Abstract
Free full text

A Crohn’s disease–associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1
Abstract
A common mutation in the gene encoding the cytoplasmic sensor Nod2, involving a frameshift insertion at nucleotide 3020 (3020insC), is strongly associated with Crohn’s disease. How 3020insC contributes to this disease is a controversial issue. Clinical studies have identified defective production of interleukin 10 (IL-10) in patients with Crohn’s disease who bear the 3020insC mutation, which suggests that 3020insC may be a loss-of-function mutation. However, here we found that 3020insC Nod2 mutant protein actively inhibited IL10 transcription. The 3020insC Nod2 mutant suppressed IL10 transcription by blocking phosphorylation of the nuclear ribonucleoprotein hnRNP-A1 via the mitogen-activated protein kinase p38. We confirmed impairment in phosphorylation of hnRNP-A1 and binding of hnRNP-A1 to the IL10 locus in peripheral blood mononuclear cells from patients with Crohn’s disease who bear the 3020insC mutation and have lower production of IL-10.
Crohn’s disease is a chronic inflammatory bowel disease that affects mainly the terminal ileum, cecum, colon and perianal area. Inflammatory bowel disease is thought to result from inappropriate and continuing activation of the mucosal immune system driven by normal luminal flora. This aberrant response is probably facilitated by defects in both the barrier function of the intestinal epithelium and the mucosal immune system. These defects are in turn caused by genetic and environmental factors1.
Interleukin 10 (IL-10; A001243) is a pleiotropic cytokine produced by T cells, B cells and macrophages. IL-10-deficient mice show enhanced development of several inflammatory and autoimmune diseases, which suggests that IL-10 serves a central function in vivo in restricting inflammatory responses2. Although they are healthy in germ-free conditions, some IL-10-deficient mouse strains develop colitis when given a normal, specific pathogen–free bowel flora, probably as a result of a poorly controlled mucosal immune response.
Innate immune responses are initiated by the detection of microbial invaders by several distinct host systems collectively called ‘pattern-recognition receptors’. The nucleotide-binding oligomerization domain (Nod)-like receptors, Toll-like receptors (TLRs), C-type lectin receptors and RIG-I-like receptors are examples of pattern-recognition receptors. Two members of the Nod-like receptor family of proteins, Nod1 and Nod2, are believed to be cytoplasmic sensors of microbial products3. Both Nod1 and Nod2 recognize peptidoglycan, but each protein senses distinct molecular motifs in peptidoglycan. Nod1 recognizes a naturally occurring muropeptide of peptidoglycan that presents a unique amino acid at its terminus called ‘diaminopimelic acid’. This amino acid is found mainly in the peptidoglycan of Gram-negative bacteria4. In contrast, Nod2 can detect the minimal bioactive fragment of peptidoglycan called ‘muramyl dipeptide’. Thus, whereas Nod1 detects mainly Gram-negative bacteria, Nod2 is a more general sensor of bacterial peptidoglycan5.
Nod2 expression is restricted to monocytes6 and is recognized as an important initiator of inflammation. Models hypothesize that after binding to muramyl dipeptide, Nod2 forms oligomers and binds to the caspase-recruitment domain–containing serine-threonine kinase RICK (also called RIP2, CARDIAK, CCK and Ripk2). RICK then forms oligomers and facilitates the ubiquitination of a site on the modulator of transcription factor NF-κB NEMO (also called IKKγ)7, which results in activation of the inhibitor of NF-κB kinase complex and NF-κB8.
The gene encoding Nod2 on human chromosome 16 (NOD2) was the first gene identified as being associated with susceptibility to Crohn’s disease9. Between 30% and 50% of patients with Crohn’s disease in the Western hemisphere carry NOD2 mutations on at least one allele. Mutations resulting in substitution of tryptophan for arginine at amino acid residue 702 (R702W) or arginine for glycine at amino acid residue 908 (G908R) and a frameshift substitution at amino acid position 1007 are estimated to represent 32%, 18% and 31%, respectively, of all disease-causing mutations in NOD2 and are independently associated with susceptibility to Crohn’s disease10. All three mutations are located in the leucine-rich repeat (LRR) domain of NOD2. The frameshift substitution at amino acid 1007 associated with Crohn’s disease stems from an insertion mutation at nucleotide 3020; this mutation (3020insC) results in partial deletion of the terminal LRR domain of the protein. The genotype relative risk for developing Crohn’s disease in people heterozygous or homozygous for this mutation alone is calculated to be 1.5–2.6 or 17.6–42.1, respectively11. Overall, patients homozygous for 3020insC demonstrate a much more severe disease phenotype than other patients with Crohn’s disease and have a higher risk for ileal stenosis and surgical intervention12. It is postulated that the LRR domain of Crohn’s disease–associated Nod variants is impaired in its ability to recognize microbial components and/or in its ability to inhibit the formation of Nod2 dimers, which results in lack of activation of NF-κB in monocytes9. However, the higher NF-κB activity in patients with Crohn’s disease and the clinical effectiveness of NF-κB inhibitors in ameliorating symptoms of Crohn’s disease1 suggest that disease-causing variants of Nod2 may promote activation of NF-κB.
IL-10-deficient mice develop a chronic enterocolitis that shares histopathological features with human Crohn’s disease13. Colitis in IL-10-deficient mice is driven by T helper type 1 (TH1) cells but not by B cells14 and is dependent on the presence of resident enteric bacteria15. In accordance with those findings, Helicobacter hepaticus triggers colitis in specific pathogen–free IL-10-deficient mice by a mechanism that depends on TH1 proinflammatory cytokines16.
The relationship between Nod2 and IL-10 is not completely understood. Peripheral blood mononuclear cells (PBMCs) from patients with Crohn’s disease who are homozygous for the 3020insC mutation show defective release of IL-10 after stimulation with enteric micro-organisms and TLR ligands17. Those findings contrast with studies of mice in which an artificially created mouse version (2939insC) of human 3020insC replaces the endogenous wild-type mouse Nod2 gene (Nod22939insC)18. In these mice, macrophages stimulated by the TLR4 ligand lipopolysaccharide (LPS), peptidoglycan or muramyl dipeptide release quantities of IL-10 resembling those released by wild-type macrophages. We hypothesized that the human 3020insC and mouse Nod22939insC alleles may have different functional properties because of species-specific physiology or evolutionary adaptation. We undertook this study to explore that possibility and the potential target(s) of the mutant Nod2 protein produced by the 3020insC mutation (called ‘3020insC Nod2’ here).
RESULTS
Suppression of IL10 production by 3020insC Nod2
To investigate the influence of endogenous Nod2 on cytokine production, we measured the production of IL-10 and IL-12p40 by mouse bone marrow–derived macrophages (BMDMs) from wild-type and Nod2−/− mice19. We detected little difference in the amount of IL-10 (Fig. 1a) or IL-12p40 (Fig. 1b) released by wild-type and Nod2−/− BMDMs after stimulation with LPS, peptidoglycan or Pam3Cys (a synthetic TLR2 ligand). Muramyl dipeptide alone stimulated little production of IL-10 or IL-12p40 in this experiment, consistent with results obtained with human PBMCs, in which muramyl dipeptide augments only TLR2 ligand–mediated induction of IL-12 and IL-10 (ref. 20). The patterns of production of IL-10 and IL-12p40 by peritoneal macrophages and splenocytes were very similar to those in BMDMs (data not shown). Furthermore, RICK (RIP2) was also dispensable for IL-10 production in primary macrophages stimulated by LPS, peptidoglycan or Pam3Cys (Fig. 1c).

Influence of Nod2 signaling on macrophage cytokine production. (a–c) ELISA of mouse IL-10 (a,c) and IL-12p40 (b) in supernatants of wild-type (WT), Nod2−/− (Nod2-KO) and Ripk2−/− (RIP2-deficient; RIP2-KO) mouse BMDMs stimulated for 24 h in vitro with medium, LPS (1 μg/ml), peptidoglycan (PGN; 10 μg/ml), muramyl dipeptide (MDP; 10 μg/ml) or Pam3Cys (1 μg/ml); results are normalized to total cellular protein. Data represent three independent experiments (error bars, s.e.m.). (d,e) ELISA of human IL-10 (d) and tumor necrosis factor (e) in supernatants of monocytes obtained from healthy people expressing wild-type Nod2 (n = 9) and patients with Crohn’s disease who were homozygous for 3020insC (n = 5) and then stimulated for 24 h with medium, muramyl dipeptide, Pam3Cys or a combination of muramyl dipeptide and Pam3Cys (MDP + Pam); results are normalized to total cellular protein. *, P < 0.05. Data are representative of one experiment with each donor sample measured in triplicate (error bars, s.d.).
Next we tested the response of primary monocytes to muramyl dipeptide, Pam3Cys or a combination of muramyl dipeptide and Pam3Cys in cells from patients with Crohn’s disease who were homozygous for the 3020insC mutation, as well as cells from control volunteers bearing the wild-type NOD2 allele. In contrast to the data obtained with mice, IL-10 production was much lower in cells from patients homozygous for 3020insC (Fig. 1d). Production of tumor necrosis factor was also significantly lower in cells from patients homozygous for 3020insC after stimulation with muramyl dipeptide alone but was similar with that of wild-type cells after stimulation with Pam3Cys or muramyl dipeptide plus Pam3Cys (Fig. 1e).
Thus, we hypothesized that the 3020insC Nod2 mutant may be able to regulate expression of the gene encoding IL-10 (IL10). To determine the effect of 3020insC Nod2 on endogenous IL10 expression, we transduced primary human monocytes with lentivirus expressing wild-type or 3020insC Nod2 (Fig. 2a). We found that 3020insC Nod2 but not wild-type Nod2 inhibited both basal as well as LPS-induced expression of IL-10 mRNA and protein (Fig. 2b,c). In contrast, we found no difference in IL-1β production by cells over-expressing wild-type or 3020insC Nod2 (Fig. 2d), and IL-12p40 production was suppressed by high expression of wild-type Nod2 but not 3020insC Nod2 (Fig. 2e). However, it is possible that the lack of influence of 3020insC Nod2 on IL-12p40 production was secondary to its suppression of IL-10 production. Thus, wild-type and 3020insC Nod2 seem to affect IL-10, IL-1β and IL-12p40 expression differently in human monocytes. We obtained similar results when we used peptidoglycan and Pam3Cys instead of LPS (data not shown).

Different effects of wild-type and 3020insC Nod2 on endogenous IL-10 expression in primary human monocytes. (a) Wild-type and 3020insC Nod2 constructs. CARD, caspase-recruitment domain. (b) RT-PCR analysis of the expression of mRNA encoding IL-10, Nod2 and β-actin by primary human monocytes transduced for 4 d with empty control vector (CV) or lentivirus encoding wild-type or 3020insC Nod2 and then stimulated for 5 d with medium or LPS. On the basis of lentiviral expression of green fluore-scent protein, an average infection rate of 38% was achieved. ‘Background’ bands represent endogenous Nod2 mRNA in infected cells. (c–e) ELISA of the secretion of IL-10 (c), IL-1β (d) and IL-12p40 (e) by human monocytes infected with 5 or 50 μl lentivirus-containing supernatant (key) and treated with medium or LPS, analyzed on day 5 after infection and normalized to cytokine produced (pg/ml) per 1 ×105 live cells. *, P < 0.03. (f) Luciferase activity in lysates of primary human monocytes transfected for 6 h with a human IL10 promoter–luciferase reporter, together with empty vector (pCDNA3) or vector encoding wild-type or 3020insC Nod2 constructs (optimized response); results are presented as the stimulation index, derived from the ratio of the luciferase activity in each experimental condition to that of cells transfected with empty vector. Mean transfection efficiency, 35%. Data are representative of three experiments with one donor each (error bars (c–f), s.d.).
To confirm our finding of an influence of Nod2 on IL10 transcription, we transfected primary human monocytes with a human IL10 promoter–luciferase reporter construct (containing the sequence between positions −1044 and +30 relative to the transcription start site)21 together with vectors expressing wild-type or 3020insC Nod2. Neither the empty vector (pCDNA3) nor wild-type Nod2 altered constitutive transcription of IL10; however, 3020insC Nod2 suppressed constitutive IL10 transcription by about 50% (Fig. 2f). These results collectively demonstrate that the 3020insC mutation conferred on Nod2 a new function, the ability to inhibit IL-10 production, and loss of the ability to repress IL-12p40 expression.
Inhibition of IL10 promoter activity by 3020insC
To further explore the molecular mechanism whereby 3020insC Nod2 inhibited IL10 expression, we transfected the IL10 promoter–luciferase reporter and the wild-type or 3020insC Nod2 construct into the RAW264.7 mouse macrophage cell line. We verified expression of each construct by RT-PCR (Supplementary Fig. 1 online). Although the IL10 promoter was constitutively active, peptidoglycan and Pam3Cys further boosted luciferase activity (Fig. 3a). Expression of 3020insC Nod2 strongly inhibited IL10 transcription in all conditions, even in the absence of any microbial stimuli, but wild-type Nod2 did not.

Inhibition of IL10 transcription by 3020insC Nod2. (a) Luciferase activity of lysates of RAW264.7 cells transfected for 1 d with the human IL10 promoter–lucifersase reporter together with empty vector (pCDNA3) or vector encoding wild-type or 3020insC Nod2 at an effector/reporter molar ratio of 0.3:1, then stimulated for 24 h with medium (Med), peptidoglycan or Pam3Cys. RLU, raw luciferase units. Expression of the transfected Nod2 and 3020insC mutant in RAW264.7 cells was verified by RT-PCR with primers selective for human but not mouse Nod2 (Supplementary Fig. 1). (b,c) Luciferase activity in lysates of cells transfected for 1 d with the human IL10 promoter–luciferase reporter, together with various molar ratios (horizontal axes) of vectors encoding wild-type and 3020insC Nod2 (b) or wild-type Nod2 and empty vector (pCDNA3; c), then stimulated for 7 h with medium or peptidoglycan. Reporter amount remained constant, set as 1. (d) Immunoassay of extracts of HEK293T cells transfected for 24 h (Input) with Flag- or Myc-tagged wild-type Nod2 (WT) or 3020insC Nod2 (frameshift (FS)), then immunoprecipitated (IP) with anti-Flag (α-Flag) or anti-c-Myc (α-Myc) and analyzed by immunoblot (IB) with anti-Flag. Arrows indicate that 3020insC Nod2 is smaller than wild-type Nod2. Data represent the pooled results of at least three independent experiments (error bars (a–c), s.d.).
To determine if wild-type and 3020insC Nod2 could functionally compete (for example, in the setting of a heterozygous person), we transfected wild-type and 3020insC Nod2 together, at various ratios, into RAW264.7 cells, along with the IL10 promoter–luciferase reporter. When wild-type Nod2 was more abundant than 3020insC Nod2, it countered the inhibitory effect of 3020insC Nod2 on IL10 transcription; this counter-inhibitory effect was lost as 3020insC Nod2 increased in abundance (Fig. 3b,c). Competition between wild-type and 3020insC Nod2 could have been due to direct interaction between the two molecules. To investigate that possibility, we generated wild-type and 3020insC Nod2 constructs tagged with Flag and c-Myc epitopes. After being transfected into human embryonic kidney (HEK293) cells, wild-type and 3020insC Nod2 constructs engaged in homotypic and heterotypic interactions (Fig. 3d). Thus, wild-type Nod2 may inhibit 3020insC Nod2–mediated suppression of IL10 transcription through direct physical interference.
Because only a fraction of patients with Crohn’s disease are homozygous carriers of the 3020insC mutation, we also analyzed the effect of the other two common Nod2 variants (R702W and G908R) on IL10 expression. Like the 3020insC Nod2 mutant, the R702W and G908R Nod2 mutants exerted an inhibitory effect on IL10 promoter activity (data not shown). In summary, these data suggest that all three LRR mutants of Nod2 are able to inhibit basal and TLR-induced IL10 transcription.
Gain of function is unique to human Nod2 and IL10
A study of Nod22939insC mice (in which an artificially created mouse version of human 3020insC replaces endogenous wild-type mouse Nod2) did not detect alteration in IL-10 expression in macrophages18. That is inconsistent with our experimental data presented above and with clinical observations of human patients with Crohn’s disease17,20. We hypothesized that the human and mouse Nod2 mutants may have different functional properties, as the latter was created artificially and thus was not subject to evolutionary adaptation in mammalian physiology. We tested our hypothesis by comparing the effects of the human mutant (3020insC Nod2) and mouse mutant (2939insC Nod2) on the expression of a human IL10 promoter–luciferase reporter construct or mouse Il10 promoter–luciferase reporter construct. Human 3020insC Nod2 suppressed basal and TLR-induced expression of the human reporter but not of the mouse reporter (Fig. 4a,b). In contrast, mouse 2939insC Nod2 did not inhibit expression of either the human or mouse reporter (Fig. 4a,b). Furthermore, retrovirus-mediated expression of 3020insC Nod2 into BMDMs derived from Nod2−/− mice had little effect on either basal or TLR-stimulated IL-10 production (Fig. 4c). These results indicate that the human and mouse Nod2 mutants have different functional properties and that only the human Nod2 mutant can inhibit expression of human IL-10.

Human and mouse Nod2 mutant proteins have different transcriptional effects. (a,b) Luciferase activity in lysates of RAW264.7 cells transfected for 1 d with a human IL10 promoter–luciferase reporter (a) or a mouse Il10 promoter–luciferase reporter (b) together with empty control vector or vector expressing human wild-type or 3020insC Nod2 (top) or mouse wild-type or 2939insC Nod2 (bottom) at a reporter/effector molar ratio of 1:1, then stimulated for 24 h with medium, muramyl dipeptide, LPS or peptidoglycan. *, P < 0.03. Data represent three to four independent experiments (mean and s.d.). (c) ELISA of mouse IL-10 in supernatants of Nod2−/− BMDMs infected twice with empty control vector (CV) or retrovirus encoding wild-type or 3020insC Nod2 and then, on day 5 after infection, stimulated for 24 h with medium, LPS, peptidoglycan or Pam3Cys. Equivalent expression of transduced human wild-type and 3020insC Nod2 mRNA in unstimulated Nod2−/− BMDMs was ensured (Supplementary Fig. 3 online). Data are representative of three experiments with one mouse each (error bars, s.d. of triplicate samples). (d) Luciferase activity in lysates of RAW264.7 cells transiently transfected with constructs encoding wild-type or 3020insC Nod2 (key) plus luciferase reporter constructs driven by the IL10 promoter fragment (positions −782 to +12) with (Mut IL10) or without (WT IL10) point mutations in the region between positions −30 and −25 and then stimulated for 7 h with medium, LPS or peptidoglycan. Data are representative of four individual experiments (mean and s.d.).
Blockade of hnRNP-A1–IL10 binding by 3020insC Nod2
Next we searched for IL10 promoter elements (3020insC Nod2–response elements (NREs)) required for 3020insC Nod2–mediated inhibition of IL-10 production. Using an extensive and systematic deletion- and mutagenesis-based search strategy, we focused on a narrow region of the IL10 promoter between positions −30 and −25 upstream of the transcription-initiation site. A version containing point mutations introduced into the region between positions −30 and −25 was not inhibited by 3020insC Nod2, in contrast to the inhibition of a construct containing a wild-type IL10 promoter fragment (Fig. 4d). Notably, the mutant also lost a substantial portion of its basal transcriptional activity. These results suggest that the TACACA sequence in the region between positions −30 and −25 may form the core of an NRE and that a nuclear DNA-binding protein may constitutively interact with this region. It is noteworthy that the NRE is predicted to be a positive functional element because its mutation resulted in less basal and microbe-stimulated IL10 transcription.
We hypothesized that the NRE may bind to transcription factors involved in IL10 transcription and that this binding may be regulated by 3020insC Nod2. To test our hypothesis, we established stable clones of RAW264.7 cells constitutively expressing wild-type or 3020insC Nod2 (Fig. 5a). We stimulated these clones with LPS or peptidoglycan, then isolated nuclear extracts and assessed by electrophoretic mobility-shift assay (EMSA) their binding to a probe containing the wild-type or mutated human IL10 NRE (Fig. 5b,c). We detected a nuclear factor bound to the wild-type but not mutant probe in unstimulated cells; this binding did not increase after stimulation with LPS or peptidoglycan (Fig. 5c). We designated this binding factor, which may represent a putative transcription factor involved in maintaining basal IL10 transcription, ‘TF-X’. Consistent with our prediction, 3020insC Nod2 resulted in much less binding of TF-X to the NRE, but wild-type Nod2 did not (Fig. 5c).
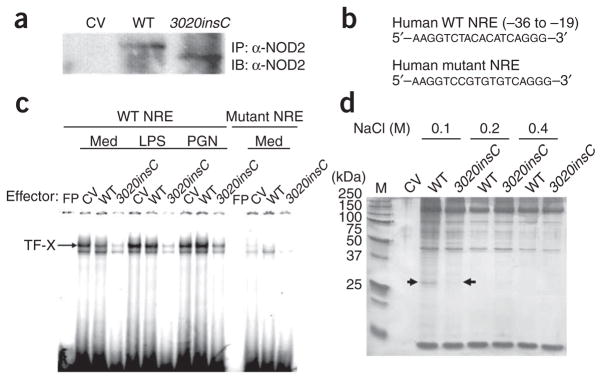
Binding of nuclear proteins to the NRE. (a) Immunoassay of lysates of RAW264.7 cells stably transfected with empty vector or vector encoding wild-type or 3020insC Nod2, then immunoprecipitated and analyzed by immunoblot with anti-Nod2. Results are representative of more than five separate experiments. (b) Sequences of probes used for EMSA. The mutant NRE probe bears the same mutations as those between positions −30 and −25 in the IL10 promoter mutant construct in Figure 4d. (c) EMSA (with the probes in b) of nuclear extracts of the stable RAW264.7 transfectants in a, stimulated with medium, LPS or peptidoglycan. FP, free probe. Results are representative of four separate experiments. (d) DNA precipitation of nuclear extracts of the stable RAW264.7 transfectants in a with biotinylated oligonucleotides encompassing the IL10 NRE, followed by elution at increasing NaCl concentrations (above gel), separation by 12% SDS-PAGE and silver staining. Arrows indicated bands that were excised and analyzed. M, molecular size markers (in kilodaltons (kDa)). Results are representative of three independent experiments with identical results.
We then did a series of biochemical experiments to identify TF-X. First we used biotinylated oligonucleotides encoding the NRE to precipitate TF-X from nuclear extracts derived from RAW264.7 clones expressing wild-type or 3020insC Nod2 (Fig. 5d). Next we excised the approximately 26-kilodalton TF-X band and analyzed it by nanoflow liquid chromatography–tandem mass spectrometry. The result with the highest score (30.98) for bands excised from cells expressing wild-type Nod2 was heterogeneous nuclear ribonucleoprotein A1 (hnRNP-A1; A001137); these hnRNP-A1 peptides (1,218.64 and 1,784.91 daltons in size, with intensities of 1.89 × 106 and 5.57 × 105, respectively), which covered 8% of the amino acid residues of the full-length protein, were undetectable for the bands excised from extracts of cells expressing 3020insC Nod2.
Activation of IL10 transcription by hnRNP-A1
The hnRNPs are among the most abundant proteins in the eukaryotic cell nucleus and are directly involved in DNA repair and telomere elongation; chromatin remodeling and transcription; RNA splicing and stability; export of mature RNA; and translation. Approximately 30 hnRNPs have been identified. Autoantibodies specific for A and B proteins of the hnRNP complex have been detected in rheumatoid arthritis, systemic lupus erythematosus and mixed connective tissue disease22,23. Moreover, hnRNP-A1 is also found as a potential auto-antigen in 38% of psoriasis patients24.
To analyze the influence of hnRNP-A1 on IL10 transcription, we transfected a mouse hnRNP-A1 expression vector (effector) together with the IL10 promoter–luciferase reporter into RAW264.7 cells. We found that hnRNP-A1 stimulated Il10 promoter activity in a dose-dependent way at fairly low ratios of reporter to effector (Fig. 6a). Moreover, hnRNP-A1 also augmented LPS- and peptidoglycan-stimulated IL10 promoter–luciferase reporter activity (data not shown). Notably, mouse hnRNP-A1 also stimulated a mouse Il10 promoter–luciferase reporter construct (Fig. 6b). However, hnRNP-A1 had little effect on luciferase reporters driven by human IL-12p40 or ‘generic’ NF-κB promoters, even at high ratios of reporter to effector (data not shown). Furthermore, the stimulatory effect of human hnRNP-A1 was mediated through the NRE in the IL10 promoter, as hnRNP-A1 had no effect on the NRE-mutant IL10 promoter–luciferase reporter (Fig. 6c).

Stimulation of IL10 transcription by hnRNP-A1. (a,b) Luciferase activity of RAW264.7 cells transfected for 48 h with a human (a) or mouse (b) IL-10 reporter together with pCDNA3 and/or mouse hnRNP-A1 at various reporter/effector ratios (horizontal axes). (c) Luciferase activity of RAW264.7 cells transfected with reporter constructs driven by a wild-type (WT NRE) or mutant (Mut NRE) IL10 NRE, together with empty vector (pcDNA3) or vector encoding mouse hnRNP-A1, presented relative to that in pCDNA3-transfected cells. (d) ELISA of IL-10 and IL-12p40 in supernatants of primary human monocytes transfected with siRNA specific for hnRNP-A1 (4 and 1–3) or mock transfected (Mock; d) or transfected with with empty vector (pCDNA3) or expression vector encoding hnRNP-A1 (e), then, 6 h after transfection, stimulated for 12 h with LPS. Below, RT-PCR analysis of the expression of hnRNP-A1 and β-actin mRNA demonstrates the efficiency of siRNA-mediated knockdown (d) or vector transfection (e). *, P < 0.05. Data represent from three to five independent experiments (mean and s.d.).
To demonstrate the physiological function of hnRNP-A1 in the regulation of endogenous IL-10 production in primary cells, we transfected human peripheral blood–derived monocytes with small interfering RNA (siRNA) specific for human hnRNP-A1. A pool of three siRNA molecules that diminished expression of hnRNP-A1 inhibited LPS-stimulated IL-10 production by about 45%; a fourth hnRNP-A1-specific siRNA resembled the mock siRNA, as it failed to suppress hnRNP-A1 and did not influence IL-10 production (Fig. 6d). These siRNAs had no effect on LPS-induced production of IL-12p40. The degree of inhibition of IL-10 production by hnRNP-A1-specific siRNA was notable, given that fewer than 40% of the monocytes were transfected with the siRNA (data not shown) and that hnRNP-A1 expression was only partially ‘knocked down’. Conversely, overexpression of hnRNP-A1 in these cells strongly enhanced LPS-stimulated production of IL-10 but had no effect on IL-12p40 secretion (Fig. 6e). These results suggest that hnRNP-A1 promotes transcription of human IL10.
Suppression of hnRNP-A1–NRE binding by 3020insC Nod2
To confirm the identification of hnRNP-A1 as an NRE-binding nuclear protein by mass spectrometry, we used EMSA and chromatin immunoprecipitation (ChIP). As shown by EMSA, overexpression of hnRNP-A1 in RAW264.7 cells resulted in binding of protein to the human NRE probe; this binding activity was diminished by an antibody to hnRNP-A1 but not by an isotype-matched control antibody (Fig. 7a). ChIP analysis of primary human monocytes showed strong and specific binding of hnRNP-A1 in the region between positions −121 and +42 of IL10, which contains the NRE, but not in an irrelevant upstream region between positions −3158 and −2947 (Fig. 7b). The binding seemed to be constitutive and was not regulated by stimulation of primary human monocytes with LPS (Fig. 7c). These data indicate that hnRNP-A1 interacts with the IL10 NRE in human monocytes.

Binding of hnRNP-A1 to the IL10 NRE. (a) EMSA of nuclear extracts of RAW264.7 cells stably transfected with empty vector (pCDNA3) or vector encoding hnRNP-A1, analyzed with a probe containing wild-type IL10 NRE. Right, extracts incubated with anti-hnRNP-A1 or control immunoglobulin (IgG) before incubation with probe. Results are representative of three independent experiments. (b) ChIP of primary human monocytes with anti-hnRNP-A1 or control immunoglobulin (Ig), followed by PCR amplification of IL10 promoter regions between positions −121 and +42 (top) or positions −3158 and −2947 (bottom) in input DNA or DNA extracted from immunoprecipitates. Results are representative of three separate experiments. (c) ChIP of unstimulated (Med) and LPS-stimulated (LPS) primary human monocytes, followed by PCR amplification of the IL10 promoter region between positions −121 and +42. Rat IgG serves as a control. Results are representative of three separate experiments. (d) ChIP analysis of unstimulated human PBMCs from healthy control subjects (Ctrl) or patients with Crohn’s disease (CD) with wild-type NOD2 (WT) or homozygous for the 3020insC mutation (FS). Results are from one representative of three experiments. (e) Real-time quantitative PCR analysis of the binding of hnRNP-A1 in each sample in d. Results for immunoprecipitated DNA are presented relative to those for genomic input DNA. Data are representative of experiments done in triplicate (mean and s.d. of three donors per group).
Next we analyzed PBMCs from three patients with Crohn’s disease who were homozygous for 3020insC and had profoundly diminished IL-10 production25. The binding of hnRNP-A1 to IL10 around the NRE was much lower in these patients than in age- and sex-matched healthy people and patients with Crohn’s disease who lacked the 3020insC mutation (Fig. 7d). The diminished hnRNP-A1 binding in the patients with Crohn’s disease who were homozygous for 3020insC was even more prominent in experiments using real-time quantitative PCR (Fig. 7e).
We further investigated the molecular basis for the impaired nuclear binding activity of hnRNP-A1 in patients with Crohn’s disease who were homozygous for 3020insC. For this, we over-expressed Flag-tagged wild-type or 3020insC Nod2 and Flag-tagged hnRNP-A1 in HEK293 cells. Wild-type and 3020insC Nod2 were expressed in the cytoplasm but not in the nucleus, whereas hnRNP-A1 was expressed in both compartments (Fig. 8a). Notably, in addition to the intact form of hnRNP-A1 (about 36 kilodaltons), we noted three shorter forms of hnRNP-A1, which have been described as products of hnRNP-A1 cleavage mediated by caspase 3 activated during apoptosis of the human Burkitt lymphoma cell line BL60 induced by antibody to immunoglobulin M (anti-IgM)26. There was less cleaved hnRNP-A1 in cells expressing 3020insC Nod2 than in cells expressing empty vector or wild-type Nod2. In addition, intact hnRNP-A1 was serine-phosphorylated exclusively in the nucleus, whereas we detected serine-phosphorylated forms of cleavage products only in the cytoplasm. Expression of 3020insC Nod2 selectively diminished the presence of phosphorylated hnRNP-A1 in the nucleus. By coimmunoprecipitation, we found evidence of direct physical interaction between endogenous hnRNP-A1 and exogenous wild-type Nod2; hnRNP-A1 interacted less efficiently with 3020insC Nod2 (Fig. 8b). This interaction seemed to take place exclusively in the cytoplasm (data not shown), presumably because Nod2 is present only in the cytoplasm.
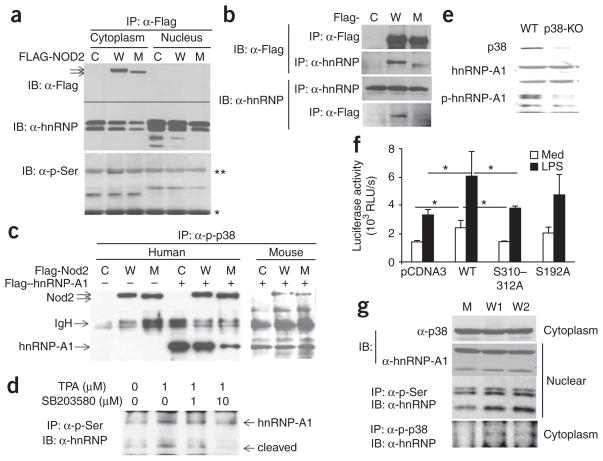
Nod2–hnRNP-A1 interaction and phosphorylation of hnRNP-A1 by p38. (a) Immunoassay of HEK293 cells transiently transfected for 1 d with empty vector (C) or with vector encoding Flag-tagged wild-type (W) or 3020insC (M) Nod2, together with vector encoding Flag-tagged hnRNP-A1, followed by immunoprecipitation of cytoplasmic and nuclear extracts with anti-Flag and immunoblot analysis with anti-Flag, anti-hnRNP-A1 (α-hnRNP) and antibody to phosphorylated serine (α-p-Ser). **, immunoglobulin heavy chain; *, immunoglobulin light chain. (b) Immunoassay of HEK293 cells transiently transfected for 1 d with the Flag-tagged constructs described in a, followed by immunoprecipitation and immunoblot analysis of whole-cell lysates with anti-Flag or anti-hnRNP-A1. (c) Immunoassay of HEK293 cells transfected with human wild-type Nod2 (W) or 3020insC Nod2 (M; left) or with mouse wild-type Nod2 (W) or 2939insC Nod2 (M; right), followed by immunoprecipitation of whole-cell lysates with antibody to serine-phosphorylated p38 (α-p-p38) and immunoblot analysis with anti-Flag. (d) Immunoassay of HEK293 cells treated for 60 min with the p38 inhibitor SB203580, followed by the addition of TPA (12-O-tetradecanoylphorbol-1,3-acetate) for 30 min (concentrations, above lanes) and immunoprecipitation and immunoblot analysis of nuclear extracts. Results in a–d represent at least three independent experiments. (e) Immunoblot analysis of whole-cell lysates of BMDMs obtained from wild-type mice (WT; n = 3) and p38α-deficient mice (p38-KO; n = 3) and then stimulated with LPS. Results are representative of two separate experiments. (f) Luciferase activity of lysates of RAW264.7 cells transfected with the human IL10 promoter–luciferase reporter together with empty vector (pCDNA3) or vector encoding wild-type, S310312A or S192A hnRNP-A1, then stimulated for 24 h with medium or LPS. *, P < 0.03. Data represent three independent experiments (mean and s.d.). (g) Immunoprecipitation and immunoblot analysis of cytoplasmic and nuclear extracts of PBMCs isolated from patients with Crohn’s disease homozygous for 3020insC (M; n = 4) or expressing wild-type Nod2 (W1; n = 4) and healthy people expressing wild-type Nod2 (W2; n = 6). Results are representative of one experiment.
Next we investigated whether the mitogen-activated protein kinase p38α (A001717) was responsible for phosphorylation of hnRNP-A1. Flag-tagged wild-type and 3020insC Nod2 interacted equally well with phosphorylated p38 (Fig. 8c). However, we detected more hnRNP-A1 in complexes of serine-phosphorylated p38 and wild-type Nod2 than in complexes of serine-phosphorylated p38 and 3020insC Nod2. Notably, the 2939insC Nod2 mouse counterpart of 3020insC Nod2 did not act like the human mutant in that it resembled wild-type Nod2 in its ability to facilitate interaction between serine-phosphorylated p38 and hnRNP-A1.
In support of the idea that p38 was responsible for hnRNP-A1 phosphorylation, we found that SB203580, a small chemical inhibitor of p38, blocked phorbol ester–induced phosphorylation of cleaved hnRNP-A1 in HEK293 cells in a dose-dependent way (Fig. 8d). As chemical inhibitors of p38 may have additional unidentified targets, we confirmed our findings in mice lacking p38α specifically in myeloid cells27. BMDMs from p38α-deficient and wild-type mice expressed similar amounts of total hnRNP-A1, but LPS-induced serine phosphorylation of hnRNP-A1 was much lower in p38α-deficient macrophages (Fig. 8e). These data support the idea that p38, particularly the α-isoform, is critical for phosphorylation of hnRNP-A1.
It has been shown that hnRNP-A1 is phosphorylated at the serine residue at position 192 (Ser192), Ser310, Ser311 and Ser312 after T cell stimulation28. To determine if these serine residues are critical for hnRNP-A1-mediated IL10 transcription, we generated mutants of hnRNP-A1 in which these serine residues were replaced with alanine (S192A and S310–312A). We transfected cells expressing the IL10 promoter–luciferase reporter with empty vector or vector encoding wild-type, S192A or S310–312A hnRNP-A1 constructs. Wild-type hnRNP-A1 enhanced both basal and LPS-stimulated IL10 promoter activity relative to the promoter activity resulting from transfection of empty vector (Fig. 8f). S192A hnRNP-A1 also induced promoter activity, albeit less efficiently than wild-type hnRNP-A1 did; in contrast, S310–312A hnRNP-A1 failed to stimulate IL10 reporter activity (Fig. 8f). These data collectively suggest that phosphorylation of hnRNP-A1 at Ser310–Ser312, possibly by p38, is important for its ability to enhance transcription of human IL10; moreover, 3020insC Nod2 may interfere with p38-mediated phosphorylation of hnRNP-A1.
In agreement with the hypothesis proposed above, we noted considerable impairment in the phosphorylation of cleaved and full-length hnRNP-A1 in the nucleus of cells from patients with Crohn’s disease who were homozygous for 3020insC, relative to that of cells from healthy subjects or patients with Crohn’s disease who lacked 3020insC (Fig. 8g). Expression of total p38 and hnRNP-A1 was similar in all samples. The average extent of phosphorylation of hnRNP-A1 in control subjects (n = 6) was 3.2-fold higher than that of patients with Crohn’s disease who were homozygous for 3020insC (n = 6; P = 0.02467). Consistent with the impaired phosphorylation of hnRNP-A1, there was less interaction between serine-phosphorylated p38 and hnRNP-A1 in these patients (Fig. 8g). These data collectively indicate that p38 phosphorylates hnRNP-A1 and that 3020insC Nod2 inhibits this event by blocking the interaction between p38 and hnRNP-A1 (Supplementary Fig. 2 online).
DISCUSSION
In this study we have investigated the controversial issue of the influence of the NOD2 mutation 3020insC on host defense and inflammation. We have shown that 3020insC Nod2 acted as an inhibitor of IL10 expression in human monocytes. Because of the critical function of IL-10 in controlling mucosal inflammation caused by intestinal microflora, this finding links 3020insC to a probable route leading to the development and pathogenesis of Crohn’s disease. Our observations indicate that 3020insC Nod2 can interfere with the steady-state intracellular signaling responsible for constitutive IL10 transcription. We have identified hnRNP-A1 as a chief target of 3020insC Nod2 in this pathway and have shown that hnRNP-A1 acted as a physiological inducer of IL10 transcription. Although 3020insC Nod2 did not inhibit the expression or nuclear localization of hnRNP-A1, it did suppress the DNA-binding activity of hnRNP-A1 by interfering with p38-mediated phosphorylation of hnRNP-A1.
Notably, 3020insC Nod2 blocked transcription of human IL10 but not mouse Il10, and the engineered mouse counterpart of the 3020insC Nod2 mutant, 2939insC Nod2, did not modulate expression of either mouse or human IL-10. Thus, the 3020insC-p38-hnRNP pathway is operative only in humans. This species specificity seems to be due to the ability of 3020insC Nod2 but not 2939insC Nod2 to block the interaction between p38 and hnRNP-A1.
We did not find evidence that 3020insC Nod2 could enhance secretion of IL-1β from primary human monocytes. That finding is inconsistent with studies of the Nod22939insC knock-in mouse, which shows exacerbated IL-1β production due to hyperactivation of NF-κB18. It suggests that the discrepancy between those results and our finding of a lack of effect on IL-1β production in primary human monocytes transfected with 3020insC Nod2 is due to the possibility that 2939insC Nod2 has a gain of activity that the human mutant does not have. The data collectively demonstrate that the mouse 2939insC Nod2 mutant is not equivalent to human 3020insC Nod2 and that lacking Nod2 is not the same as expressing 3020insC. These findings call for caution in comparisons of mouse models and human disease.
As 3020insC Nod2 blocked steady-state as well as microbe-induced IL10 expression, people with the 3020insC mutation may have chronically lower IL-10 production. This diminution in IL-10, when combined with other contributing factors over a lengthy period of time, could cumulatively cause persistent inflammation and homeostatic imbalance in the intestinal mucosa, leading to the development and pathogenesis of Crohn’s disease. In people heterozygous for 3020insC, wild-type Nod2 can presumably directly bind to 3020insC Nod2 and may thereby diminish its IL-10-inhibiting and disease-causing activities. This interaction could explain the much lower susceptibility to Crohn’s disease of heterozygous patients. Emphasizing the influence of other contributing factors, not all people with the 3020insC mutation on both chromosomes suffer from Crohn’s disease29. Moreover, none of the three common mutations in the LRR domain of NOD2 have been found in Japanese patients with Crohn’s disease30. That observation provides evidence for the presence of genetic heterogeneity among patients of different ethnic and racial backgrounds.
It should be emphasized that published research documenting lower defensin expression31, dysfunction of dendritic cells32 and diminished production of other cytokines25,33 supports the idea that 3020insC represents a loss-of-function mutation. It is likely that the cumulative phenotypic effect of the 3020insC variant in patients with Crohn’s disease may be attributable to other alterations in addition to diminished IL-10 production. Whether the 3020insC Nod2 mutant represents a loss of function or gain of function may depend on the response being measured. For example, because the 3020insC mutation is in the LRR domain of NOD2, the 3020insC Nod2 mutant may not be able to bind muramyl dipeptide; thus, all muramyl dipeptide–induced responses could be lost. In that context, 3020insC Nod2 is a loss-of-function mutant. In agreement with that conclusion, monocytes derived from patients with Crohn’s disease who bear other mutations in the LRR domain, like those from patients with the 3020insC mutation, show impaired responses to muramyl dipeptide and several TLR ligands in terms of secretion of IL-8 and tumor necrosis factor, respectively33. However, our evidence suggests that 3020insC Nod2 and other Nod2 variants bearing mutations in the LRR domain have also acquired the ability to inhibit IL-10 transcription.
In summary, our study challenges the present paradigms about the influence of the 3020insC mutation on Crohn’s disease. Notably, 3020insC is also strongly associated with graft-versus-host disease34, which is similar to Crohn’s disease in that both disorders are thought to be exacerbated by TH1 responses and suppressed by IL-10. Thus, our findings might be useful in efforts to identify therapeutic targets for the treatment of Crohn’s disease and other TH1-mediated auto-immune diseases associated with the 3020insC mutation.
METHODS
Mice
Nod2-knockout and control littermate mice were from P.J. Murray. RICK-knockout (RIP2-knockout) and control mice were from E. Pamer. Mice with conditional knockout of p38α were provided by J.M. Park27.
Antibodies and reagents
Antibody to phosphorylated serine (AB1603) was from Chemicon International; anti-Flag (M2) was from Sigma; anti-p38 (9212) and antibody to phosphorylated p38 (c9211) were from Cell Signaling; and other antibodies for immunoblot analysis, EMSA and ChIP (anti-hnRNP-A1 (10030), anti-Nod2 (30199)) were from Santa Cruz Biotechnology. Recombinant human and mouse interferon-γ were from Genzyme. Muramyl dipeptide, peptidoglycan and LPS from Escherichia coli were from Sigma. The peptidoglycan, muramyl dipeptide and Pam3Cys (tripalmitoyl cysteinyl seryl tetralysine) used in all experiments were from the same source and were used at the concentrations described before35. They were free of endotoxin contamination, as shown by analysis with polymixin B (data not shown).
Expression vectors
The mouse Nod2 expression vector and the mouse 2939insC Nod2 mutant were provided by G. Nunez. The 3020insC and other human Nod2 mutants were generated by overlapping PCR. Human hnRNP-A1 cDNA was amplified by RT-PCR of total RNA from human monocytes. The sequences of all vectors were verified and all plasmids were isolated with an EndoFree Maxi kit (Qiagen).
DNA affinity binding assay
The DNA affinity binding assay was done essentially as described36 with 2 μg biotinylated DNA oligonucleotides encompassing the IL10 NRE conjugated to 100 μl streptavidin-bound Dynabeads. Bound proteins were eluted by increasing the strength of the elution buffer and were separated by 10% SDS-PAGE gel and visualized by silver staining.
Retroviral and lentiviral packaging and macrophage transduction
GP2-293 packaging cells (Clontech Laboratories) at a confluency of about 50–70% in 100-mm culture plates were transfected, using Lipofectamine (Invitorogen), with 5 μg each of plasmids (pVSV-G; Clontech Laboratories) encoding human wild-type Nod2 or 3020insC Nod2. Then, 2 d after transfection, supernatants were cleared by centrifugation for 10 min at 1,000g and were pelleted for 90 min at 50,000g. Pelleted viruses were resuspended at 4 °C in DMEM. Bone marrow cells were infected with retroviral particles on day 2 and again on day 4 during their maturation in the presence of 20% (vol/vol) conditioned medium from mouse L cells. For lentivirus generation, HEK293TN cells (System Biosciences) at a confluency of 50% were transfected, using FuGENE 6 (Roche), with 3 μg of plasmid encoding viral envelope protein (G glycoprotein of vesicular stomatitis virus), 5 μg of the pMDLg/pRRE lentivirus packaging plasmid, 2.5 μg pRSV-REV (expressing human immunodeficiency virus 1 reverse transcriptase under control of the Rous sarcoma virus U3 promoter), and 10 μg Nod2-MA1 or 3020insC Nod2–MA1 expression plasmid (the bidirectional vector MA1). Then, 2 d after transfection, supernatants were cleared by centrifugation for 10 min at 1,000g and were pelleted for 90 min at 50,000g. Pelleted viruses were resuspended at 4 °C in DMEM. Human monocytes (0.5 × 106 cells per condition) were infected with 5 μl or 50 μl of lentivirus-containing supernatant on day 1 or day 3, respectively, before being stimulated with LPS on day 4.
Patients and genotyping of NOD2 mutations
Blood was collected from patients with Crohn’s disease (n = 74) and healthy volunteers (n = 20). NOD2 gene fragments containing the 3020insC site were amplified by PCR in 50-μl reaction volumes containing 100–200 ng genomic DNA and the appropriate primers in various concentrations (Supplementary Table 1 online) in 10 mM Tris-HCl pH 9.0, 50 mM KCl, 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, 0.1% (vol/vol) Triton X-100, 0.35 mM dNTPs and 2 U Taq DNA polymerase (Invitrogen). Samples were denatured at 92 °C for 5 min and then were amplified in a PTC-200 thermal cycler (MJ Research; Biozym) by 35 cycles of 92 °C for 1 min, 59.8 °C for 1 min and 72 °C for 1 min, followed by a final extension step of 72 °C for 3 min. The 3020insC polymorphism was analyzed by Genescan on an ABI Prism 3100 Genetic Analyzer according to the manufacturer’s protocol (Applied Biosystems). Three patients with Crohn’s disease homozygous for the 3020insC mutation were selected for further studies. Three patients with Crohn’s disease bearing the wild-type NOD2 allele and three healthy people, matched for age and sex (23–38 years of age; male), served as controls. All patients with Crohn’s disease were in remission and were not treated with steroid drugs or antibiological therapy (such as anti–tumor necrosis factor) during the 2 months before the study. None of the people selected had the NOD2 mutations of C to T at position 2104 (R702W) or G to C at position 2722 (G908R), also known to be associated with Crohn’s disease (data not shown)37.
Additional methods
Information on cell culture, cytokine measurement, reporter plasmids, preparation of nuclear extracts, immunoblot and immuno-precipitation, ChIP assay, siRNA, nanoflow liquid chromatography–tandem mass spectrometry analysis and the database search of tandem mass spectrometry data for the identification of peptide sequences is available in the Supplementary Methods online.
Statistical analysis
Student’s t-test (one-tailed) was used for data analysis where appropriate.
Accession codes
UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A001243, A001137 and A001717.
Acknowledgments
We thank P.J. Murray (St. Jude Children’s Research Hospital) for Nod2-knockout and control littermate mice; E. Pamer (Memorial Sloan-Kettering Cancer Center) for RICK-knockout (RIP2-knockout) and control mice; J.M. Park (Harvard University School of Medicine) for mice with conditional knockout of p38α; and G. Nunez (University of Michigan) for the mouse Nod2 expression vector and mouse 2939insC Nod2. Supported by the Broad Medical Research Program (IBD-210R2 to X.M.).
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
E.N. contributed to the work in Figures 3,,44,,66,,8;8; Y.H. contributed to the work in Figures 1–7; X.K. contributed to Figure 4; M.G.N. contributed to Figures 1,,77,,8;8; and X.M. contributed to the overall project.Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ni.1722
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2928218?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Selected Cytokines and Metalloproteinases in Inflammatory Bowel Disease.
Int J Mol Sci, 25(1):202, 22 Dec 2023
Cited by: 1 article | PMID: 38203373 | PMCID: PMC10779120
Review Free full text in Europe PMC
Immunologic and Genetic Contributors to CD46-Dependent Immune Dysregulation.
J Clin Immunol, 43(8):1840-1856, 21 Jul 2023
Cited by: 2 articles | PMID: 37477760 | PMCID: PMC10661731
Intestinal Barrier Dysfunction in Inflammatory Bowel Disease: Underpinning Pathogenesis and Therapeutics.
Dig Dis Sci, 68(12):4306-4320, 29 Sep 2023
Cited by: 10 articles | PMID: 37773554
Review
Role of Hydrogen Sulfide in Inflammatory Bowel Disease.
Antioxidants (Basel), 12(8):1570, 06 Aug 2023
Cited by: 6 articles | PMID: 37627565 | PMCID: PMC10452036
Review Free full text in Europe PMC
Role of Heterogeneous Nuclear Ribonucleoproteins in the Cancer-Immune Landscape.
Int J Mol Sci, 24(6):5086, 07 Mar 2023
Cited by: 4 articles | PMID: 36982162 | PMCID: PMC10049280
Review Free full text in Europe PMC
Go to all (135) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
NOD2 3020insC mutation and the pathogenesis of Crohn's disease: impaired IL-1beta production points to a loss-of-function phenotype.
Neth J Med, 63(8):305-308, 01 Sep 2005
Cited by: 45 articles | PMID: 16186640
Crohn's disease-associated Nod2 mutants reduce IL10 transcription.
Nat Immunol, 10(5):455-457, 01 May 2009
Cited by: 19 articles | PMID: 19381138
Crohn's disease patients homozygous for the 3020insC NOD2 mutation have a defective NOD2/TLR4 cross-tolerance to intestinal stimuli.
Immunology, 123(4):600-605, 20 Nov 2007
Cited by: 39 articles | PMID: 18028374
Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn's disease.
Immunol Rev, 260(1):249-260, 01 Jul 2014
Cited by: 62 articles | PMID: 24942694
Review
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R01 AI072149-01A2
Grant ID: R01 AI072149





