Abstract
Free full text

Characterization of the Butyrivibrio fibrisolvens glgB gene, which encodes a glycogen-branching enzyme with starch-clearing activity.
Abstract
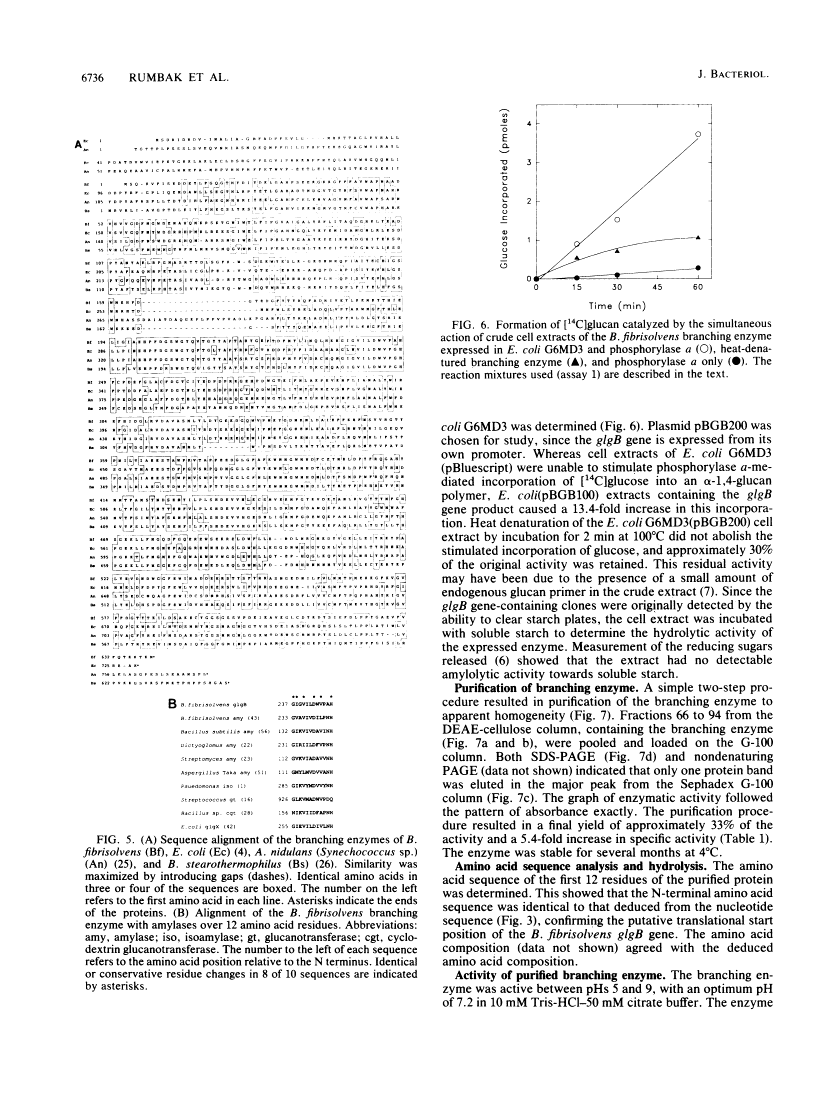
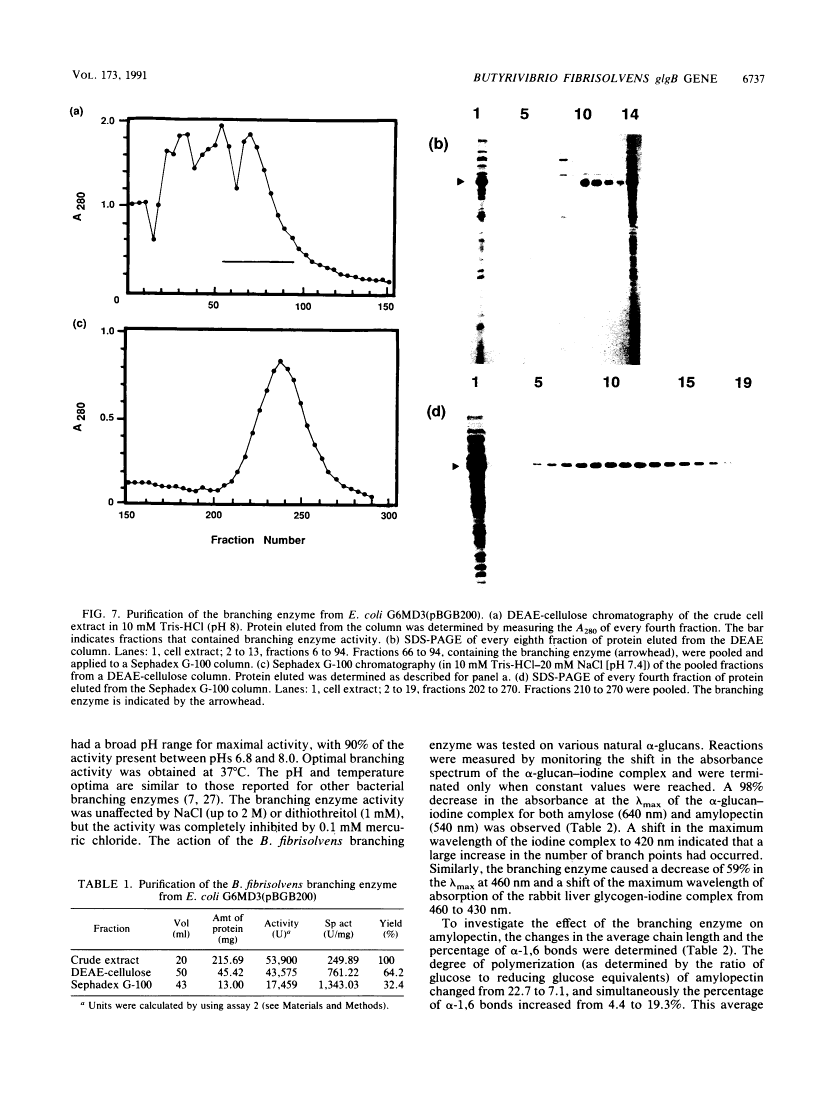
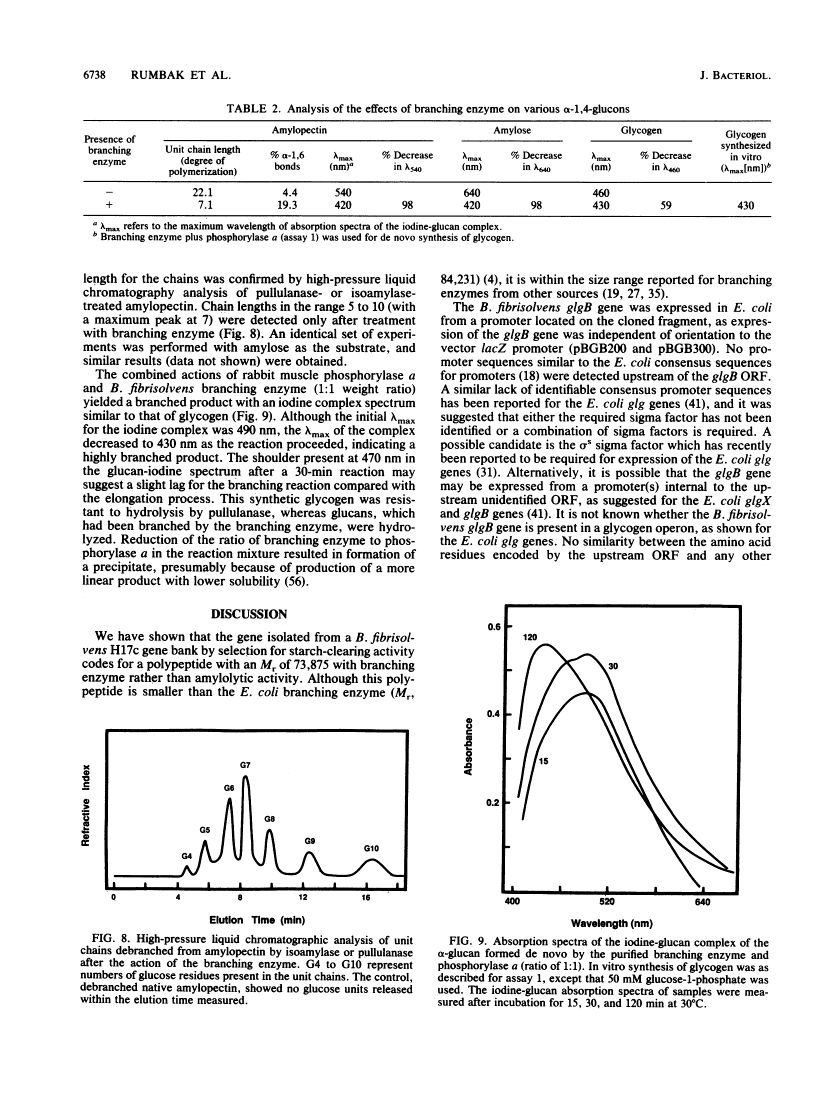
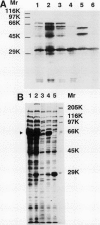
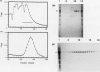
A Butyrivibrio fibrisolvens H17c glgB gene, was isolated by direct selection for colonies that produced clearing on starch azure plates. The gene was expressed in Escherichia coli from its own promoter. The glgB gene consisted of an open reading frame of 1,920 bp encoding a protein of 639 amino acids (calculated Mr, 73,875) with 46 to 50% sequence homology with other branching enzymes. A limited region of 12 amino acids showed sequence similarity to amylases and glucanotransferases. The B. fibrisolvens branching enzyme was not able to hydrolyze starch but stimulated phosphorylase alpha-mediated incorporation of glucose into alpha-1,4-glucan polymer 13.4-fold. The branching enzyme was purified to homogeneity by a simple two-step procedure; N-terminal sequence and amino acid composition determinations confirmed the deduced translational start and amino acid sequence of the open reading frame. The enzymatic properties of the purified enzyme were investigated. The enzyme transferred chains of 5 to 10 (optimum, 7) glucose units, using amylose and amylopetin as substrates, to produce a highly branched polymer.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amemura A, Chakraborty R, Fujita M, Noumi T, Futai M. Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J Biol Chem. 1988 Jul 5;263(19):9271–9275. [Abstract] [Google Scholar]
- Baecker PA, Furlong CE, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose synthetase as deduced from the nucleotide sequence of the glg C gene. J Biol Chem. 1983 Apr 25;258(8):5084–5088. [Abstract] [Google Scholar]
- Baecker PA, Greenberg E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli 1,4-alpha-D-glucan:1,4-alpha-D-glucan 6-alpha-D-(1, 4-alpha-D-glucano)-transferase as deduced from the nucleotide sequence of the glg B gene. J Biol Chem. 1986 Jul 5;261(19):8738–8743. [Abstract] [Google Scholar]
- Berger E, Jones WA, Jones DT, Woods DR. Cloning and sequencing of an endoglucanase (end1) gene from Butyrivibrio fibrisolvens H17c. Mol Gen Genet. 1989 Oct;219(1-2):193–198. [Abstract] [Google Scholar]
- Boyer C, Preiss J. Biosynthesis of bacterial glycogen. Purification and properties of the Escherichia coli b alpha-1,4,-glucan: alpha-1,4-glucan 6-glycosyltansferase. Biochemistry. 1977 Aug 9;16(16):3693–3699. [Abstract] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. [Abstract] [Google Scholar]
- Cattaneo J, Chambost JP, Creuzet-Sigal N. Combined action of Escherichia coli glycogen synthase and branching enzyme in the so-called "unprimed" polyglucoside synthesis. Arch Biochem Biophys. 1978 Sep;190(1):85–96. [Abstract] [Google Scholar]
- Ceska M, Birath K, Brown B. A new and rapid method for the clinical determination of alpha-amylase activities in human serum and urine. Optimal conditions. Clin Chim Acta. 1969 Dec;26(3):437–444. [Abstract] [Google Scholar]
- Cheng KJ, Brown RG, Costerton JW. Characterization of a Cytoplasmic Reserve Glucan from Ruminococcus albus. Appl Environ Microbiol. 1977 Mar;33(3):718–724. [Europe PMC free article] [Abstract] [Google Scholar]
- Dehority BA. Characterization of several bovine rumen bacteria isolated with a xylan medium. J Bacteriol. 1966 May;91(5):1724–1729. [Europe PMC free article] [Abstract] [Google Scholar]
- Ferretti JJ, Gilpin ML, Russell RR. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J Bacteriol. 1987 Sep;169(9):4271–4278. [Europe PMC free article] [Abstract] [Google Scholar]
- Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. [Europe PMC free article] [Abstract] [Google Scholar]
- Hawker JS, Ozbun JL, Ozaki H, Greenberg E, Preiss J. Interaction of spinach leaf adenosine diphosphate glucose alpha-1,4-glucan alpha-4-glucosyl transferase and alpha-1,4-glucan, alpha-1,4-glucan-6-glycosyl transferase in synthesis of branched alpha-glucan. Arch Biochem Biophys. 1974 Feb;160(2):530–551. [Abstract] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. [Abstract] [Google Scholar]
- Holmes E, Boyer C, Preiss J. Immunological characterization of Escherichia coli B glycogen synthase and branching enzyme and comparison with enzymes from other bacteria. J Bacteriol. 1982 Sep;151(3):1444–1453. [Europe PMC free article] [Abstract] [Google Scholar]
- Horinouchi S, Fukusumi S, Ohshima T, Beppu T. Cloning and expression in Escherichia coli of two additional amylase genes of a strictly anaerobic thermophile, Dictyoglomus thermophilum, and their nucleotide sequences with extremely low guanine-plus-cytosine contents. Eur J Biochem. 1988 Sep 15;176(2):243–253. [Abstract] [Google Scholar]
- Hoshiko S, Makabe O, Nojiri C, Katsumata K, Satoh E, Nagaoka K. Molecular cloning and characterization of the Streptomyces hygroscopicus alpha-amylase gene. J Bacteriol. 1987 Mar;169(3):1029–1036. [Europe PMC free article] [Abstract] [Google Scholar]
- Kawaguchi K, Fox J, Holmes E, Boyer C, Preiss J. De novo synthesis of Escherichia coli glycogen is due to primer associated with glycogen synthase and activation by branching enzyme. Arch Biochem Biophys. 1978 Oct;190(2):385–397. [Abstract] [Google Scholar]
- Kiel JA, Boels JM, Beldman G, Venema G. Nucleotide sequence of the Synechococcus sp. PCC7942 branching enzyme gene (glgB): expression in Bacillus subtilis. Gene. 1990 Apr 30;89(1):77–84. [Abstract] [Google Scholar]
- Kiel JA, Elgersma HS, Beldman G, Vossen JP, Venema G. Cloning and expression of the branching enzyme gene (glgB) from the cyanobacterium Synechococcus sp. PCC7942 in Escherichia coli. Gene. 1989 May 15;78(1):9–17. [Abstract] [Google Scholar]
- Kimura K, Kataoka S, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the beta-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. strain 1011 and similarity of its amino acid sequence to those of alpha-amylases. J Bacteriol. 1987 Sep;169(9):4399–4402. [Europe PMC free article] [Abstract] [Google Scholar]
- Kumar A, Larsen CE, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose:alpha-1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J Biol Chem. 1986 Dec 5;261(34):16256–16259. [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):49–59. [Abstract] [Google Scholar]
- Lin LL, Rumbak E, Zappe H, Thomson JA, Woods DR. Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a beta-glucosidase. J Gen Microbiol. 1990 Aug;136(8):1567–1576. [Abstract] [Google Scholar]
- MARGHERITA SS, HUNGATE RE. SEROLOGICAL ANALYSIS OF BUTYRIVIBRIO FROM THE BOVINE RUMEN. J Bacteriol. 1963 Oct;86:855–860. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsumoto A, Kamata T, Matsuda K. Biosynthesis of glycogen in Neurospora crassa. Purification and properties of the branching enzyme. J Biochem. 1983 Aug;94(2):451–458. [Abstract] [Google Scholar]
- Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. [Abstract] [Google Scholar]
- Okita TW, Rodriguez RL, Preiss J. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J Biol Chem. 1981 Jul 10;256(13):6944–6952. [Abstract] [Google Scholar]
- Orpin CG, Mathiesen SD, Greenwood Y, Blix AS. Seasonal changes in the ruminal microflora of the high-arctic Svalbard reindeer (Rangifer tarandus platyrhynchus). Appl Environ Microbiol. 1985 Jul;50(1):144–151. [Europe PMC free article] [Abstract] [Google Scholar]
- Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. [Abstract] [Google Scholar]
- Preiss J, Romeo T. Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv Microb Physiol. 1989;30:183–238. [Abstract] [Google Scholar]
- Romeo T, Kumar A, Preiss J. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene. 1988 Oct 30;70(2):363–376. [Abstract] [Google Scholar]
- Rumbak E, Rawlings DE, Lindsey GG, Woods DR. Cloning, nucleotide sequence, and enzymatic characterization of an alpha-amylase from the ruminal bacterium Butyrivibrio fibrisolvens H17c. J Bacteriol. 1991 Jul;173(13):4203–4211. [Europe PMC free article] [Abstract] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Schwartz M. Location of the maltose A and B loci on the genetic map of Escherichia coli. J Bacteriol. 1966 Oct;92(4):1083–1089. [Europe PMC free article] [Abstract] [Google Scholar]
- Steiner KE, Preiss J. Biosynthesis of bacterial glycogen: genetic and allosteric regulation of glycogen biosynthesis in Salmonella typhimurium LT-2. J Bacteriol. 1977 Jan;129(1):246–253. [Europe PMC free article] [Abstract] [Google Scholar]
- Stewart CS, Paniagua C, Dinsdale D, Cheng KJ, Garrow SH. Selective isolation and characteristics of Bacteriodes succinogenes from the rumen of a cow. Appl Environ Microbiol. 1981 Feb;41(2):504–510. [Europe PMC free article] [Abstract] [Google Scholar]
- Strange RE. Bacterial "glycogen" and survival. Nature. 1968 Nov 9;220(5167):606–607. [Abstract] [Google Scholar]
- Verhue W, Hers HG. A study of the reaction catalysed by the liver branching enzyme. Biochem J. 1966 Apr;99(1):222–227. [Europe PMC free article] [Abstract] [Google Scholar]
- Walker GJ, Builder JE. Metabolism of the reserve polysaccharide of Streptococcus mitis. Properties of branching enzyme, and its effect on the activity of glycogen synthetase. Eur J Biochem. 1971 May 11;20(1):14–21. [Abstract] [Google Scholar]
- Wallace RJ. Cytoplasmic reserve polysaccharide of Selenomonas ruminantium. Appl Environ Microbiol. 1980 Mar;39(3):630–634. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang M, Galizzi A, Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu F, Jen Y, Takeuchi E, Inouye M, Nakayama H, Tagaya M, Fukui T. Alpha-glucan phosphorylase from Escherichia coli. Cloning of the gene, and purification and characterization of the protein. J Biol Chem. 1988 Sep 25;263(27):13706–13711. [Abstract] [Google Scholar]
- Zabeau M, Stanley KK. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1(10):1217–1224. [Europe PMC free article] [Abstract] [Google Scholar]
- ZEVENHUIZEN LP. BRANCHING ENZYME OF ARTHROBACTER GLOBIFORMIS. Biochim Biophys Acta. 1964 Mar 9;81:608–611. [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.173.21.6732-6741.1991
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/173/21/6732.full.pdf
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/173/21/6732
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/173/21/6732
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.173.21.6732-6741.1991
Article citations
Characterization of the Glucan-Branching Enzyme GlgB Gene from Swine Intestinal Bacteria.
Molecules, 28(4):1881, 16 Feb 2023
Cited by: 1 article | PMID: 36838868 | PMCID: PMC9960391
Biochemical characteristics and potential application of a thermostable starch branching enzyme from Bacillus licheniformis.
AMB Express, 13(1):8, 20 Jan 2023
Cited by: 0 articles | PMID: 36662316 | PMCID: PMC9859979
Metagenomic reconstructions of gut microbial metabolism in weanling pigs.
Microbiome, 7(1):48, 26 Mar 2019
Cited by: 52 articles | PMID: 30914068 | PMCID: PMC6436221
Distribution of glucan-branching enzymes among prokaryotes.
Cell Mol Life Sci, 73(14):2643-2660, 03 May 2016
Cited by: 13 articles | PMID: 27141939 | PMCID: PMC11108348
Review Free full text in Europe PMC
Improved yields of cyclic nigerosylnigerose from starch by pretreatment with a thermostable branching enzyme.
J Biosci Bioeng, 109(4):381-387, 21 Oct 2009
Cited by: 2 articles | PMID: 20226381
Go to all (15) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cloning, nucleotide sequence, and enzymatic characterization of an alpha-amylase from the ruminal bacterium Butyrivibrio fibrisolvens H17c.
J Bacteriol, 173(13):4203-4211, 01 Jul 1991
Cited by: 14 articles | PMID: 2061294 | PMCID: PMC208071
The glgB gene from the thermophile Bacillus caldolyticus encodes a thermolabile branching enzyme.
DNA Seq, 3(4):221-232, 01 Jan 1992
Cited by: 14 articles | PMID: 1296817
Cloning and characterization of the glycogen branching enzyme gene existing in tandem with the glycogen debranching enzyme from Pectobacterium chrysanthemi PY35.
Biochem Biophys Res Commun, 300(1):93-101, 01 Jan 2003
Cited by: 3 articles | PMID: 12480526
[Key enzymes in starch synthesis in plants].
Yi Chuan, 28(1):110-116, 01 Jan 2006
Cited by: 1 article | PMID: 16469726
Review