Abstract
Free full text

Lin28 Enhances Tumorigenesis and is Associated With Advanced Human Malignancies
Abstract
Multiple members of the let-7 family of miRNAs are often repressed in human cancers1,2, thereby promoting oncogenesis by de-repressing the targets K-Ras, c-Myc, and HMGA2 3,4. However, the mechanism by which let-7 miRNAs are coordinately repressed is unclear. The RNA-binding proteins Lin28 and Lin28B block let-7 precursors from being processed to mature miRNAs5–8, suggesting that over-expression of Lin28/Lin28B might promote malignancy via repression of let-7. Here we show that LIN28 and LIN28B are over-expressed in primary human tumors and human cancer cell lines (overall frequency ~15%), and that over-expression is linked to repression of let-7 family miRNAs and de-repression of let-7 targets. Lin28/Lin28B facilitate cellular transformation in vitro, and over-expression is associated with advanced disease across multiple tumor types. Our work provides a mechanism for the coordinate repression of let-7 miRNAs observed in a subset of human cancers, and associates activation of LIN28/LIN28B with poor clinical prognosis.
Human tumors display a global reduction in miRNA expression1,2. Inhibition of the miRNA processing pathway via knockdown of miRNA processing enzymes promotes cellular transformation and tumorigenesis; this effect can be attenuated by ectopic expression of let-7g, suggesting that inhibition of let-7 processing is likely to be central to promoting the transformed state3. Low levels of let-7 correlate with increased transformation capacity in vitro3, and have been observed in various malignancies, including lung cancer,9,10 hepatocellular carcinoma11, melanoma12, and ovarian cancer13. Over-expression of let-7 represses cellular proliferation pathways, inhibits cell growth, and impairs tumor development in both xenograft models and in an autochthonous model of non-small cell lung cancer3,14,15,16. Copy number loss3,17–19 and epigenetic silencing20 of individual let-7 family members have been reported in human cancers, and such events have been suggested to contribute to the disease state. However, in tissues where let-7 family members are co-expressed, inactivation of individual let-7 miRNAs would be expected to only modestly decrease let-7 dosage, given the functional redundancy among family members. Therefore, we hypothesized that activation of LIN28/LIN28B could serve as a physiological mechanism by which levels of all let-7 family miRNAs are coordinately repressed in the context of human malignancy.
We initially sought to determine whether over-expression of Lin28 could promote cellular transformation in experimental settings. We observed that over-expression of Lin28 in NIH/3T3 cells potently depletes levels of multiple mature let-7 family member miRNAs (Fig. 1a), leading to a concomitant increase in protein levels of c-Myc, a let-7 target (Fig. 1b). NIH/3T3 cells overexpressing Lin28 formed colonies in soft agar (Fig 1c–d) at a frequency comparable to BCR-ABL (Fig. S1) and slightly lower than that reported for c-Myc (1–2%), both of which are weakly transforming in this assay21. Expression of Lin28 together with BCR-ABL resulted in an increase in both colony size and number as compared with either Lin28 or BCR-ABL alone (Fig. S1). NIH/3T3 cells expressing either Lin28 or LIN28B formed tumors in nude mice (Fig. S1b and Table S1). Although the tumors were small and of delayed latency, they displayed evidence of local invasion (Fig. S1b), similar to what has been observed upon global inhibition of miRNA processing3, and consistent with a recent report that Lin28 enhances metastasis22. Co-expression of Lin28 and 7S21L (a let-7 loop mutant capable of being processed to mature let-7 in the presence of Lin2823) abrogated the ability to form colonies in semisolid medium (Fig 1c–d); thus, Lin28 transforms NIH/3T3 cells via the de-repression of let-7 target genes. We also observed that Lin28 over-expression enhanced transformation of a mouse lung adenocarcinoma cell line in which impairment of miRNA processing had been previously reported to be transforming3 (Fig. S2).
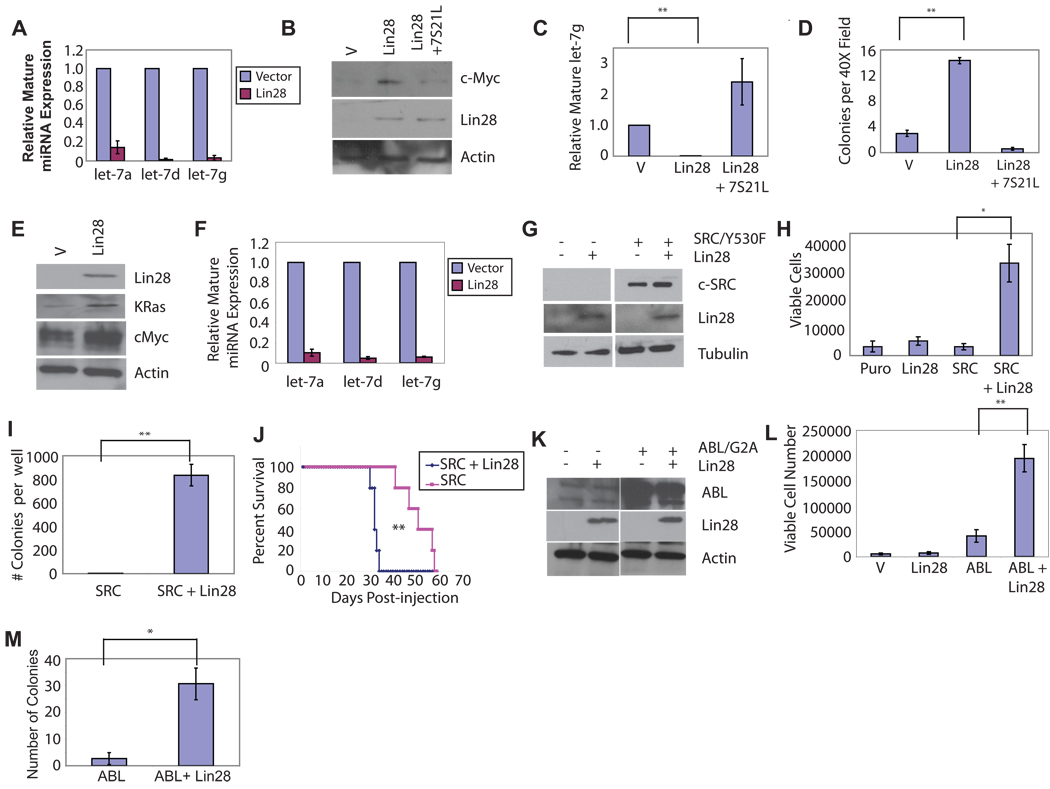
(A) Levels of mature miR species in representative infection determined by quantitative PCR in NIH/3T3 cells infected with pMSCV.Neo or pMSCV.Neo.Lin28, and selected on G418. (B) Western blot analysis of extracts from NIH/3T3 cells infected with MSCV.Neo/pBabe.Puro, pMSCV.Neo.Lin28/pBabe.Puro, or pMSCV.Neo.Lin28/pBabe.Puro.7S21L, selected with Puromycin and G418. (C) Levels of mature let-7g in NIH/3T3 cells infected with MSCV.Neo/pBabe.Puro,pMSCV.Neo.Lin28/pBabe.Puro, or pMSCV.Neo.Lin28/pBabe.Puro.7S21L, selected with Puromycin and G418. (D) Colony numbers in semisolid media for 25,000 NIH/3T3 cells infected with pMSCV.Neo/pBabe.Puro, pMSCV.Neo.Lin28/pBabe.Puro, or pMSCV.Neo.Lin28/pBabe.Puro.7S21L, selected with Puromycin and G418. Colonies were counted after 4 weeks with five random fields counted per well. Results are plotted as average colony number +/− S.E.M., N=3. (E) Western blot analysis on cell extracts from Ba/F3 cells infected with pMSCV.Neo and pMSCV.Neo.Lin28, selected on G418. (F) Levels of mature miR species in representative infection determined by quantitative PCR in Ba/F3 cells infected with pMSCV.Neo or pMSCV.Neo.Lin28, and selected on G418. (G) Western blot analysis on cell extracts from Ba/F3 cells infected with pEYK.Puro.SrcY530F and either pMSCV.Neo or pMSCV.Neo.Lin28 and selected on puromycin and G418 (H) Ba/F3 lines established in (C) were plated in media without IL-3 at 10,000 cells per well. Cells were stained with Trypan blue and viable cells were counted 6d after plating. (I) Colony formation of Ba/F3 cells expressing Src/Y530F or Src/Y530F + Lin28 in semisolid medium, plated at 50,000 cells per well in medium without IL-3. Quantitation of colony number from soft agar assay after 21d of growth. Results are plotted as average colony number per well +/− S.E.M., N=3. (J) Survival of immunocompromised mice injected with 107 Ba/F3 cells expressing Src-Y530F or Src-Y530F+Lin28. Average survival time 51.4d vs. 33.2d vs. respectively, p=0.00276 (K) Western blot analysis of extracts from Ba/F3 cells infected with pBabe.Puro or pBabe.Puro.Lin-28, selected on Puromycin. (L) Ba/F3 lines established in (C) were plated in media without IL-3 at 10,000 cells per well. Cells were stained with Trypan blue and counted 3d after plating. (M) Quantitation of colony number from soft agar assay. Results are plotted as average colony number per well +/− S.E.M., N=3. *, p<0.05; **, p<0.01.
We next tested Lin28 for its ability to transform cytokine-dependent BaF/3 cells to factor independence. Over-expression of Lin28 in BaF/3 cells potently depleted mature let-7 family members and led to the upregulation of the let-7 targets K-Ras and c-Myc (Fig. 1e–f). Co-expression of Lin28 and a weakly transforming variant of c-SRC (SRC/Y530F) conferred factor independence upon Ba/F3 cells (Fig. 1g–h) and strongly promoted growth in semisolid medium (Fig. 1i). Mice injected with Ba/F3 cells co-expressing Lin28 and Src/Y530F succumbed to leukemic disease more rapidly than mice injected with Ba/F3 cells expressing Src/Y530F alone (Fig. 1j). Lin28 cooperated similarly with a weakly transforming variant of c-Abl (Abl/G2A) to transform Ba/F3 cells (Fig. 1k–m). Notably, BCR-ABL, v-abl, and v-src have all been suggested to transform lymphoid cells via a similar pathway24. Together, these experiments demonstrate that over-expression of Lin28 facilitates cellular transformation in multiple in vitro assays and accelerates disease progression in a murine model.
To determine whether LIN28 and LIN28B are aberrantly expressed in the setting of human cancer, we examined LIN28 expression by microarray analysis of a large panel of human cancer cell lines; we observed expression in 3.2% of cell lines (17/527; Fig. S3a), including lines derived from germ cell tumors, Wilms’ tumor (WT), and hepatocellular carcinomas (HCC). Analysis of microarray data available for the National Cancer Institute panel of 60 cell lines (NCI-60) revealed LIN28 expression in 5% (3/60) and LIN28B expression in 10% (6/60) of the lines (Fig. S3b and Table S2). We also performed quantitative PCR analysis on a select panel of cancer cell lines and found several that express LIN28B, including multiple cell lines derived from patients in the blast crisis phase of chronic myeloid leukemia (CML) (Table S2). Examination of LIN28 expression in a range of primary tumor types by immunohistochemistry on a multi-tumor tissue microarray revealed that LIN28 was highly overexpressed in diverse tumor types including colon cancer, breast cancer, lung cancer, and cervical cancer (Fig. S4 and Table S3). Together, these data show that LIN28/LIN28B are aberrantly expressed in ~15% of surveyed cancer samples.
We surmised that activation of LIN28/LIN28B could represent a physiologic means by which let-7 levels are coordinately repressed in a subset of human cancers, and that over-expression of these factors might modify disease severity. Given that we observed over-expression in cell lines from patients with CML, we examined levels of LIN28 and LIN28B in peripheral blood from patients in either chronic phase (CP) or blast crisis (BC) of CML. LIN28 expression was detected more frequently in patients with BC-CML (42.8%, 6/14) or accelerated phase CML (40%, 4/10) than in patients with CP-CML (9.1%, 1/11) (Fig. 2a). Furthermore, analysis of 13 paired CP-BC samples showed activation of LIN28B upon progression to BC in three patients (23%, 3/13) (Table S4). Therefore LIN28/LIN28B expression is associated with disease progression in a subset of CML patients. Interestingly, we observed that expression of HMGA2, a let-7 target gene, shows a similar relationship with disease progression in CML, as has also been reported elsewhere25 (Fig. S5). This supports the notion that LIN28/LIN28B activation drives progression in CML via let-7 pathways. To determine whether cell lines derived from patients with BC-CML might be dependent on LIN28/LIN28B for growth, we inhibited LIN28B expression in K562 cells by RNA interference. Infection of K562 cells with an shRNA against LIN28B restored levels of mature let-7 miRNAs (Fig. 2b–c), decreased levels of c-MYC (Fig. 2d), impaired proliferative capacity and the ability to form colonies in soft agar (Fig. 2e–f), and resulted in morphological changes suggestive of differentiation (Fig. 2g)26. Similar results were obtained in a second CML-BC cell line and in a lung adenocarcinoma cell line (Fig. S6–Fig. S7), both of which express LIN28B.

(A) Lin28 expression as determined by quantitative PCR in peripheral blood from patients in either chronic phase, accelerated phase, or myeloid blast crisis. (B) Levels of LIN28B expression determined by quantitative PCR in K562 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA, and selected on Puromycin. (C) Levels of mature miR species determined by quantitative PCR in K562 cells were infected with pLKO.controlshRNA or pLKO.LIN28BshRNA, and selected on Puromycin. (D) Western blot for K-Ras and c-Myc on cell extracts from K562 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA and selected on Puromycin. (E) Cell proliferation of K562 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA, selected on Puromycin, and seeded at 10000 cells per well. Results are plotted as average cell number per well +/− S.E.M. N=3. (F) Colony formation for K562 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA and plated in semisolid medium. 5000 cells were plated per well and colonies were counted after 21d. (G) Wright-Giemsa stain on cytospin preparation of K562 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA, selected on Puromycin. *, p<0.05; **, p<0.01.
We next sought to determine whether LIN28/LIN28B might be similarly associated with advanced disease in HCC. We performed genome-wide mRNA and microRNA profiling on 89 HCC samples. LIN28B was highly expressed in a subset of HCCs, and these tumors displayed coordinate repression of all let-7 family miRNAs (Fig. 3a). Gene set enrichment analysis revealed that expression of let-7 target genes and myc target genes were significantly enriched in LIN28B-expressing tumors (Fig. 3b), suggesting that LIN28B represses let-7 and functionally activates let-7 targets in the setting of HCC. Tumors with high LIN28B expression accumulated in a molecular subclass defined by a meta-analysis of published transcriptome datasets (YH and TRG, data not shown), which is characterized by high serum alpha-fetoprotein level, higher tumor grade, and c-MYC activation (p=0.006, Fisher’s exact test). Consistently, LIN28B expression was observed in 66% (4/6) of tumors from patients with serum alpha-fetoprotein (AFP) > 10,000 ng/ml as compared to 16.3% (15/92) (p=0.00981, Fisher’s Exact Test). LIN28B-expressing tumors also fell predominantly within a subclass characterized by a proliferative nature as defined by unsupervised clustering27 (p=0.004), and most LIN28B-expressing tumors were of advanced stage (Fig. 3d). Moreover, LIN28B expression was associated with a significantly increased incidence of early recurrence (within 2 years of resection, p=0.02, log-rank test), but was not associated with late recurrence (Fig. 3c). Early recurrence is an indicator of malignant disease while late recurrence is thought to occur as a de novo event from the carcinogenetic microenvironment of the cirrhotic liver28. Knockdown of LIN28B in HepG2 cells (an HCC cell line expressing LIN28B) led to a marked decrease in the number of colonies formed in semisolid medium (Fig. 3e). Together, these results demonstrate that LIN28B is associated with advanced disease and poor clinical outcome in HCC.

(A) Expression of let-7 family miRNAs measured as part of genome-wide microRNA profile in 89 human HCC Tumor samples were ordered according by decreasing expression level of LIN28B as measured on an mRNA expression microarray on the same samples. (B) Induction of experimentally-defined MYC target gene signatures (left and middle) and let-7 target gene expression (right) was evaluated in LIN28B-high tumors using Gene Set Enrichment Analysis (C) LIN28B-high tumors showed significantly higher incidence of early recurrence (within 2 years after surgical treatment, p=0.02, log-rank test), but not late recurrence (p=0.22), as compared with LIN28B-low tumors. (D) The percentage of tumors expressing LIN28B is higher in BCLC-C stage HCC compared to BCLC-A or BCLC-B stage HCC. [Barcelona Clinic Liver Cancer (BCLC) staging system: A:early, B : intermediate C: advanced, D: end –stage53] (E) Serum AFP values were plotted against log2-normalized LIN28B expression values for patients with log2N signal > 6.00. (F) Colony formation for HepG2 cells infected with pLKO.controlshRNA or pLKO.LIN28BshRNA and plated in semisolid medium. 50,000 cells were plated per well and colonies were counted after 14d. *, p<0.05; **, p<0.01.
To determine whether LIN28/LIN28B are associated with advanced malignancy in other settings, we interrogated published microarray data from multiple tumor types and found high levels of LIN28 and/or LIN28B expression in a discrete subset of human tumors including WT (Fig. S8), ovarian carcinoma (Fig. S9), and germ cell tumors (West et al., submitted). Strikingly, LIN28/LIN28B expression was specifically associated with advanced disease in all of these tumor types. In WT, LIN28/LIN28B expression was only observed in Stage 3 or Stage 4 tumors (p= 0.00361, Fisher’s exact test), but not in lower stage tumors (Fig. S8). Among ovarian carcinomas, LIN28/LIN28B expression was observed predominantly in tumors of histological Grade 2 or 3, rather than Grade 1 (Fig. S9).
Various mechanisms may explain LIN28/LIN28B expression in human cancers, including genomic amplification, aberrant hypomethylation, and expression in a rare cell of origin that acquires additional malignant lesions. Amplification of regions containing the LIN28 (1p36) 29 and LIN28B (6q21) 30,31 loci in human cancers have been reported. We surveyed SNP array data from 3330 primary tumors and cell lines to check for focal copy number changes at the LIN28 and LIN28B loci. Although we observed no appreciable copy number changes at the LIN28 locus, a small subset of cancers showed focal copy number increases of LIN28B (1.7% overall) (Table S5 and Figure S10). However, most amplification events were rare and typically broad, involving both LIN28B and numerous neighboring genes. Analysis of SNP array data in 31 CML-BC samples (22 myeloid, 9 lymphoid) revealed no focal alterations at either the LIN28 or LIN28B loci (data not shown). Thus, while genomic amplification may explain LIN28B expression in a small number of tumors, it is unlikely to be a dominant mechanism of activation. We also assessed methylation status at a CpG island located within the LIN28B locus, and observed relative hypomethylation in LIN28B-expressing cell lines (Fig. S11), suggesting that aberrant hypomethylation may poise the LIN28B locus for transcriptional activation in some tumors. Interestingly, this region of aberrant hypomethylation overlaps with a recently defined c-Myc binding site within the LIN28B locus32. This, together with our observation that LIN28B-expressing tumors cluster in a high-Myc expression subclass, suggests that Myc may transcriptionally activate LIN28B in some contexts.
Notably, there have been several reported cases of WT with a translocation involving 6q2133–37, the band on which LIN28B is located. To determine whether these translocations may be associated with altered expression of LIN28B, we analyzed a case of sporadic WT with a previously described chromosomal translocation t(6;15)(q21;q21)33,38. Previous mapping by FISH analysis had narrowed the 6q21 breakpoint in this case to a region intergenic to two 6q21 genes, HACE1 and LIN28B, and had demonstrated that loss of HACE1 expression in a cohort of WT cases39 was associated with altered methylation of several CpG islands in the region, including CpG-29 directly adjacent to the LIN28B gene (Fig S12a). To further map the 6q21 breakpoint in this case, we performed Southern blot assay using probes from the intergenic region. A probe covering CpG-29 and flanking sequences lying ~3–4 kb upstream of the LIN28B transcriptional start site demonstrated rearrangement using PstI-digested genomic DNA from the index WT case compared to matching normal kidney and other controls (Figure S12a–c). Moreover, quantitative RT-PCR showed dramatically increased expression of LIN28B in the index case compared to patient-matched normal kidney (Figure S12d). A second WT cause carrying a t(6;15) (Fig. S12e) also displayed dramatically increased expression of LIN28B in tumor sample compared with normal kidney (Fig. S12f). Therefore, LIN28B may be activated by translocation, although the frequency of such events remains to be determined.
Lin28 and Lin28b are expressed in various stem cell populations8,40,41, although there is scarce expression in most terminally differentiated somatic tissues42. Although our data suggest that altered gene expression of LIN28 or LIN28B can promote tumorigenesis and is associated with advanced malignancy, we cannot rule out the possibility that in some settings, LIN28 or LIN28B may be endogenously expressed in a rare cell of origin that develops into an aggressive tumor. We have observed relatively strong LIN28 expression within cells of the normal ovarian surface epithelium, thought to be the cells of origin of epithelial ovarian cancer (Fig. S9)43. Some poor-prognosis HCCs have also been suggested to arise from a primitive cell of origin that shares gene expression patterns with fetal hepatoblasts44.
Recently, LIN28 was used together with OCT4, NANOG, and SOX2 to reprogram human somatic fibroblasts to pluripotency45. Over-expression of other stem cell factors, such as BMI146 and OCT447,48, has been reported to promote oncogenesis by driving self-renewal and proliferation. Moreover, poorly differentiated, aggressive human tumors have recently been shown to have an embryonic stem cell-like gene expression signature49; other embryonic factors have also been reported to have roles in tumor progression50,51. Our data add LIN28/LIN28B to a growing list of embryonic genes involved in oncogenesis, and suggest a mechanism whereby repression of let-7 by LIN28/LIN28B may help to confer self-renewal capacity on cancer stem cells52. Antagonizing LIN28 and/or LIN28B function in tumors that overexpress these genes may provide a means to reactivate the expression of let-7 family tumor suppressors, and may thus be therapeutically beneficial.
Methods
Cell Culture
Cell lines were originally obtained from ATCC and cultured under standard conditions. BaF3/AblG2A and Baf3/SrcY530F lines have been described elsewhere54. LKR cells were derived from a mouse adenocarcinoma55 and were a kind gift of Carla Kim.
Antibodies
The following antibodies were used for immunoblotting: c-myc (Santa Cruz, sc-764), KRas (Santa Cruz, sc-30), Abl-K12 (Santa Cruz, sc-131), c-Src (Cell Signaling, #2110). Anti-Lin28 (Proteintech group) was used for immunohistochemistry.
Cloning and Plasmid Construction
Mammalian Lin28 and LIN28B cDNA were subcloned into pBabe.Puro and pMSCV.Neo retroviral vectors. pMSCV.Neo.let-7g construct was previously described3. LIN28B shRNA in lentiviral plasmid was purchased from Sigma-Aldrich (TRCN0000122599). Control shRNA was commercially purchased (SHC002V, Sigma-Aldrich)
Immunohistochemistry
Sections of tissues were deparaffinized with xylene and rehydrated with graded series of ethanol (absolute, 95%, 80% and 50%, respectively, and distilled water), followed by two washes of 5 min each in PBS-T. Antigen retrieval was performed for 20 min in sodium citrate buffer (10mM pH 6) at 90–100°C followed by wash with PBST 1× 5 min. Tissue sections were then incubated for 10 min in 3% (v/v) hydrogen peroxide in methanol to block endogenous peroxidase activity. Sections were then washed for 5 min in PBS-T and blocked at room temperature for 1 h by using 2% normal goat serum, 2% bovine serum albumin (BSA) and 0.1% triton-X in PBS. Tissue sections were then incubated in humidified chamber for overnight incubation at 4°C with primary antibody (1/200 in TBST). Sections were subsequently washed with PBS-T (3× 5min) and incubated at room temperature for 1 h with secondary antibody (goat anti rabbit). After a wash with PBS-T (3× 5min), sections were incubated with ready to use streptavidin peroxidase (Lab Vision, Fremont, CA) for 10 min at room temperature and then color was developed by using a DAB kit (Vector laboratories, Burlingame, CA). Sections were counterstained with hematoxylin.
Viral Production
For ecotropic viral production, retroviral plasmid DNA and pCL-Eco were transfected into 293T cells in a 1:1 mass ratio and virus harvested after 48h. For VSV-G pseudotyped lentivirus, viral plasmid, lentiviral gag/pol, and VSV-G were transfected in a mass ratio 1:0.9:0.1, and virus was harvested after 72 hrs. 1 mL of unconcentrated viral supernatant was used to infect 50,000 cells. Infected cells were selected on antibiotic prior to subsequent analysis.
Bisulfite Sequencing
Bisulfite sequencing was performed as described56 with the commercially available EZ DNA Methylation Kit (Zymo Research). Primers were designed using MethPrimer to target a region 1 kb downstream of the LIN28B transcriptional start site.
Transformation and Growth Assays
Soft agar assays were performed essentially as described previously with 50,000 cells seeded per well57. Colonies were stained with 0.005% crystal violet and counted after 3–4 weeks of growth.
Quantitative RT-PCR
quantitative RT-PCR was used for detection of mature miRNA species as described previously8. mRNA expression analysis was performed by quantitative PCR using SYBR green dye with fold-changes calculated by the ΔΔCt method or using LIN28B Taqman probe on patient samples.
Microarray Data Analysis
The following are Gene expression Omnibus accession numbers for publically available microarray datasets analyzed for LIN28 and LIN28B expression in this paper: GSE11024, GSE6222, GSE9843, GSE9891, GSE4170. Log2 transformed expression data were normalized to the average expression signal for normal tissue (where available), or row-normalized for the GSE9843 and GSE9891 datasets. Cutoff for over-expression was taken as a normalized expression value of 2 or greater unless stated otherwise. For microarray profiling of cancer cell lines, the samples run were from the Sanger Cell line collection as described at http://www.sanger.ac.uk/genetics/CGP/CellLines/ using an HT-HG_U133A chip, miRNAs were profiled using our previously described platform58. The HCC samples were stratified into LIN28B-high and -low groups based on a cut-off of 200U. LIN28B amplification status was determined in a panel of 1,103 cancer DNA samples – consisting of breast cancer (n = 242), chronic myelogenous leukemia (n = 6), lung non-small cell cancer (n=734), and hepatocellular carcinoma (n=121) – hybridized to the Affymetrix 250K StyI SNP Array (Beroukhim et al, Manuscript in preparation). Copy number estimates for LIN28B were normalized using a panel of population-matched controls and segmented using the Gain and Loss of DNA (GLAD) algorithm59.
Human tissue samples
The expression levels of let-7 species and LIN28B were assessed in 89 HCV-related hepatocellular carcinoma (HCC) samples including 77 clinically early HCC (25 pathologically early and 52 pathologically advanced cases) and 12 very advanced cases. All the samples were obtained from patients undergoing liver resection in the three hospitals of the HCC Genomic Consortium: Mount Sinai School of Medicine, NY (US), Hospital Clinic, Barcelona (Spain) and Istituto Nazionale dei Tumori, Milan (Italy). We obtained approval from the Institutional Review Board and patients were properly consented.
Quantitative PCR detection of LIN28 and LIN28B on CML samples
Total RNA was isolated from cell lines and patient samples (a mixture of both peripheral blood and bone marrow) using Trizol reagent (Invitrogen; Carlsbad, CA). cDNA was generated from 1mg of total RNA using Cloned AMV Reverse Transcriptase (Invitrogen). The cDNA synthesis was performed under the following thermocycler conditions: 25°C for 10 minutes, 42°C for 60 minutes, 99°C for 5 minutes and 4°C hold. Gene specific primers and probes were designed using Primer Express Version 3 (ABI; Foster City, CA). All of the QPCR primers and florescent probes were manufactured by Integrated DNA Technologies (Coralville, IA). Real-time quantitative PCR (QPCR) was performed on 100ng of synthesized cDNA in a 50ml reaction using the ABI 7900HT Real-Time PCR System under the following conditions: stage 1, 95°C for 10 minutes: and stage 2, 95°C for 15 seconds, 60°C for 1 minute for a total of 40 cycles.
Fluorescence in-situ hybridization (FISH)
FISH was performed using standard protocols on metaphase nuclei as previously described.1 The RP-770C15 and RP-809N15 BAC probes were obtained from the RPCI-11 human BAC library and directly labelled with SpectrumGreen and SpectrumRed, respectively, via a nick translation kit (Vysis), hybridized in a humidified chamber overnight at 37°C, and counterstained with DAPI. Detection was performed using a Zeiss Axioplan epifluorescence microscope equipped with a COHU-CCD camera and Quips software.
Acknowledgements
We thank Carla Kim for providing LKR cells and the MGH Gyn Tissue Repository for ovarian tissue samples. Thanks to Dieter Fink and Natalya Melnyk for technical assistance. This work was partially supported by grants from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (J.M.L; 1R01DK076986-01), the Samuel Waxman Cancer Research Foundation (J.M.L.), the Spanish National Health Institute (J.M.L; SAF-2007-61898). This work was supported by funds from the National Cancer Institute of Canada (NCIC) (P.S.). This work was supported by grants from the NIH, the NIH Director's Pioneer Award of the NIH Roadmap for Medical Research, Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukemia and Lymphoma Society (GQD), and the Howard Hughes Medical Institute (GQD).
References List
Full text links
Read article at publisher's site: https://doi.org/10.1038/ng.392
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2757943
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Epstein-Barr virus reactivation induces divergent abortive, reprogrammed, and host shutoff states by lytic progression.
PLoS Pathog, 20(10):e1012341, 24 Oct 2024
Cited by: 0 articles | PMID: 39446925 | PMCID: PMC11563402
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in hematological diseases.
Mol Med, 30(1):165, 28 Sep 2024
Cited by: 0 articles | PMID: 39342091 | PMCID: PMC11439276
Review Free full text in Europe PMC
Development of miRNA-based PROTACs targeting Lin28 for breast cancer therapy.
Sci Adv, 10(38):eadp0334, 18 Sep 2024
Cited by: 0 articles | PMID: 39292784 | PMCID: PMC11409961
RNA binding protein-directed control of leukemic stem cell evolution and function.
Hemasphere, 8(8):e116, 22 Aug 2024
Cited by: 0 articles | PMID: 39175825 | PMCID: PMC11339706
Review Free full text in Europe PMC
DROSHA Regulates Mesenchymal Gene Expression in Wilms Tumor.
Mol Cancer Res, 22(8):711-720, 01 Aug 2024
Cited by: 1 article | PMID: 38647377
Go to all (585) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (3)
- (1 citation) GEO - GSE9843
- (1 citation) GEO - GSE6222
- (1 citation) GEO - GSE11024
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells.
Int J Biochem Cell Biol, 45(5):973-978, 16 Feb 2013
Cited by: 106 articles | PMID: 23420006
Review
Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma.
Carcinogenesis, 31(9):1516-1522, 04 Jun 2010
Cited by: 78 articles | PMID: 20525879
Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer.
Mol Cancer, 14:125, 30 Jun 2015
Cited by: 101 articles | PMID: 26123544 | PMCID: PMC4512107
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NHLBI NIH HHS (1)
Grant ID: T32-HL 66987
NICHD NIH HHS (2)
Grant ID: R01 HD052701-02
Grant ID: R01 HD052701
NIDDK NIH HHS (2)
Grant ID: 1 R01 DK076986-01
Grant ID: R01 DK076986
NIGMS NIH HHS (1)
Grant ID: T32 GM007753
NIH HHS (2)
Grant ID: DP1 OD000256-01
Grant ID: DP1 OD000256





