over the last years, 17 Y-STRs included in the commercially available AmpFlSTR® Yfiler® polymerase chain reaction (PCR) amplification kit (Applied Biosystems, Inc., Foster City, CA, USA) have become widely used in the forensic genetic community as well as for evolutionary anthropological studies. Establishing a reliable knowledge on the mutation rates and characteristics of these particular 17 Y-STRs included in the kit are important for particular forensic and anthropological applications. In forensics, mutation rates are needed when STRs are applied to paternity testing, and Y-STRs are especially powerful in deficiency cases of disputed paternity involving male offspring where the alleged father is not available for DNA analysis but is replaced by any of his male paternal relatives. In such applications, the knowledge on Y-STR mutation rates needs to be considered in the paternity probabilities, and mutations are more likely the more generations the son is separated from its putative male paternal relative [1]. There are also other forensic applications where Y-STR mutation rates have to be considered, i.e., all those that include different members of the same male lineage. In evolutionary anthropological studies, Y-STRs are usually applied to unveil the local and temporal origin of a given Y-SNP based haplogroup, and Y-STR mutation rates are used for time estimations as well as (often) for weighted network constructions [2]. In addition, Y-STRs are useful in genealogical studies where mutation data are needed as well [3].
There are several approaches to establish Y-STR mutation rates including genotyping father–son pairs from trio cases of autosomal DNA confirmed paternity [4], males from deep-rooted pedigrees [5], single sperm cells or small pools of sperm cells [6], or using Y-STR population data in combination with known historical events for time calibration [7]. Of these, studying DNA-confirmed father–son pairs is the most reliable approach but only if the number of father–son pairs investigated is large enough to reveal reliable mutation rate estimates. This is because mutation rates of STRs, including Y-STRs, are expected to be small (about one mutation in 1,000 generations per locus). It is therefore important to further increase the number of father–son pairs typed for the specific Y-STR loci intended to be applied for forensic and evolutionary analyses to provide more reliable knowledge about their mutability and thus to further gain certainty in Y-STR data interpretation.
Several studies have investigated mutation rates and characteristics of Y-STR loci widely used in forensic, genealogical, and evolutionary studies [4,5,8–23]. However, the mutation information for some of the Y-STRs included in the Yfiler kit is still very limited as most of the Y-STR mutation rate studies so far were conducted on a subset of markers included in Yfiler kit (e.g., the nine Y-STRs defining the so-called minimal haplotype). Only six previous studies investigated the complete set of 16 Yfiler Y-STR loci (DYS385a/b was considered jointly) in father–son pair analyses covering all together only 1,624 meiotic transfers per single locus [16,18,19,21–23]. In this paper, we report mutation data for the 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR amplification kit from analyzing 1,730–1,764 father–son pairs per locus, comprising a total of 29,792 meiotic transfers (mutations at DYS385a and DYS385b were analyzed separately) and representing the largest single Yfiler mutation study available thus far. We additionally provide summarized mutation data from our study and previously published data for the same 16 Y-STR loci (DYS385a/b considered as combined system) comprising 3,531–11,900 meiotic transfers per each of the Y-STR loci (all together 135,212 meiotic transfers).
Materials and methods
Father–son pair samples used in this study were confirmed in their family relationship by DNA analysis using various sets of DNA markers before this study, and all had paternity probabilities of >99.9%. Samples came from five sampling regions: Cologne, Leipzig, and Berlin in Germany as well as Warsaw and Wroclaw in Poland. Individuals came from the named cities as well as their surrounding regions, i.e., provinces/counties these cities are part of. Although the vast majority will have originated from the respective geographic regions, we cannot exclude some migrants from other regions. If known, individuals of origin from countries other than those considered in the respective regional sample sets were excluded from the study. There is no sample overlap between the present study and our previously published mutation study [4]. Because of very low DNA quantities available for the Leipzig samples, a whole genome amplification procedure was performed before Yfiler PCR analysis using the GenomiPhi DNA Amplification Kit (GE Healthcare, Little Chalfont, UK). One or 5 µl (depending on DNA concentration) genomic DNA were added to 9 µl of sample buffer and denatured at 95°C for 3 min, then cooled on ice. Subsequently, 9-µl reaction buffer plus 1 µl of enzyme mix were added to the cooled sample and incubated at 30°C for 16–18 h, then heat inactivated at 65°C for 10 min. Afterwards, the whole-genome-amplified DNA was purified using Invisorb® 96 Filter Microplates (Invitek GmbH, Berlin, Germany).
The Y-STRs included in the AmpFlSTR® Yfiler® PCR amplification kit (Applied Biosystems, Inc.): DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS348, DYS439, DYS448, DYS456, DYS458, DYS635, and Y-GATA-H4 were genotyped according to the instructions provided by the manufacturer and using a gold-plated silver block GeneAmp®PCR System 9700 (Applied Biosystems, Inc.). All PCRs, except for the Berlin samples, were carried out at the Department of Forensic Molecular Biology, Erasmus MC Rotterdam (The Netherlands), and after quality control, PCR products were shipped on dry ice to Applied Biosystems at Foster City (USA), where fragment length analyses was performed using the 3130xl genetic analyzer according to the guidelines in the AmpFlSTR® Yfiler® PCR amplification kit user manual. Yfiler profiles were generated using Genemapper ID v3.2 software (Applied Biosystems Inc.), and generated profiles were manually inspected by experienced technicians in Rotterdam for quality control. The Berlin samples were genotyped at the Abteilung für Forensische Genetik, Institut für Rechtsmedizin und Forensische Wissenschaften, Charité (Germany) according to the manufacturer’s instructions. Genotype differences between respective fathers and sons were identified using in-house developed MATLAB®-scripts using version 7.6.0.324 (The MathWorks, Inc., Natick, MA, USA).
All mutations were confirmed by DNA sequence analysis of the respective father and son DNA sample at the respective Y-STR locus in Rotterdam. Mutations at the DYS385a/b system were sequenced separately for DYS385a and DYS385b as described elsewhere [24]. Before DNA sequence analysis, PCR was carried out using the following conditions: 10–20 ng genomic DNA was used in a total volume of 20 μl PCR reaction. Final concentrations were 1× PCR GeneAmp PCR gold buffer and 0.5–1 unit AmpliTaq Gold (Applied Biosystems Inc.), 1 mM deoxyribonucleotide triphosphates (dNTPs; Roche Diagnostics GmbH, Mannheim, Germany), 250 nM of each primer (see Supplementary Table S1 for primer sequences used for sequencing as well as for PCR before sequence analysis) and 1.5–2.5 mM MgCl2 depending on the marker. DYS393, DYS439 (2.5 mM MgCl2), GATA-H4, DYS385a, and DYS385b (1.5 mM MgCl2) were amplified using a 60–50-touchdown protocol: 95°C 10 min, ten cycles, 94°C 30 s, 60–1°C 30 s, 72°C 45 s; 25 cycles, 94°C 30 s, 50°C 30 s, 72°C 45 s, and final extension at 72°C 10 min. The combined DYS389I/II fragment was amplified using a 60–55 touchdown protocol: 95°C 10 min, five cycles, 94°C 30 s, 60–1°C 30 s, 72°C 45 s; 30 cycles, 94°C 30 s, 55°C 30 s, 72°C 45 s, and final extension at 72°C 10 min. DYS437, DYS392, DYS438, DYS19, DYS456 (all 2.0 mM MgCl2), and DYS390 (2.5 mM MgCl2) were amplified with a 65–50 touchdown protocol; 95°C 10 min, 15 cycles, 94°C 30 s, 65–1°C 30 s, 72°C 45 s; 20 cycles 94°C 30 s, 50°C 30 s, 72°C 45 s, and final extension at 72°C 10 min. DYS635 (1.5 mM MgCl2) and DYS391 (2.0 mM MgCl2) were amplified using a 70–50 touchdown protocol: 95°C 10 min, 20 cycles, 94°C 30 s, 70–1°C 45 s, 72°C 1 min; 15 cycles, 94°C 30 s, 50°C 45 s, 72°C 1 min, and a final extension at 72°C 10 min. DYS458 (1.5 mM MgCl2) was amplified using a fixed annealing temperature of 60°C; 95°C 10 min, 35 cycles, 94°C 30 s, 60°C 30 s, 72°C 45 s, then a final extension at 72°C 10 min. DYS385a and DYS385b were amplified separately as described elsewhere [24]. Excess of PCR primers and dNTP was removed via enzymatic treatment of exonuclease I (Exo) and shrimp alkaline phosphatase (SAP) using the ExoSAP-IT™ Kit (USB Corporation, Cleveland, OH, USA) where 5 μl PCR product was incubated with 2 μl ExoSap-IT mix for 15 min at 37°C and inactivated at 80°C for 15 min, then cooled to 15°C for 5 min. DNA sequence analysis was performed via cycle sequencing in a total volume of 10 μl using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc.) and the following conditions: 1 μl ExoSAP-IT-treated PCR product, 1.5 μl sequencing buffer (Applied Biosystems Inc.), 1.0 μl BigDyeTerminator v1.1 (Applied Biosystems Inc.), 5 pmol of sequencing primer (see Supplementary Table S1 for sequences) and LiChrosolv water (Merck KGaA, Darmstadt, Germany). The cycle sequencing was performed in an MJ-Research PTC-200 (Bio-Rad, Hercules, CA, USA) by heating to 96°C for 1 min, then 25 cycles of 96°C 10 s, 50°C for 5 s and 60°C for 4 min and subsequent cooling to 15°C. The sequencing products were purified using 96-well multiscreen plates (Millipore, Billerica, MA) filled with Sephadex G-50 superfine (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) absorbed with LiChrosolv water (Merck KGaA). After spinning the column for 5 min at 2900 rpm, 10 μl sequencing product was added to the column and collected in a clean 96-well PCR plate after centrifugation for 5 min at 2900 rpm. To the purified product, 5 μl HiDi formamide (Applied Biosystems Inc.) was added and loaded on the ABI 3100 Genetic Analyzer (Applied Biosystems Inc.). Separation of the purified sequencing products was performed using capillary electrophoresis under standard conditions. DNA sequences were aligned using the DNAstar software (DNASTAR, Inc., Madison, WI, USA). Since Y-STR typing was performed by Yfiler chemistry using labeled primers and therefore DNA sequencing was performed from an independent PCR reaction, our confirmation procedure thus included two independent analyses: one Yfiler fragment-length analysis and one sequence analysis. Y-STR mutations were only accepted as such if the repeat counts from the DNA sequence analysis matched the repeat-based allele nomenclature of the Yfiler fragment length analysis. For additional confirmation, we included for all Y-STRs sequenced control DNA samples that had known size and repeat-based alleles from multiple Yfiler fragment length analyses as well as known repeat counts from multiple sequence analyses as performed previously.
Mutation rates were estimated by means of two different approaches: a frequentist approach and a Bayesian approach. Frequentist estimation of the mutation rates was conducted by dividing the number of sequence-confirmed mutations by the number of father–son pairs for every Y-STR locus and for every sampling region separately. Ninety-five percent confidence intervals of the mutation rates were established by using a binomial model given the total number of working father–son pairs and the estimated mutation rate and obtained via the website http://statpages.org/confint.html. To test for locus-specific differences in the mean of the mutation rates between sampling regions (Cologne, Leipzig, Berlin, Warsaw, and Wroclaw), a permutation analysis was carried out. In each iteration, each father–son pair was assigned at random to each sampling region, keeping the original population sample size. The average mutation rate computed for the permutated populations was then compared with the observed rate, and the number of times that the permutated averaged mutation rate was larger than the observed one was recorded. The one tail p value was obtained by dividing such numbers by the 100,000 iterations that were conducted for each locus. Overall mutation rate distributions collected from the present as well as previous studies were estimated by means of a binomial hierarchical Bayesian model [25] by using the Marcov Chain Monte Carlo (MCMC) Gibbs sampling implemented in WinBUGS [26]. A non-informative prior normal distribution (μ =
= 0, σ
0, σ =
= 1.0E−06) was specified to estimate the logit of the overall mutation rate and a prior gamma distribution with parameters α
1.0E−06) was specified to estimate the logit of the overall mutation rate and a prior gamma distribution with parameters α =
= 1.0E−5, and β
1.0E−5, and β =
= 1.0E−5 for the parameter tau as suggested in WinBUGS. Three different Gibbs MCMC chains were generated when estimating the mutation rate for each locus, and 100,000 runs were performed for each chain. Mean, median, and 95% credible intervals (CI) were estimated from the three chains after discarding the first 50,000 runs and performing a thinning of 15 in order to reduce the amount of autocorrelation (representing a final number of 9,999 retained runs). Bayesian estimations of DYS385a and DYS385b separately (as only available from our own study) were performed by using a binomial model with a uniform prior, which led to a posterior Beta distribution [25] with parameters α
1.0E−5 for the parameter tau as suggested in WinBUGS. Three different Gibbs MCMC chains were generated when estimating the mutation rate for each locus, and 100,000 runs were performed for each chain. Mean, median, and 95% credible intervals (CI) were estimated from the three chains after discarding the first 50,000 runs and performing a thinning of 15 in order to reduce the amount of autocorrelation (representing a final number of 9,999 retained runs). Bayesian estimations of DYS385a and DYS385b separately (as only available from our own study) were performed by using a binomial model with a uniform prior, which led to a posterior Beta distribution [25] with parameters α =
= m
m +
+ 1 and β
1 and β =
= n
n +
+ 1, where m is the number of mutant father–son pairs and n is the number of non-mutant father–son pairs. The ratio of repeat gains versus losses and the ratio of single- versus multi-repeat changes were estimated using a multinomial-logistic Bayesian model. For the individual studies, the relatively low number of observed counts of each class required using informative priors, which highly skewed the posterior distributions towards the prior distributions, and credible intervals tended to be large, including the 1:1 ratio (results not shown). Therefore, we did not use the Bayesian approach for such estimations. The ages (at the time of son’s birth) of fathers with and without mutations were compared with a Mann–Whitney U test. The estimation of the effect on the mutation rate of the age of the father was calculated by means of a Bayesian approach. Mutation rate was modeled as a function of each age class using a Poisson distribution:
1, where m is the number of mutant father–son pairs and n is the number of non-mutant father–son pairs. The ratio of repeat gains versus losses and the ratio of single- versus multi-repeat changes were estimated using a multinomial-logistic Bayesian model. For the individual studies, the relatively low number of observed counts of each class required using informative priors, which highly skewed the posterior distributions towards the prior distributions, and credible intervals tended to be large, including the 1:1 ratio (results not shown). Therefore, we did not use the Bayesian approach for such estimations. The ages (at the time of son’s birth) of fathers with and without mutations were compared with a Mann–Whitney U test. The estimation of the effect on the mutation rate of the age of the father was calculated by means of a Bayesian approach. Mutation rate was modeled as a function of each age class using a Poisson distribution:

where θ is the mutation rate, y
i is the number of mutations, and x
i is the number of father–son pairs for the age class i. θ is assumed to be dependent on the age of the father, with 

Results and discussion
Y-STR mutation characteristics
We investigated all together 29,792 meiotic events from analyzing 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR amplification kit (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a, DYS385b, DYS437, DYS348, DYS439, DYS448, DYS456, DYS458, DYS635, and Y-GATA-H4) in 1,730–1,764 (per locus) father–son pairs of DNA-confirmed biological paternity. Note that, although DYS385a/b was genotyped jointly as part of the Yfiler kit, mutation confirmation was performed separately for DYS385a and DYS385b (see Materials and methods), providing mutation rates separately for both DYS385 loci. We identified 84 mutations that were all confirmed by DNA sequence analysis (Table 1, Supplementary Table S2). These 84 mutations were found among 16 Y-STRs, and no mutation was observed for DYS448 among 1,746 meiotic transfers studied. Single-repeat changes were observed for 83 (98.8%) mutations, whereas one (1.2%) mutation (at DYS438) was a double-repeat change [ratio =
= 1:0.01; 95% binomial confidence interval (CIL), <0.006–1:0.037]. Among the 84 mutations, about the same number of repeat gains with 43 (51.2%) and repeat losses with 41 (48.8%) were found (ratio
1:0.01; 95% binomial confidence interval (CIL), <0.006–1:0.037]. Among the 84 mutations, about the same number of repeat gains with 43 (51.2%) and repeat losses with 41 (48.8%) were found (ratio =
= 1:0.95; 95% CIL 1:1.47–1:0.61; Table 1, Supplementary Table S2).
1:0.95; 95% CIL 1:1.47–1:0.61; Table 1, Supplementary Table S2).
Table 1
Mutation data from father–son pair analysis of 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR amplification kit
| Y-STR Locus | Number of meiotic transfers | Number of mutationsa | Number of gains | Number of losses | Number of single-repeat changes | Number of multi-repeat changes | Mutation rate | Binomial 95% confidence interval limits | Gains versus losses | Single- versus multi-repeat changes | Median mutation rate | 95% Credible interval |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequentist approach | Bayesian approach | |||||||||||
| DYS19 | 1,757 | 7 | 4 | 3 | 7 | 0 | 0.0040 | 0.0016–0.0082 | 1:0.75 | 1:0 | 0.0029 | 0.0017–0.0055 |
| DYS389I | 1,756 | 9 | 4 | 5 | 9 | 0 | 0.0051 | 0.0023–0.0097 | 1:1.25 | 1:0 | 0.0041 | 0.0021–0.0077 |
| DYS389IIb | 1,745 | 6 | 2 | 4 | 6 | 0 | 0.0034 | 0.0013–0.0075 | 1:2 | 1:0 | 0.0031 | 0.0017–0.0053 |
| DYS390 | 1,760 | 2 | 0 | 2 | 2 | 0 | 0.0011 | 0.0001–0.0041 | 0:1 | 1:0 | 0.0015 | 0.0004–0.0033 |
| DYS391 | 1,762 | 5 | 3 | 2 | 5 | 0 | 0.0028 | 0.0009–0.0066 | 1:0.67 | 1:0 | 0.0029 | 0.0015–0.0046 |
| DYS392 | 1,730 | 1 | 1 | 0 | 1 | 0 | 0.0006 | 0.0000–0.0032 | 1:0 | 1:0 | 0.0005 | 0.00004–0.0017 |
| DYS393 | 1,752 | 3 | 2 | 1 | 3 | 0 | 0.0017 | 0.0004–0.0050 | 1:0.5 | 1:0 | 0.0010 | 0.0004–0.0024 |
| DYS385ac | 1,764 | 3 | 2 | 1 | 3 | 0 | 0.0017 | 0.0003–0.0050 | 1:0.5 | 1:0 | 0.0020 | 0.0006–0.0050c |
| DYS385bc | 1,762 | 6 | 6 | 0 | 6 | 0 | 0.0034 | 0.0013–0.0074 | 1:0 | 1:0 | 0.0038 | 0.0016–0.0074c |
| DYS438 | 1,753 | 1 | 0 | 1 | 0 | 1 | 0.0006 | 0.0000–0.0032 | 0:1 | 0:1 | 0.0005 | 0.0001–0.0014 |
| DYS439 | 1,737 | 6 | 2 | 4 | 6 | 0 | 0.0035 | 0.0013–0.0075 | 1:2 | 1:0 | 0.0048 | 0.0023–0.0074 |
| DYS437 | 1,762 | 2 | 2 | 0 | 2 | 0 | 0.0011 | 0.0001–0.0041 | 1:0 | 1:0 | 0.0011 | 0.0003–0.0026 |
| DYS448 | 1,746 | 0 | 0 | 0 | 0 | 0 | 0 | 0–0.0021 | 0 | 0 | 0.0003 | 0.00003–0.0015 |
| DYS456 | 1,760 | 8 | 6 | 2 | 8 | 0 | 0.0045 | 0.0020–0.0089 | 1:0.33 | 1:0 | 0.0043 | 0.0022–0.0076 |
| DYS458 | 1,758 | 14 | 7 | 7 | 14 | 0 | 0.0080 | 0.0044–0.0133 | 1:1 | 1:0 | 0.0074 | 0.0044–0.0117 |
| DYS635 | 1,733 | 6 | 1 | 5 | 6 | 0 | 0.0035 | 0.0013–0.0075 | 0.2:1 | 1:0 | 0.0036 | 0.0017–0.0063 |
| Y-GATA-H4 | 1,755 | 5 | 1 | 4 | 5 | 0 | 0.0028 | 0.0009–0.0066 | 0.25:1 | 1:0 | 0.0029 | 0.0013–0.0053 |
| Total | 29,792 | 84 | 43 | 41 | 83 | 1 | 0.0028 | 0.0022–0.0035 | 1:0.95 d | 1:0.01 e | 0.0025 | 0.0016–0.0034 |
aAll mutations were confirmed by DNA sequence analysis, see Supplementary Table S2 for mutation details
bExcludes DYS389I separately sequenced and separately analyzed via the Bayesian approach (see “Materials and methods” for details)
cDYS385a and DYS385b separately sequenced and separately analyzed via the Bayesian approach (see “Materials and methods” for details)
dNinety-five percent binomial confidence interval limits [CIL] 1:1.47 to 1:0.61
eNinety-five percent CIL <1:0.006 to 1:0.037
Double-copy alleles either in the father or in the son were involved in two of the 84 mutations (see Supplementary Table S2). For the only mutation found at DYS438, we observed a slippage mutation from two equal-sized alleles (12) in the father to two alleles with two repeat differences (ten and 12) in the son. For one of the mutations at DYS635, there was a slippage mutation from one of two differently sized alleles (23 and 24) in the father to two equal-sized alleles (23) in the son. However, although these two father–son pairs were sequenced at both loci to confirm the mutations (as all other 82 mutations were confirmed by DNA sequence analysis), which allowed identification of the two partially overlaying sequences in the two different-sized alleles per individual, this confirmation test cannot rule out the possibility of an alternative deletion polymorphism in the case of the DYS635 mutation. In addition, double-copy alleles in both father and son as result of a locus duplication with subsequent slippage mutation in previous generations were found at DYS19 in three of the 1,757 father–son pairs investigated (two pairs with 15,16 and one pair with 15,17) but not at any other Y-STR locus investigated in this study. Double alleles at Y-STRs that usually exist in single copies were previously observed especially for DYS19 but also for several other Y-STR loci included in the Yfiler kit [23,27–29]. They represent larger duplication events, including the respective Y-STR locus with subsequent Y-STR slippage mutations that length-differentiate the two (or more) Y-STR alleles. A recent study investigated the structural basis and phylogenetic relationship of DYS19 duplications in detail [30].
Inherited null alleles in both father and son were observed in three cases and at two Y-STRs (DYS448, one out of 1,746 pairs; DYS456, two out of 1,760 pairs) as a consequence of a locus deletion or, alternatively, mutation(s) in the primer-binding sites. Null alleles at these and several other Yfiler Y-STRs were also observed in previous studies [23,27–29,31,32] and were especially investigated recently for DYS448 where both phenomena, mutations in the primer binding sites as well as deletions (including small deletions that caused apparent double alleles at another YSTR, which we did not observe in the DYS448 null allele observed in this study) were found to provide the molecular explanation [33].
Y-STR mutation rates
It seems to be the convention that mutation counts and father–son pair counts are used for simple frequency estimation of mutation rates and characteristics (“frequentist approach”) in individual studies but, moreover, also when considering data from several independent studies [10,17,21,23,34]. However, there is an alternative way of modeling such data in order to incorporate the uncertainty of the estimation obtained by each study and also to estimate the meta-parameters of interest (i.e., the mutation rate) when considering data from multiple studies. This is a general issue in meta-analysis, which has been successfully solved in areas outside the forensic mutation field [25]. For a more realistic consideration of the uncertainty of the data, we have applied such an approach for mutation rate estimation from our own data as well as to combine the data from our study with those from the 18 previous studies [4,5,8–23] using a hierarchical Binomial Bayesian model (“Bayesian approach”; see “Materials and methods” for details). In our new data, medians from Bayesian estimation of the locus-specific mutation rates ranged from 0.0003 (95% CI, 0.00003–0.0015) for DYS448 to 0.0074 (95% CI, 0.0044–0.0117) for DYS458, with a median mutation rate across all 17 Y-STRs of 0.0025 (95% CI, 0.0016–0.0034; Table 1). These estimates are based on pooled data per Y-STR locus, as we did not find any statistically significant differences in the locus-specific mutation rates between the five sampling regions (P >
> 0.05).
0.05).
To provide overall locus-specific mutation rates and characteristics that can be applied to forensic and evolutionary studies, we collected mutation data for the same 16 Y-STRs from 18 previously published studies that analyzed DNA-confirmed male families [4,5,8–23] and combined those with our new data (Table 2). Note that since previous studies did not separate DYS385a from DYS385b, we considered in this study the combined DYS385a/b locus (hence 16 loci in total). Noteworthy, only six of the 18 studies included complete Yfiler Y-STR data considering all together only 1,624 meiotic transfers per single locus [16,18,19,21–23], whereas all others included subsets of the markers analyzed in this study. Combining the data from the 18 previous studies with those presented in this study comprises all together 135,212 meiotic transfers and revealed 331 mutations (Table 2). Of the 331 mutations, 189 (57.1%) were repeat gains and 141 (42.6%) were repeat losses (ratio 1:0.75; 95% CIL, 1:0.93–1:0.59); 322 (97.3%) were single repeat changes, but only nine (2.7%) were multi-repeat changes (ratio 1, 0.027; 95% CIL, 1:0.05–1:0.012; Table 2). Medians from Bayesian estimation of the locus-specific mutation rates considering all available data ranged from 0.0002 (95% CI, 0.00002–0.0008) for DYS448 to 0.0065 (95% CI, 0.0023–0.0126) for DYS458, with a median rate across all 16 Y-STRs of 0.0022 (95% CI, 0.0019–0.0026). Although the Bayesian-based median mutation rates differed only slightly from those obtained via the simple frequentist approach (Tables 1 and and2),2), the 95% CI from the Bayesian approach are usually somewhat wider compared with the binomial confidence interval limits of the frequentist approach (Table 2), reflecting the uncertainties of the data available thus far. Therefore, Bayesian-based median mutation rates reported in this study shall be used rather than simple frequency-derived rates considering mutations in paternity probability estimations, e.g., in deficiency cases with male offspring as well as in genealogical and evolutionary studies.
Table 2
Combined mutation data from the present and 18 previous family-based studies [4,5,8–23] of 16 Y-STRs included in the AmpFlSTR® Yfiler® PCR amplification kit
| Y-STR Locus | Number of meiotic transfers | Number of mutations | Number of gains | Number of losses | Number of single-repeat changes | Number of multi-repeat changes | Mutation rate | Binomial 95% confidence interval limits | Gains versus losses | Single- versus multi-repeat changes | Median mutation rate | 95% Credible interval |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequentist approach | Bayesian approach | |||||||||||
| DYS19 | 11,900 | 32 | 19 | 13 | 32 | 0 | 0.0027 | 0.0018–0.0038 | 1:0.68 | 1:0 | 0.0026 | 0.0016–0.0039 |
| DYS389I | 10,103 | 28 | 12 | 16 | 28 | 0 | 0.0028 | 0.0018–0.0040 | 1:1.33 | 1:0 | 0.0024 | 0.0011–0.0040 |
| DYS389IIa | 10,079 | 31 | 17 | 14 | 29 | 2 | 0.0031 | 0.0021–0.0044 | 1:0.82 | 1:0.07 | 0.0030 | 0.0018–0.0045 |
| DYS390 | 11,385 | 25 | 13 | 12 | 25 | 0 | 0.0022 | 0.0014–0.0032 | 1:0.92 | 1:0 | 0.0019 | 0.0008–0.0032 |
| DYS391 | 11,336 | 34 | 20 | 14 | 33 | 1 | 0.0030 | 0.0021–0.0042 | 1:0.70 | 1:0.03 | 0.0028 | 0.0016–0.0041 |
| DYS392 | 11,268 | 7 | 4 | 2 | 7 | 0 | 0.0006 | 0.0002–0.0013 | 1:0.50 | 1:0 | 0.0004 | 0.00001–0.0011 |
| DYS393 | 10,079 | 10 | 6 | 4 | 10 | 0 | 0.0010 | 0.0005–0.0018 | 1:0.67 | 1:0 | 0.0009 | 0.0003–0.0017 |
| DYS385a/bb | 19,108 | 40 | 33 | 7 | 38 | 2 | 0.0021 | 0.0015–0.0029 | 1:0.21 | 1:0.05 | 0.0020 | 0.0013–0.0029 |
| DYS438 | 6,947 | 4 | 1 | 3 | 1 | 3 | 0.0006 | 0.0002–0.0015 | 0.33:1 | 0.33:1 | 0.0005 | 0.0001–0.0013 |
| DYS439 | 6,908 | 39 | 19 | 20 | 39 | 0 | 0.0056 | 0.0040–0.0077 | 1:1.05 | 1:0 | 0.0055 | 0.0035–0.0079 |
| DYS437 | 6,919 | 9 | 7 | 2 | 9 | 0 | 0.0013 | 0.0006–0.0025 | 1:0.29 | 1:0 | 0.0011 | 0.0003–0.0023 |
| DYS448 | 3,531 | 2 | 2 | 0 | 2 | 0 | 0.0006 | 0.0001–0.0020 | 1:0 | 1:0 | 0.0002 | 0.00002–0.0008 |
| DYS456 | 3,384 | 15 | 9 | 6 | 15 | 0 | 0.0044 | 0.0025–0.0073 | 1:0.67 | 1:0 | 0.0043 | 0.0017–0.0095 |
| DYS458 | 3,382 | 24 | 15 | 9 | 24 | 0 | 0.0071 | 0.0045–0.0105 | 1:0.60 | 1:0 | 0.0065 | 0.0023–0.0126 |
| DYS635 | 4,349 | 17 | 7 | 10 | 17 | 0 | 0.0039 | 0.0023–0.0063 | 1:1.43 | 1:0 | 0.0037 | 0.0015–0.0066 |
| Y-GATA-H4 | 4,534 | 14 | 5 | 9 | 13 | 1 | 0.0031 | 0.0017–0.0052 | 1:1.8 | 1:0.08 | 0.0029 | 0.0013–0.0055 |
| Total | 135,212 | 331 | 189 c | 141 c | 322 | 9 | 0.0024 | 0.0022–0.0027 | 1:0.75 d | 1:0.03 e | 0.0022 | 0.0019–0.0026 |
aExcludes mutations at DYS389I
bDYS385a/b considered jointly
cOne mutation from [5] could not be assigned
dNinety-five percent binomial confidence interval limits [CIL] 1:0.93–1: 0.59
eNinety-five percent CIL 1:0.05–1:0.012
As seen from Tables 1 and and2,2, our study comprising 1,730–1,764 meiotic transfers for each of the 17 Yfiler markers reflects a considerable increase in the knowledge of mutation rates and characteristics. This is most evident not only for Y-STR loci such as DYS448, DYS456, and DYS458, where previous mutation data were limited and our new data represent more than a 100% increase of information in respect of the meiotic transfers analyzed, but also for loci such as DYS635 and Y-GATA-H4, where our data reflect about a 65% increase of data. By combining our data with those from 18 previously published studies we are able to provide highly reliable locus-specific mutation rates for at least eight Yfiler Y-STRs: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and the combined DYS385a/b system with more than 10,000 meiotic transfers studied per locus (for the combined DYS385a/b almost 20,000 meiotic transfers were considered). Also for DYS437, DYS438, and DYS439, the obtained mutation rates are now expected to be much closer to reality with about 7,000 meiotic transfers investigated thus far. However, currently available mutation data for five Yfiler Y-STRs, DYS448, DYS456, DYS458, DYS635, and Y-GATA-H4 still need to be considered somewhat less reliable before additional data become available, as they are based on considerably less meiotic transfers (about 3,500–4,500 per locus) studied thus far.
Multiple Y-STR mutations and implications
Three father–son pairs were identified with mutations at more than one out of the 17 Yfiler Y-STR loci (see Supplementary Tables S2 and S3). For these three pairs, we assured individual sample authenticity by a second analysis of the ten or 15 autosomal STRs, respectively, in the six DNA aliquots in Rotterdam (the same that were used for Yfiler analysis) and compared the results with those obtained independently for the same individuals at the three laboratories initially involved in the paternity testing, with no allelic inconsistencies observed. Pair 1 (from Warsaw) had mutations at three Y-STRs, Y-GATA-H4, DYS393, and DYS391 confirmed by an independent Yfiler analysis as well as by independent DNA sequence analyses. On basis of 15 autosomal STRs included in the PowerPlex® 16 System (Promega), a high paternity index (PI) for the complete trio of 7.3 ×
× 107 was calculated. Additional evidence in favor of paternity is provided by matching pattern in one multilocus probe (33.15) and four single-locus probes (MS31, MS43a, YNH24, and MS205) not considered in the PI provided. Two other pairs had sequence-confirmed mutations at two Y-STRs each. Pair 2 (from Cologne) showed mutations at DYS456 and DYS389II with a PI for the complete trio of 7.75
107 was calculated. Additional evidence in favor of paternity is provided by matching pattern in one multilocus probe (33.15) and four single-locus probes (MS31, MS43a, YNH24, and MS205) not considered in the PI provided. Two other pairs had sequence-confirmed mutations at two Y-STRs each. Pair 2 (from Cologne) showed mutations at DYS456 and DYS389II with a PI for the complete trio of 7.75 ×
× 109 from analyzing ten autosomal STRs using the AmpFlSTR® SGMPlus® PCR amplification kit (Applied Biosystems, Inc.) together with three minisatellites (MS31, MS43a, and MS205). Pair 3 (from Wroclaw) showed mutations at DYS439 and DYS635 with a PI for the complete trio of 7.44
109 from analyzing ten autosomal STRs using the AmpFlSTR® SGMPlus® PCR amplification kit (Applied Biosystems, Inc.) together with three minisatellites (MS31, MS43a, and MS205). Pair 3 (from Wroclaw) showed mutations at DYS439 and DYS635 with a PI for the complete trio of 7.44 ×
× 107 from analyzing 15 autosomal STRs using the AmpFlSTR® Identifiler® PCR amplification kit (Applied Biosystems, Inc.) and with additional evidence in favor of paternity from matching pattern from one multi-loci probe (33.15 with one mutation band), two single-locus probes (MS1 and MS8), and the hypervariable PCR system D1S80 not considered in the PI mentioned.
107 from analyzing 15 autosomal STRs using the AmpFlSTR® Identifiler® PCR amplification kit (Applied Biosystems, Inc.) and with additional evidence in favor of paternity from matching pattern from one multi-loci probe (33.15 with one mutation band), two single-locus probes (MS1 and MS8), and the hypervariable PCR system D1S80 not considered in the PI mentioned.
According to recent ISFG recommendations on biostatistics in paternity testing [35], the weight of the genetic evidence of Y-chromosome markers shall be combined with the genetic weight from independent autosomal markers. However, this is recommended only in cases where no other family members in the paternal lineage are relevant for alternative paternity hypotheses, a knowledge that in practice is difficult, if not impossible, to access a priori [35]. In a recent study, Amorim [36] demonstrated that, while the genetic evidence obtained from autosomal loci, reshuffle at every meiosis, is appropriate for individual probability calculations, data from mitochondrial DNA (mtDNA) and Y-chromosome, which escape recombination, are not. Consequently, it was concluded that joining the evidential value of bi-parentally inherited autosomal and uni-parentally-inherited Y-chromosomal/mtDNA markers is generally inconsistent and should thus be avoided [36]. This notwithstanding, we calculated a Y-STR PI and joined it with the autosomal PI to demonstrate the effect of multiple mutations on the evidential value. The PI obtained from the Y-STR results was calculated according to Rolf et al. [37], with the mutation rates for each locus in question multiplied to give a frequency of the occurrence of the observed double or triple mutation. We used Y-STR haplotype frequencies as obtained from the German and Polish datasets in the Y Chromosome Haplotype Reference Database (YHRD) database (build 3.0, R27), respectively. The combined paternity indices based on autosomal and Y-chromosomal STRs were 1.86 ×
× 101 for pair 1 (with 3 Y-STR mutations), as well as 4.94
101 for pair 1 (with 3 Y-STR mutations), as well as 4.94 ×
× 106 for pair 2 and 2.11
106 for pair 2 and 2.11 ×
× 104 for pair 3 (both with two Y-STR mutations, respectively). Expectedly, these results emphasize the strong impact of mutations on the outcome of a paternity suit, but the overwhelming evidence provided by the autosomal DNA markers typed in the respective three trio cases would allow most paternity testing labs to conclude in favor of paternity. Furthermore, it might be interesting to mention that we see additional Y-chromosomal evidence in favor of paternity in all three father–son pairs from having analyzed in an extended study additional 161 polymorphic Y-STRs in the three pairs with three additional mutations in pair 1 (overall six of 178 Y-STRs mutated), one additional mutation in pair 2 (overall three of 178 Y-STRs mutated), and no additional mutations in pair 3 (overall two of 178 Y-STRs mutated; M.K. et al. unpublished data). All Y-STRs involved in the additional mutations have mutation rates considerably higher than usually observed for Y-STRs, e.g., those included in Yfiler (M.K. et al. unpublished data).
104 for pair 3 (both with two Y-STR mutations, respectively). Expectedly, these results emphasize the strong impact of mutations on the outcome of a paternity suit, but the overwhelming evidence provided by the autosomal DNA markers typed in the respective three trio cases would allow most paternity testing labs to conclude in favor of paternity. Furthermore, it might be interesting to mention that we see additional Y-chromosomal evidence in favor of paternity in all three father–son pairs from having analyzed in an extended study additional 161 polymorphic Y-STRs in the three pairs with three additional mutations in pair 1 (overall six of 178 Y-STRs mutated), one additional mutation in pair 2 (overall three of 178 Y-STRs mutated), and no additional mutations in pair 3 (overall two of 178 Y-STRs mutated; M.K. et al. unpublished data). All Y-STRs involved in the additional mutations have mutation rates considerably higher than usually observed for Y-STRs, e.g., those included in Yfiler (M.K. et al. unpublished data).
Observing mutations at up to three out of 17 Yfiler Y-STRs in the same father–son pair is of great forensic relevance and updates previous conclusions on the threshold for the number of allelic differences to conclude exclusion constellations, based on findings of two mutations in the same father–son pairs observed among nine Y-STRs [4,38] or 16 Yfiler Y-STRs [23] or in line with our new data from 17 Yfiler Y-STRs. Three mutations in the same father–son pair, as obtained in this study for 17 Y-STRs, were also found previously for autosomal DNA markers in a trio case analyzed with 30 autosomal DNA markers, where paternity was established without any reasonable doubt (PI >1010) [39]. This previous observation has led to the practical consequence of giving the excluding opinion only in case of four or more observed DNA inconsistencies in some laboratories (T.D., personal communication) since 2003, which would be consistent with the Y-STR findings obtained in this study. However, as the number of forensically evaluated and applied STR loci on the autosomes as well as the Y-chromosome steadily increases, it is difficult to recommend an absolute upper limit of allelic differences that inevitably support the exclusion constellation. Instead, we emphasize that recommendations should refer to the mutational characteristics of each marker, the number of markers involved, and the case assumptions. For instance, the threshold for testing members of multi-generation families needs to be higher than for analyzing trio/father–son cases as the number of meioses with potential mutation events is increased, e.g., as is relevant in deficiency paternity cases. All available evidence suggests that (Y-)STR markers currently applied in forensic and paternity testing have a mutation rate in the range of 10−3 or lower, providing solid tools for solving paternity cases with high evidentiary power. However, even with such relatively low mutation rates, in rare instances, several mutations may occur in the same father–son pair or trio as shown in this study and elsewhere [4,23,39]. Assumptions in such cases must be clearly and explicitly stated, and their acceptance must finally be left to the court decision. Moreover, some (Y-)STR markers that may become applied in the future may have elevated mutation rates. Preliminary evidence for this notion comes from a recent pedigree study that highlights two Y-STRs (DYS570 and DYS576), which seem to mutate about 10× faster than (Y-)STRs usually applied in forensic and familial testing [40], currently further investigated by analyzing the set of father–son pairs used in the present study together with a large number of additional Y-STRs (M.K., unpublished work).
Father’s age and Y-STR mutation rates
The average age (at the time of son’s birth) of fathers without a mutation was 30.32 (±10.22) years, in comparison to the average age of fathers with at least one mutation at 34.40 (±11.63) years, a difference that is highly statistically significant (p <
< 0.001). The relationship between fathers age and mutation rate is illustrated in Fig. 1, and data are provided in Supplementary Table S4. The effect of the father’s age on the mutation rate was modeled using a Poisson distribution, where the mutation rate was estimated as an exponential function of the age of the father. This showed that the mutation rate increased with increasing age of the father (α
0.001). The relationship between fathers age and mutation rate is illustrated in Fig. 1, and data are provided in Supplementary Table S4. The effect of the father’s age on the mutation rate was modeled using a Poisson distribution, where the mutation rate was estimated as an exponential function of the age of the father. This showed that the mutation rate increased with increasing age of the father (α =
= 0.0294, 2.5% quantile
0.0294, 2.5% quantile =
= 0.0001), suggesting that age is a factor that should be taken into account not only when estimating Y-STR mutation rates but also when comparing estimated mutation rates from different studies. Several previous studies investigated the age effect on the mutation rate for all or some of the Y-STRs studied here with conflicting results. Some studies found the average age of the fathers with mutations being older than that of fathers without [13,34]; others observed the reverse effect [4,18], and some found no age difference between mutated and non-mutated fathers [10,19,23]. Two studies report a higher mutation rate for older fathers compared to younger ones [17,34], but in both studies, this effect was only seen when deliberately excluding mutated fathers of medium age from the analysis. Therefore, our study provides the first evidence for a statistically significant increase of mutation rate with increased father’s age not deliberately excluding data. This may be due to the fact that our study represents the largest single study available thus far, hence being somewhat less biased toward younger fathers (which usually are frequent) and against older ones (which usually are rare). This is also reflected in the relatively old average age of mutated fathers in our study, which is considerably older than all previous studies that did not observe mutated fathers being older than non-mutated ones [4,10,13,17–19,23,34]. Notably, the average age of the mutated father from our study is only marginally older or somewhat younger than that of the two studies also reporting mutated fathers to be older than non-mutated ones [13,34], of which the latter was only marginally smaller in size compared to ours. This clearly shows the effect of sampling bias when investigating age effects of Y-STR mutation rates in limited sized studies, a notion that should be considered for future investigations.
0.0001), suggesting that age is a factor that should be taken into account not only when estimating Y-STR mutation rates but also when comparing estimated mutation rates from different studies. Several previous studies investigated the age effect on the mutation rate for all or some of the Y-STRs studied here with conflicting results. Some studies found the average age of the fathers with mutations being older than that of fathers without [13,34]; others observed the reverse effect [4,18], and some found no age difference between mutated and non-mutated fathers [10,19,23]. Two studies report a higher mutation rate for older fathers compared to younger ones [17,34], but in both studies, this effect was only seen when deliberately excluding mutated fathers of medium age from the analysis. Therefore, our study provides the first evidence for a statistically significant increase of mutation rate with increased father’s age not deliberately excluding data. This may be due to the fact that our study represents the largest single study available thus far, hence being somewhat less biased toward younger fathers (which usually are frequent) and against older ones (which usually are rare). This is also reflected in the relatively old average age of mutated fathers in our study, which is considerably older than all previous studies that did not observe mutated fathers being older than non-mutated ones [4,10,13,17–19,23,34]. Notably, the average age of the mutated father from our study is only marginally older or somewhat younger than that of the two studies also reporting mutated fathers to be older than non-mutated ones [13,34], of which the latter was only marginally smaller in size compared to ours. This clearly shows the effect of sampling bias when investigating age effects of Y-STR mutation rates in limited sized studies, a notion that should be considered for future investigations.
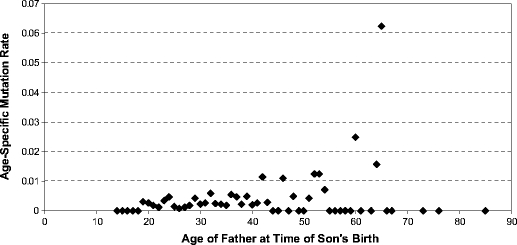
Relationship between age of fathers at the time of son’s birth and Y-STR mutation rate considering 29,792 meiotic transfers with 84 mutations from analyzing the 17 Y-STR loci included in the AmpFlSTR® Yfiler® PCR amplification kit in DNA-confirmed father–son pairs (see text for model-based statistical testing and Supplementary Table S4 for data)
Conclusions
From considering all currently available data, we can conclude that none of the 16 Yfiler Y-STRs (DYS385a/b considered as one combined system) had a mutation rate of >1% (although for DYS458 the 97.5% confidence interval is >1.0%), 12 Yfiler Y-STRs had mutation rates >0.1%, whereas four loci had mutation rates <0.1% (DYS392, DYS393, DYS438, and DYS448, of which the latter has to be seen as somewhat preliminary given the somewhat limited number of meiotic transfers studied thus far). Additionally, we can conclude that at least for 15 of the Y-STRs included in the Yfiler kit (except DYS438 and considering DYS385a/b as one system), there is convincing evidence that single-repeat changes are strongly favored over multiple-repeat changes. Multi-repeat changes only seemed to be more frequent than single-repeat ones at DYS438, although only a small number of mutations were observed thus far at this locus. In contrast, considerable heterogeneity was observed in the ratio of repeat gains versus repeat losses between Yfiler Y-STR loci: for ten Y-STR loci, repeat gains were clearly favored over repeat losses (DYS19, DYS389II, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS448, DYS456, and DYS458), whereas for two Y-STR loci, the ratio was about equal (DYS390 and DYS439), and for four Y-STR loci (DYS389I, DYS438, DYS635, and Y-GATA-H4), considerably more repeat losses were found compared with repeat gains. Our observation of up to three Y-STR mutations in the same father–son pair shall be recognized in the interpretation of Yfiler Y-STR profiles when determining the threshold of allelic differences for concluding exclusion constellations in future paternity and genealogical testing and in applications that involve multiple members of the same male lineage. We recommend that the mutational features described in this study for the Yfiler Y-STRs, including multiple events and age dependency, together with the overall locus-specific median mutation rates, shall be considered in future studies relying on Yfiler information. In addition to their provision in the Supplementary Table S3, the complete Yfiler Y-STR haplotype data of unrelated individuals investigated in the course of this study are made publicly available via two public Y-STR reference databases, the YHRD (http://www.yhrd.org) as well as the YFiler Haplotype Database (http://www.appliedbiosystems.com/yfilerdatabase/), for future haplotype frequency estimations in forensic case work as well as for genealogical and evolutionary applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Primers for Y-STR PCR prior to DNA sequence analysis and for DNA sequence analysis (DOC 36 kb)
Details of 84 mutations observed among 29,792 (1730 to 1764 per locus) meiotic transfers from analyzing the 17 Y-STR loci included in the AmpFlSTR® Yfiler® PCR amplification kit in DNA-confirmed father-son pairs (DOC 115 kb)
Complete Yfiler Y-STR haplotypes of unrelated males included in the study with their sampling origin (DOC 3359 kb)
Age-specific Y-STR mutation rates considering 29,792 meiotic transfers with 84 mutations from analyzing the 17 Y-STR loci included in the AmpFlSTR® Yfiler® PCR amplification kit in DNA-confirmed father-son pairs. (DOC 64 kb)
Acknowledgements
We thank Damla Arslantunali, Tomas Petten, Ying Choi, and Kate van Duijn for technical assistance. Pieter van Oers and his colleagues at Applied Biosystems are greatly acknowledged for technical support. This study was supported by funds from the Netherlands Forensic Institute. None of the conclusions in this study were influenced by the supporting organizations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
Full text links
Read article at publisher's site: https://doi.org/10.1007/s00414-009-0342-y
Read article for free, from open access legal sources, via Unpaywall:
https://link.springer.com/content/pdf/10.1007/s00414-009-0342-y.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1007/s00414-009-0342-y
Article citations
Bibliometric analysis of kinship analysis from 1960 to 2023: global trends and development.
Front Genet, 15:1401898, 06 Jun 2024
Cited by: 0 articles | PMID: 38903754 | PMCID: PMC11187311
Review Free full text in Europe PMC
Delineating the dispersal of Y-chromosome sub-haplogroup O2a2b-P164 among Austronesian-speaking populations.
Sci Rep, 14(1):2066, 24 Jan 2024
Cited by: 0 articles | PMID: 38267477 | PMCID: PMC10808098
Improving the strategy to identify historical military remains: a literature review and Y-STR meta-analysis.
Forensic Sci Res, 9(1):owad050, 25 Dec 2023
Cited by: 0 articles | PMID: 38562552 | PMCID: PMC10982847
Review Free full text in Europe PMC
The sequence of the repetitive motif influences the frequency of multistep mutations in Short Tandem Repeats.
Sci Rep, 13(1):10251, 24 Jun 2023
Cited by: 2 articles | PMID: 37355683 | PMCID: PMC10290632
Development and validation of a new multiplex for upgrading Y-STRs population databases from 12 to 23 markers and its forensic casework application.
Sci Rep, 12(1):21734, 16 Dec 2022
Cited by: 2 articles | PMID: 36526709 | PMCID: PMC9758231
Go to all (69) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[Application of 17 Y-chromosome specific STR loci in paternity testing].
Zhongguo Shi Yan Xue Ye Xue Za Zhi, 16(3):699-703, 01 Jun 2008
Cited by: 1 article | PMID: 18549658
Analysis of Mutation Rate of 17 Y-Chromosome Short Tandem Repeats Loci Using Tanzanian Father-Son Paired Samples.
Genet Res Int, 2018:8090469, 06 Aug 2018
Cited by: 1 article | PMID: 30174958 | PMCID: PMC6106792
A comprehensive mutation study in wide deep-rooted R1b Serbian pedigree: mutation rates and male relative differentiation capacity of 36 Y-STR markers.
Forensic Sci Int Genet, 41:137-144, 04 May 2019
Cited by: 3 articles | PMID: 31082622
Population and segregation data on 17 Y-STRs: results of a GEP-ISFG collaborative study.
Int J Legal Med, 122(6):529-533, 24 Jul 2008
Cited by: 24 articles | PMID: 18651159

 1
1



