Abstract
Free full text

β-Arrestin-dependent signaling and trafficking of 7-transmembrane receptors is reciprocally regulated by the deubiquitinase USP33 and the E3 ligase Mdm2
Abstract
β-Arrestins are multifunctional adaptors that mediate the desensitization, internalization, and some signaling functions of seven-transmembrane receptors (7TMRs). Agonist-stimulated ubiquitination of β-arrestin2 mediated by the E3 ubiquitin ligase Mdm2 is critical for rapid β2-adrenergic receptor (β2AR) internalization. We now report the discovery that the deubiquitinating enzyme ubiquitin-specific protease 33 (USP33) binds β-arrestin2 and leads to the deubiquitination of β-arrestins. USP33 and Mdm2 function reciprocally and favor respectively the stability or lability of the receptor β-arrestin complex, thus regulating the longevity and subcellular localization of receptor signalosomes. Receptors such as the β2AR, previously shown to form loose complexes with β-arrestin (“class A”) promote a β-arrestin conformation conducive for binding to the deubiquitinase, whereas the vasopressin V2R, which forms tight β-arrestin complexes (“class B”), promotes a distinct β-arrestin conformation that favors dissociation of the enzyme. Thus, USP33–β-arrestin interaction is a key regulatory step in 7TMR trafficking and signal transmission from the activated receptors to downstream effectors.
The trafficking of seven-transmembrane receptors (7TMRs), also known as G protein-coupled receptors (GPCRs), is a tightly regulated process that modulates cell signaling and defines various physiological responses (1). Two homologous mammalian proteins, β-arrestin1 and β-arrestin2, have been established as primary endocytic and signaling adaptors for a large number of 7TMRs as well as other types of cell surface receptors (2). Agonist-stimulated 7TMRs first couple to and activate the heterotrimeric G proteins, but they are immediately phosphorylated by specialized serine-threonine kinases called G protein-coupled receptor kinases (GRKs) (2). This phosphorylation is an important determinant for the efficient plasma-membrane recruitment of the β-arrestin proteins to the receptor cytoplasmic domains and desensitization of G protein signaling.
Phosphorylation of a serine triplet on the carboxyl tail of receptors, such as the vasopressin V2 receptor (V2R), engenders their tight interaction with β-arrestins, facilitating cotrafficking and localization of receptor–arrestin complexes to endocytic vesicles (3). These stable β-arrestin-binders, also known as “class B” receptors, are differentiated from other 7TMRs, such as the β2-adrenergic receptors (β2AR) that are transient β-arrestin-binders (also known as “class A” receptors), which form a loose complex with translocated arrestins only at the plasma membrane (3). In addition to receptor phosphorylation, agonist-stimulated ubiquitination occurring on β-arrestins also governs the stability of receptor–β-arrestin interactions (4–6). β-Arrestin ubiquitination is crucial for both its endocytic and signaling functions (7, 8), and the kinetics of β-arrestin deubiquitination correlates with the dissociation of β-arrestins from activated receptors, suggesting that ubiquitin-specific protease(s) [USP(s)] might play specific regulatory roles in 7TMR endocytosis and signal transduction. These observations encouraged us to seek deubiq-uitinating enzyme(s) that might bind β-arrestin and regulate its functions.
Results
In a yeast 2-hybrid screen using β-arrestin1 as the bait, we obtained a truncated portion of ubiquitin-specific protease 33 (USP33) as a potential β-arrestin partner. This interaction was confirmed in mammalian cells by coimmunoprecipitation assays using HA-USP33 (ATCC clone, IMAGE:3491447) and β-arrestin1 and 2 isoforms (Fig. 1A, and Fig. S1A). Because USP33 binds both isoforms of β-arrestin equally, and β-arrestin2 ubiquitination has been extensively documented, we focused on the effects of USP33–β-arrestin2 interaction. HA-USP33 binds to both the N and C domains of β-arrestin2, and increased binding was observed with either domain alone compared to the WT (Fig. S1 B and C), suggesting that disrupting β-arrestin's basal conformation facilitates USP33 binding.
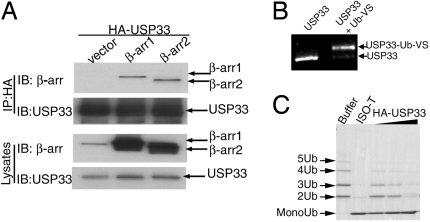
Identification of USP33 as a β-arrestin-binding deubiquitinase. (A) HA-USP33 transiently expressed in COS-7 cells along with β-arrestin1-Flag, β-arrestin2-Flag, or vector was immunoprecipitated (IP) with anti-hemagglutinin epitope (HA)-agarose conjugate, and the amount of bound β-arrestin1 and -2 was assessed by probing with an anti-β-arrestin antibody (Upper). IB, immunoblot. Expression levels of USP33 and β-arrestins in cell extracts are also displayed (Lower). Data shown are from 1 of 4 independent experiments. (B) The bands displayed represent purified USP33 that is either unmodified or covalently linked to ubiquitin-vinyl sulfone (Ub-VS), separated on an SDS/polyacrylamide gel (6%), and detected by Sypro ruby red staining. (C) Lys-63-linked polyubiquitin chains, which lack monoubiquitin, were incubated with buffer (lane 1), Isopeptidase T (lane 2), or increasing amounts of purified USP33 (lanes 3–5). The appearance of a monoubiquitin band corresponds to the depolymerizing activity of USPs. Data displayed in B and C are representative of 3 or 4 experiments performed with 3 separate USP33 purifications.
Next, we purified HA-USP33 from transfected COS-7 cells by using HA-affinity beads (Fig. S2A) and determined if it had the predicted deubiquitinating enzymatic activity. Incubation of the commonly used active site probe of deubiquitinases (DUBs), ubiquitin-vinyl sulfone (Ub-VS) (9) causes an expected mobility shift of HA-USP33 corresponding to formation of the HA-USP33-Ub-VS adduct (Fig. 1B). Additionally, when increasing amounts of USP33 (0.1–1 μg) are incubated with Lys-63-linked polyubiquitin chains, we find a corresponding increase in the appearance of monoubiquitin due to the depolymerizing activity of USP33, which is similar to that of a generic DUB, Isopeptidase T (USP5) (Fig. 1C). USP33 also depolymerizes Lys-48-linked polyubiquitin chains (Fig. S2B). Because there is no accumulation of diubiquitin in these assays, USP33 likely deubiquitinates monoubiquitinated proteins as well.
Detection of β-arrestin ubiquitination requires prior inhibition of DUBs (e.g., with 10 mM N-ethylmaleimide), therefore an effect of coexpression of USP33 on β-arrestin ubiquitination could not be assessed by previously used cellular immunoprecipitation assays (4). We therefore tested the DUB activity of HA-USP33 on ubiquitinated β-arrestin in an in vitro set-up. We transfected HEK-293 or COS-7 cells with vector or Flag-β-arrestin2, immunoprecipitated β-arrestin with or without agonist stimulation, removed N-ethylmaleimide during wash steps, and treated the isolated sample with purified HA-USP33 (Fig. 2A). USP33 efficiently deubiquitinates β-arrestin2 as evidenced by the disappearance of ubiquitination signals only in the USP33-containing lanes (Fig. 2A).
USP33 inhibits vasopressin-stimulated β-arrestin ubiquitination, endosomal trafficking, and ERK activation. (A) Flag immunoprecipitates isolated before or after 1 μM [Arg]vasopressin (AVP) stimulation from HEK-293 cells that transiently express the V2R with either pcDNA3 or β-arrestin2-Flag were treated with a mock purification (first 4 lanes in each gel) or purified HA-USP33 (last 4 lanes in each gel). After incubation, samples were subjected to Western analyses with an anti-ubiquitin antibody (Left) and reprobed with an anti-Flag antibody (Right). (B) HEK-293 cells with stable HA-V2R expression were transiently transfected with β-arrestin2-GFP and RFP-USP33 and stimulated with 1 μM AVP for 20 min. Two cells are shown, one with no detectable RFP-USP33 that has the expected endosomal recruitment of β-arrestin2, the other that expresses USP33 in which β-arrestin recruitment to the endosomes is inhibited. (C) Confocal micrographs show endosomal distribution of β-arrestin2-GFP upon AVP stimulation in cells overexpressing USP33Cys:His. (D) HEK-293 cells transiently expressing HA-V2R with either vector or USP33 were stimulated with 1 μM AVP for the indicated times and cell lysates were analyzed for ERK and phosphorylated ERK (p-ERK) by immunoblotting. (E) The graph represents quantification of p-ERK stimulated by a time course of AVP in HEK-293 transiently transfected with HA-V2R along with vector or USP33 from 4 independent experiments. *, P < 0.01 by 2-way ANOVA, n = 4.
To test if USP33-mediated accelerated deubiquitination could affect the stability of receptor–β-arrestin complexes (5), we overexpressed USP33 in HEK-293 cells along with the V2R and β-arrestin-2-GFP and examined the distribution of each protein by confocal microscopy. The subcellular distribution of GFP-USP33 or HA-USP33 when immunostained alone (see Methods) is shown in Fig. S3A. USP33 is localized throughout the membrane network including vesicles and is abundant in the perinuclear compartments (10). In cells expressing HA-V2R and β-arrestin2-GFP along with USP33 (Fig. S3B) or RFP-USP33 (Fig. 2B, lower HEK cell), we do not observe endosomal recruitment of β-arrestin upon agonist stimulation. In these transiently transfected cells, most cells coexpress both β-arrestin and USP33. However, some cells contain only β-arrestin2-GFP, and here we observe normal endosomal localization of β-arrestin2(Fig. 2B, upper HEK cell). DUBs are cysteine proteases in which a cysteine, a histidine, and an aspartate constitute the catalytic triad and are essential for enzymatic activity (11). The inhibitory effect of USP33 on trafficking requires its enzymatic activity because overexpression of a catalytic site mutant, USP33Cys:His, does lead to endosomal recruitment of β-arrestin2 upon V2R stimulation (Fig. 2C). As expected, this mutant does not depolymerize either ubiquitinated β-arrestin or polyubiquitin chains in vitro. USP33 coexpression does not alter β-arrestin recruitment to the plasma membrane upon either V2R or β2AR activation (Fig. 2B and Fig. S3B). Additionally, β-arrestin2-V2R trafficking is unaffected when another DUB, GFP-USP4 (12), is coexpressed (Fig. S4). Earlier studies on the temporal aspects of 7TMR-stimulated ERK signaling have indicated that the late-ERK component is β-arrestin dependent, whereas early transient ERK activity is due to G protein-dependent mechanisms (2). When we compared the time course of phospho-ERK1/2 (p-ERK) stimulated by vasopressin in vector- versus HA-USP33-transfected cells, we observed a marked diminution in p-ERK signals after 5-min agonist stimulation (Fig. 2 D and E) in the USP33-transfected cells. These findings suggest that an increase in USP33 activity prevents sustained ubiquitination of β-arrestin2, thereby destabilizing V2R-β-arrestin interaction and diminishing β-arrestin-dependent ERK signaling.
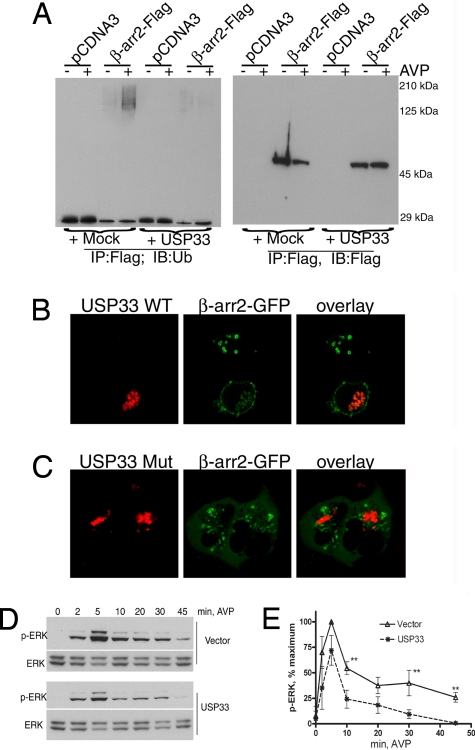
The above effects of a DUB prompted us to investigate the consequences of augmenting the reverse process (i.e., β-arrestin ubiquitination) by overexpressing an E3 ubiquitin ligase. Mdm2 is a β-arrestin-binding partner that specifically ubiquitinates β-arrestin2 upon β2AR stimulation (4). However, its role in β2AR-stimulated β-arrestin-dependent signaling and trafficking was not known. Upon coexpression of Mdm2 in HEK-293 cells, activated β2AR and β-arrestin2-GFP formed stable complexes that predominantly localized on endosomes (Fig. 3A, middle row). This pattern of endosomal cotrafficking of β2AR–β-arrestin complexes suggests that the presence of excess Mdm2 promotes sustained ubiquitination of β-arrestin and stabilizes receptor–β-arrestin interaction. In cells with endogenous levels of Mdm2 (end.), we detected only membrane recruitment of β-arrestins due to the transient nature of both β-arrestin ubiquitination and β-arrestin–receptor interaction (Fig. 3A, bottom row). Immunostaining of Mdm2-transfected cells suggests only a modest enhancement in Mdm2 levels compared with vector-transfected cells (compare the third column across Fig. 3A). However, Western blot analyses of cell lysates revealed an 8-fold overexpression above endogenous levels of Mdm2. Overexpression of Mdm2 also leads to an increase in p-ERK detected in whole-cell lysates upon isoproterenol stimulation of HEK-293 cells for more than 5 min (Fig. 3B). The p-ERK response in whole-cell lysates at 5 min of isoproterenol stimulation is composed of both G protein- and β-arrestin-dependent signals (2), and not surprisingly an Mdm2 effect is not apparent in these assays. In contrast, there is a marked Mdm2-dependent enhancement of ERK activity in β2AR-β-arrestin immunoprecipitates isolated at 5 min of agonist treatment (Fig. S5). In addition, the binding of p-ERK is significantly reduced upon coexpression of an Mdm2 deletion mutant (Mdm21–400) that lacks the RING E3 ligase domain (Fig. S5). Mdm2 coexpression also prevents rapid deubiquitination of β-arrestin2 in response to β2AR stimulation, whereas coexpression of Mdm21–400 prevents β-arrestin ubiquitination (4). Thus, an increase in Mdm2 activity promotes robust β-arrestin ubiquitination, stabilizes β2AR-β-arrestin binding, and extends the longevity of signalosomes, allowing their localization in endosomal compartments.
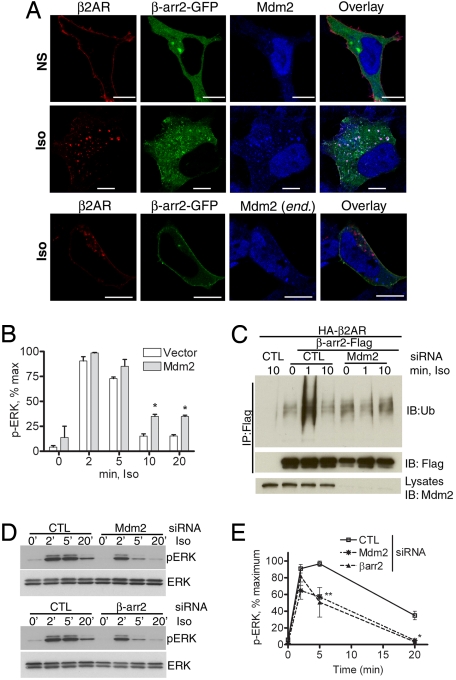
Mdm2 promotes β-arrestin recruitment to endosomes and enhances signaling. (A) HEK-293 cells transiently expressing HA-β2AR and Mdm2 without stimulation (NS, Top) or with 20-min isoproterenol (Iso, Middle) stimulation are fixed and immunostained to detect the receptor (anti-β2AR, H-20) shown in red and Mdm2 (Ab-1) displayed in blue. Distribution of β-arrestin2-GFP is shown in green. In Bottom, confocal pictures of agonist-stimulated HEK-293 cells expressing HA-β2AR, but with endogenous (end.) Mdm2 are shown. (Scale bars, 10 μm.) (B) Bar graph showing quantification (n = 3) of p-ERK in whole-cell extracts in HEK-293 cells with or without exogenous Mdm2 expression in response to isoproterenol stimulation for indicated times. *, P < 0.05, 2-way ANOVA. (C) (Top) HEK-293 cells were transiently transfected with HA-β2AR and β-arrestin2-Flag along with siRNA that targets nothing (control, CTL) or Mdm2; 48–60 hr posttransfection, β-arrestin2 was immunoprecipitated after agonist treatment for the indicated times and the immunoprecipitate was probed with an anti-ubiquitin antibody. (Middle) The amount of β-arrestin in the immunoprecipitate. (Lower) The levels of Mdm2 in cell extracts. (D) HEK-293 cells with or without either Mdm2 or β-arrestin2 depletion are analyzed for isoproterenol-stimulated p-ERK and ERK. (E) Graph represents quantification of time course of p-ERK from 4 independent experiments ± SEM. Two-way ANOVA, **, P < 0.01; *, P < 0.05, CTL versus Mdm2, and CTL versus β-arr2, 5- and 20-min signals, respectively.
We next evaluated the effects of Mdm2 knockdown on β-arrestin ubiquitination and its influence on endocytosis and signaling. Transfection of HEK-293 cells with Mdm2 siRNA leads to >98% knockdown of Mdm2 protein (Fig. 3C Bottom). Under these conditions, there is complete elimination of the 1-min β-arrestin ubiquitination induced by isoproterenol, which is observed as a robust signal in cells transfected with control siRNA (Fig. 3C). Because β-arrestin functions as a pleiotropic adaptor and because we are isolating the entire cellular pool of β-arrestin, we also detect basal signals unaffected by Mdm2 depletion, which are likely from β-arrestin modification by other E3 ligases independent of β2AR activation. Mdm2 depletion also dramatically reduces the isoproterenol-stimulated p-ERK induced at 5- and 20-min time points (Fig. 3 D and E). ERK activities at these time points are also sensitive to β-arrestin2 depletion (Fig. 3E) (2). Notably, ERK activation at the 2-min time point that is β-arrestin independent is unaffected by Mdm2 depletion (Fig. 3E). These data indicate that Mdm2 and its mediation of β-arrestin ubiquitination are required for β-arrestin-dependent signaling stimulated by the β2AR.
In contrast to Mdm2 knockdown, depletion of USP33 led to a complete inhibition of β-arrestin deubiquitination that normally occurs at 10-min stimulation (Fig. 4A). This sustained β-arrestin ubiquitination in USP33-depleted cells correlates with an accentuated stability of β-arrestin–β2AR interaction promoting their colocalization on endosomes as displayed in the confocal micrographs (Fig. 4B Upper). The plasma membrane recruitment of β-arrestin observed with endogenous USP33 levels is shown in Fig. 4B Lower. Moreover, USP33 depletion prolonged ERK activity upon isoproterenol stimulation (Fig. 4 C and D). Three separate siRNAs that knock down USP33 with equivalent efficiency (Fig. 4E) produced identical effects on β-arrestin ubiquitination and trafficking and on isoproterenol-induced ERK activation. Thus USP33 specifically deubiquitinates β-arrestin, prevents its cointernalization with the β2ARs, curtails β-arrestin-dependent signaling, and acts as an endogenous molecular “antagonist” for the β-arrestin pathway.
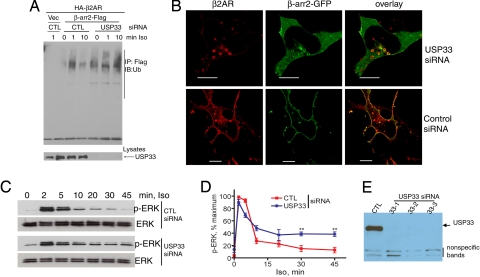
USP33 knockdown prevents β-arrestin deubiquitination, promotes endosomal trafficking, and prolongs ERK signaling. (A) HEK-293 cells were transfected with HA-β2AR, β-arrestin2-Flag, and either control or USP33 siRNA. Flag immunoprecipitates were isolated after isoproterenol stimulation for the indicated times and probed with an anti-ubiquitin antibody (Upper). (Lower) USP33 levels in control and USP-knockdown samples. (B) Confocal micrographs show distribution of Flag-β2AR (red) and β-arrestin2-GFP (green) in USP33-depleted cells (Upper) and control (Lower) cells in agonist-stimulated HEK-293 cells. (Scale bars, 10 μm.) (C) Western blots of p-ERK and ERK in response to a time course of isoproterenol (100 nM) activation, detected in HEK-293 cells with stable β2AR expression (2 pmol/mg of protein) and with indicated siRNA transfections. (D) Quantification of p-ERK signals from 3 independent experiments. **, P < 0.01, control versus USP33, 2-way ANOVA. (E) A representative blot for USP33 levels showing Western analysis of 25 μg of lysate protein from each siRNA transfection.
To address the question of why USP33 rapidly deubiquitinates β-arrestin2 upon β2AR stimulation, but not during V2R stimulation, we analyzed the kinetics of β-arrestin–USP33 interaction in response to agonist stimulation of both these receptors. Flag-β-arrestin2 immunoprecipitates isolated from COS-7 cells (Fig. 5A and Fig. S6A) or HEK-293 cells (Fig. S6 B and C) transfected with HA-β2AR contain a small amount of bound endogenous USP33 in the absence of agonist activation. However, within 1–5 min of isoproterenol stimulation, the binding increased by at least 2-fold and a robust interaction was observed up to 20 min (Fig. 5A, Fig. S6 A–C). In sharp contrast, stimulation of V2R-transfected cells leads to an agonist-promoted decrease in β-arrestin-USP33 interaction within 1 min of stimulation (Fig. 5A, Fig. S6 A–C).
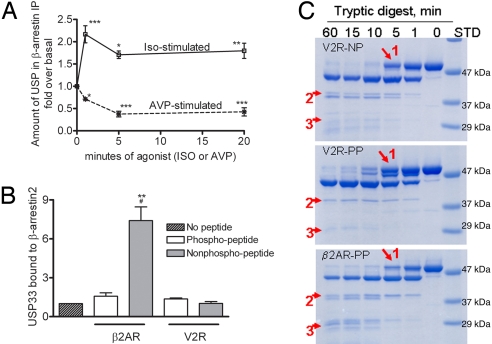
β-Arrestin–USP33 interaction displays different kinetics upon stimulation of different 7TMRs and is dependent on distinct conformational changes. (A) COS-7 cells transiently expressing either HA-β2AR or HA-V2R along with β-arrestin2-Flag were stimulated with respective agonists for the indicated times, β-arrestins were immunoprecipitated, and the immunoprecipitate was probed with USP33 antisera. The graphs show the quantification of USP33 bound to isolated β-arrestin obtained from 4 independent experiments. The 4 time points within each binding curve are analyzed by 1-way ANOVA. In each case, stimulated samples are significantly different from unstimulated samples; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Purified HA-USP33 was incubated alone or with β-arrestin2-His6 without or with indicated receptor peptides (see Methods). USP33 binding from 4 separate experiments is quantified and normalized to β-arrestin levels and plotted as bar graphs; #, P < 0.05, β2AR-PP versus β2AR-NP; **, P < 0.01, β2AR-PP versus all other samples, 1-way ANOVA, Bonferoni comparison. (C) SDS/PAGE analyses of the limited tryptic proteolytic products of β-arrestin2 with indicated receptor peptides (see Methods). Red arrows indicate the significant differences in the limited proteolysis patterns.
To define the basis for these marked differences in response to β2AR and V2R stimulation, we performed in vitro binding experiments using purified β-arrestin2, HA-USP33, and either phosphorylated or nonphosphorylated receptor peptides corresponding to the C termini of β2AR and V2R. Phosphorylated V2R peptides have been previously shown to induce an active conformation of β-arrestins (13, 14). In the absence of any receptor peptide, β-arrestin2 specifically binds to HA-USP33 (Fig. S6C). This binding is unaffected by the addition of β2AR-nonphosphopeptide or V2R phospho/nonphosphopeptides. However, a phosphorylated form of the β2AR peptide dramatically increases the interaction (Fig. 5B, Fig. S6C).
Interestingly, binding of these different phosphopeptides, which mimic the phosphorylated C termini of V2R and β2AR, induced different limited tryptic proteolysis patterns of β-arrestin2, suggesting that different phosphorylation patterns on different receptors may induce distinct conformational changes in the β-arrestin molecule (Fig. 5C). The tryptic digestion pattern of β-arrestin2 in the presence of nonphosphorylated peptides of V2R (Fig. 5C) (14) and β2AR are identical. The proteolysis patterns in the presence of a V2R phosphopeptide (Fig. 5C) (14) and β2AR phosphopeptide (Fig. 5C) are markedly different, indicating distinct conformational changes induced by different phosphopeptides mimicking the phosphorylated C termini of different receptors. These data suggest a model in which phosphorylation on distinct sites at the C termini of different 7TMRs may function as a “code” that translates to specific “active conformations” in recruited β-arrestins (Fig. 6). The conformationally active β-arrestin associated with a particular receptor is presumably modified with ubiquitin moieties/chains at distinct sites, which then promote or prevent recruitment of USP33. Thus 7TMR-induced conformations in β-arrestin2 would dictate the subsequent timing of its deubiquitination by regulating the pattern of recruitment of deubiquitinating enzymes. By dictating the formation of labile or stable receptor signalosomes, the nature of β-arrestin ubiquitination would act as a code (Fig. 6) to provide spatial and temporal resolution as well as specificity in 7TMR signal transduction.
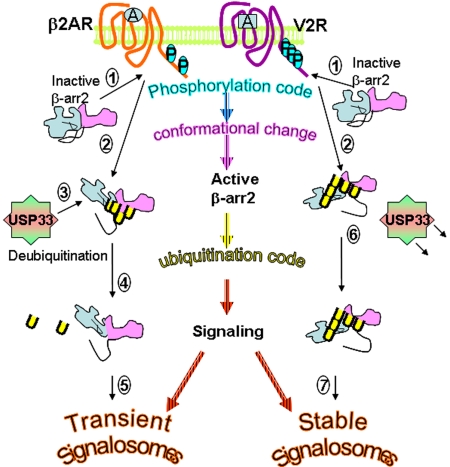
Schematic showing the effects of posttranslational modifications in 7TMR signaling. Step 1, β-arrestin2 resides in a basal state in the cytoplasm and is recruited to the plasma membrane and binds phosphorylated C termini of 7TMRs. The sites of phosphorylation differ among the 2 representative receptors shown. Step 2, upon binding to each receptor, β-arrestin2 undergoes a distinct conformational reorientation, thus allowing distinct regions to become modified by ubiquitination. Step 3, the β2AR-induced conformation promotes β-arrestin2–USP33 interaction. Step 4, USP33 deubiquitinates β-arrestin, leading to the dissociation of β-arrestin from the β2AR. Step 5, β2AR–β-arrestin2 signalosomes are short-lived and promote transient ERK activity that is predominantly nonendosomal. Step 6, β-arrestin2 conformation induced by the V2R activation prevents USP33 binding, thus protecting β-arrestin ubiquitination, allowing tight binding to activated receptors. Step 7, V2R–β-arrestin2 signalosomes are stable and result in robust ERK activity that is predominantly localized on endosomes.
Discussion
We report the discovery of a molecular mechanism that controls the rate of β-arrestin deubiquitination and delineates the reciprocal roles of ubiquitination and deubiquitination of β-arrestin2 in regulating trafficking of β-arrestin/7TMR complexes and signaling. Stimulation of the β2AR (class A or transient β-arrestin-binder) induces transient ubiquitination of β-arrestin2 and subsequently promotes association of β-arrestin with the DUB USP33. This interaction facilitates the deubiquitination of β-arrestin, leading to its dissociation from the β2AR. In contrast, stable β-arrestin-binders, such as the V2R (class B), promote a β-arrestin conformation that does not favor the association of USP33 with β-arrestin. By curtailing β-arrestin ubiquitination, USP33 destabilizes β2AR-β-arrestin binding, decreases the extent of ERK activation via the β-arrestin-dependent pathway, and thus functions to inhibit or desensitize β-arrestin-dependent signaling in response to β2AR activation. In contrast, ubiquitination of β-arrestin by Mdm2 promotes reciprocal events (i.e., stabilization of receptor–arrestin complexes and augmentation of β-arrestin-dependent ERK activation).
Posttranslational modification by ubiquitin is now appreciated for its noncanonical roles in regulating pathways other than those leading to protein degradation (15–17). In general, proteins modified with Lys-48-linked ubiquitin chains are directed to the 26S proteasomes, whereas the Lys-63 chains are suggested to play a role in endocytosis (18, 19). However, as with phosphorylation and other posttranslational modifications, ubiquitination is reversible, and the functional diversity and regulatory roles of various DUBs are just beginning to be appreciated (11). The human genome contains about 80 DUBs divided into 5 subfamilies based on their catalytic domains, of which the USP subclass includes the majority of the DUBs (11). Although several DUBs display a preferential activity in cleaving specific ubiquitin chain linkages (11), USP33 depolymerizes both Lys-48 and Lys-63 linkages and might function to deubiquitinate substrate proteins in both endocytic and proteasomal pathways analogous to the role played by Doa4 in yeast (20). Unlike other proteases, DUBs do not exist as precursors, and their cellular activity and substrate specificity are thought to be regulated by subcellular localization and protein interactions (11). Although earlier studies indicate that β-arrestins serve as crucial adaptors for escorting E3 ubiquitin ligase activity in various systems (2, 21, 22), this study further expands their scope to function as adaptors for DUBs, thus allowing substrate recognition and specificity.
Upon USP33 overexpression, the pattern of β-arrestin recruitment to the V2R and the extent of ERK signaling mimic those of receptors that transiently interact with β-arrestins and induce only transient β-arrestin ubiquitination, such as the β2ARs. Because of the increased activity of USP33, β-arrestins are prevented from being stably ubiquitinated upon V2R activation. On the other hand, depletion of USP33 prolongs the interaction between β-arrestin and a class A receptor allowing sustained signaling. This implies that during β2AR signaling, USP33 plays a regulatory role by dissolving the receptor–arrestin signalosome by promoting deubiquitination and dissociation of β-arrestin2. In this context, USP33 functions much like an “antagonist” to inhibit β-arrestin-dependent ERK activation and regulates the extent of downstream signaling.
Why is the class A system susceptible to USP33-dependent regulation, but not the class B? This results from differences in the receptor-induced conformational changes in the β-arrestin molecule, which subsequently tune the interaction between “activated” β-arrestin and the DUB. Previous studies using various biochemical and mutagenesis approaches have demonstrated that β-arrestins undergo conformational changes in response to 7TMR activation (23, 24); herein we show that distinct conformational changes in β-arrestins may be induced by different receptors. Our findings suggest an interesting form of cross-talk between different posttranslational modifications (25) on different proteins along the 7TMR signaling pathway. Thus the “phosphorylation code” on the receptor carboxyl tail produces distinct conformational changes in the bound β-arrestins. This might lead to ubiquitin modifications on distinct regions of β-arrestin resulting in an “ubiquitination code” that tunes signal strength, localization, and cellular functions (Fig. 6) (7, 8). Although data from our in vitro assays (Fig. 5C) suggest that conformational changes occur in β-arrestin2 before its ubiquitination, it is also likely that ubiquitination serves to further embellish and modify the active conformations of β-arrestin to provide a suitable platform for recruiting various endocytic and signaling proteins. It is estimated that ≈300 proteins constitute the “β-arrestin-interactome” (26), and it is plausible that many of these interacting proteins, either directly or indirectly, bind ubiquitinated β-arrestin. Our assays involving polyubiquitin chains indicate that USP33-mediated Lys-48 depolymerization is less efficient than what we observe for the Lys-63-linked chains (Fig. S2B). Future studies should reveal if the specific type of ubiquitination induced by the β2AR and the V2R involve different types of chain linkage (e.g., Lys-48 versus Lys-63), which could also determine the rate of deubiquitination and/or the affinity for USP33.
The N-domain in β-arrestin2 carries the binding site for E3 ligase Mdm2 (27, 28), whereas USP33 binds both N and C domains. Mdm2-β-arrestin binding occurs constitutively (29, 30) and does not persist beyond 10–15 min of β2AR activation (30). Mdm2 modifies β-arrestin when receptor interaction promotes an “active conformation” and likely targets specific lysine residues that are revealed on β-arrestin conformational change. Mdm2 and USP33 binding to β-arrestin2 are sequential events, and the release of Mdm2 on β-arrestin ubiquitination is followed by USP33 binding leading to β-arrestin deubiquitination. β-Arrestin binding to these and other regulatory proteins appear to involve dynamic and timed interactions choreographed by receptor activation. Our findings thus underscore the crucial regulatory significance of β-arrestin ubiquitination and deubiquitination carried out by the reciprocal activities of the E3 ligase Mdm2, which is coupled to endocytosis and signaling and of the deubiquitinase USP33, which is coupled to signal termination and perhaps receptor recycling.
Methods
Reagents, Cell Culture, Plasmids, and Transfection.
COS-7 and HEK-293 cells were purchased from the American Type Culture Collection and grown under recommended optimal conditions. Anti-Flag affinity agarose, anti-HA affinity agarose, anti-Flag-M2, Sypro-Ruby stain, N-ethylmaleimide, ovalbumin, and TPCK-treated trypsin were from Sigma. Rabbit polyclonal antiserum raised against a custom peptide of a mouse-USP33 sequence, EEEPQTLTSEETVEEEKSQSDVD, was used for immunostaining and detection of USP33.
Other Methods.
The remaining methods section is provided as SI Methods.
Acknowledgments.
We thank Donna Addison and Elizabeth Hall for excellent secretarial assistance. Grants from the National Institutes of Health (HL 080525 to S.K.S.; HL 16037 and HL 70631 to R.J.L.; GM 030308 to K.D.W.) and the American Heart Association (0530014N to S.K.S.) supported this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901083106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0901083106
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/106/16/6650.full.pdf
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/16/6650
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/16/6650.pdf
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/16/6650
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0901083106
Article citations
Arrestins: A Small Family of Multi-Functional Proteins.
Int J Mol Sci, 25(11):6284, 06 Jun 2024
Cited by: 0 articles | PMID: 38892473 | PMCID: PMC11173308
Review Free full text in Europe PMC
Beneath the surface: endosomal GPCR signaling.
Trends Biochem Sci, 49(6):520-531, 19 Apr 2024
Cited by: 1 article | PMID: 38643023
Review
Transcriptional response of the heart to vagus nerve stimulation.
Physiol Genomics, 56(2):167-178, 04 Dec 2023
Cited by: 0 articles | PMID: 39071113
Transcriptional response of the heart to vagus nerve stimulation.
Physiol Genomics, 56(2):167-178, 04 Dec 2023
Cited by: 0 articles | PMID: 38047311 | PMCID: PMC7616044
β-adrenergic receptor signaling mediated by β-arrestins and its potential role in heart failure.
Curr Opin Physiol, 37:100723, 18 Nov 2023
Cited by: 0 articles | PMID: 38094036
Go to all (99) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Receptor-specific ubiquitination of beta-arrestin directs assembly and targeting of seven-transmembrane receptor signalosomes.
J Biol Chem, 280(15):15315-15324, 07 Feb 2005
Cited by: 103 articles | PMID: 15699045
Trafficking patterns of beta-arrestin and G protein-coupled receptors determined by the kinetics of beta-arrestin deubiquitination.
J Biol Chem, 278(16):14498-14506, 06 Feb 2003
Cited by: 148 articles | PMID: 12574160
Deubiquitinases and their emerging roles in β-arrestin-mediated signaling.
Methods Enzymol, 535:351-370, 01 Jan 2014
Cited by: 4 articles | PMID: 24377933
Arrestins and protein ubiquitination.
Prog Mol Biol Transl Sci, 118:175-204, 01 Jan 2013
Cited by: 28 articles | PMID: 23764054
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NHLBI NIH HHS (7)
Grant ID: R01 HL080525
Grant ID: HL 080525
Grant ID: R01 HL070631
Grant ID: HL 16037
Grant ID: HL 70631
Grant ID: R01 HL016037
Grant ID: R01 HL080525-04
NIGMS NIH HHS (2)
Grant ID: R01 GM030308
Grant ID: GM 030308





