Abstract
Free full text

Epistasis between RET and BBS mutations modulates enteric innervation and causes syndromic Hirschsprung disease
Abstract
Hirschsprung disease (HSCR) is a common, multigenic neurocristopathy characterized by incomplete innervation along a variable length of the gut. The pivotal gene in isolated HSCR cases, either sporadic or familial, is RET. HSCR also presents in various syndromes, including Shah–Waardenburg syndrome (WS), Down (DS), and Bardet–Biedl (BBS). Here, we report 3 families with BBS and HSCR with concomitant mutations in BBS genes and regulatory RET elements, whose functionality is tested in physiologically relevant assays. Our data suggest that BBS mutations can potentiate HSCR predisposing RET alleles, which by themselves are insufficient to cause disease. We also demonstrate that these genes interact genetically in vivo to modulate gut innervation, and that this interaction likely occurs through complementary, yet independent, pathways that converge on the same biological process.
Hirschsprung disease (HSCR, MIM 164761) is the most common (1/5,000 live births) form of structural intestinal obstruction. It is defined by the absence of neural crest (NC)-derived enteric ganglia along a variable length of the bowel, invariably involving the recto-anal junction. This phenotype has been attributed to defects in migration, proliferation, and/or survival of the NC cells (NCC) that normally give rise to all neurons and supporting cells of the enteric nervous system (ENS), defining HSCR as a neurocristopathy. HSCR is also a useful model oligogenic disorder; it displays non-Mendelian modes of inheritance with low, sex-dependant penetrance in isolated HSCR cases. Although oligogenic multiplicative models have been proposed, mutations in the RET proto-oncogene have emerged as pivotal (1–4). Almost all HSCR patients harbor either a heterozygous mutation in the RET coding sequence or, more frequently, a hypomorphic allele in a conserved noncoding element in intron 1 that acts as a spatially restricted transcriptional enhancer (4–6).
In addition to HCSR, some 30% of patients also exhibit other congenital anomalies as the result of chromosomal rearrangements [mostly Down syndrome (DS)], monogenic Mendelian disorders regardless of the mode of inheritance, or undiagnosed associations (6). In such cases, penetrance for the HSCR trait is 5 to 70%, suggesting additional predisposing genetic factor(s). Interestingly, alleles at the RET locus can have a role in modifying the risk of HSCR to be associated with several HSCR predisposing syndromes [congenital central hypoventilation, DS, Waardenburg (WS) type IV due to EDNRB mutations, and Bardet-Biedl (BBS)], but not all (Mowat-Wilson and Waardenburg type IV due to SOX10 mutations) (6–8). These observations are suggestive of RET-dependent and RET-independent HSCR cases (9); consistent with RET acting as a modifier gene in some HSCR predisposing syndromes, no correlation between the genotype for the syndrome disease causing gene and the HSCR trait could be drawn in monogenic HSCR syndromes (9, 10).
Surprisingly, the greatest RET dependence has been observed in the group of patients with both BBS and HSCR. No interactions between RET and BBS proteins are known; also, whereas RET is a cell-surface tyrosine kinase receptor essential for enteric neuron population of the gut (11), BBS is a ciliopathy caused by mutations in at least 14 genes (ref. 12 and references therein), with all BBS proteins examined to date localizing in the basal body and the ciliary axoneme where they likely affect various signaling processes (13). Nonetheless, despite any obvious biochemical or subcellular links between RET and the BBS proteins, a zebrafish model of BBS was shown recently to manifest a Shh-dependent defect in the migration of neural crest cells with concomitant defects in the innervation of the ENS (14), suggesting that the observed enrichment of RET hypomorphic alleles in BBS-HSCR patients might underlie a synthetic interaction at a higher level of systems organization.
Here, we report 2 families and 1 sporadic case with BBS and HSCR, each with variations at both a BBS locus and RET. Using both in vitro and in vivo models, we show that each individual variation has deleterious effects. Importantly, the interaction of these genetic lesions is tissue dependent, modulating the extent of intestinal aganglionosis during the development of the zebrafish ENS.
Results
BBS Mutations in BBS+HSCR Patients.
To investigate a possible genetic interaction between BBS and HSCR, we searched for BBS patients also diagnosed with HSCR. We found 2 informative BBS families, one of Caucasian origin (F1), and one of Moroccan origin (F2) (Fig. 1). F1 had 3 sibs, all affected with short segment HSCR as confirmed by histological criteria that included absence of enteric plexuses and increased acetylcholinesterase histochemical staining in nerve fibers. F2 consisted of consanguineous parents (f = 1/16) of 5 BBS sibs, 2 of which also had confirmed HSCR. We also identified a third sporadic case (SB25) born to healthy unrelated parents of Caucasian origin and presenting both BBS and HSCR (Fig. 1).
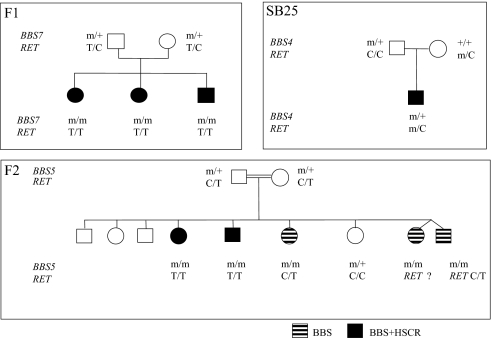
RET and BBS mutations found in BBS+HSCR cases. Segregation of BBS and RET mutations in 2 BBS families with BBS±HSCR and 1 BBS+HSCR sporadic case (SB25).
To determine the genetic lesion contributing to these cases, we first investigated all known BBS loci. We found all affected sibs in F1 to harbor 2 heterozygous missense mutations in BBS7; a paternal c.691T>C, p.W230R, and a maternal c.1512 G>A, p.R504K. Both mutations altered a conserved amino acid and were not found in 250 control chromosomes. In F2, all 5 sibs had a homozygous 1-bp deletion in exon 15 (c.1909-1910delT) of BBS5, causing a frameshift (p.K41fsX52). Both parents were heterozygous for the mutation, which was also not detected in controls. Last, the sporadic case SB25 bore a heterozygous missense BBS4 mutation (1372 C>T; S457I), which was inherited from the father and not found in 195 controls.
Genotyping of RET in Patients.
Previous reports have shown that BBS patients with HSCR are more frequent carriers of a RET hypomorphic allele than expected from population frequency (9). Therefore, we hypothesized that the BBS+HSCR patients in our families might also have a RET variant. To test this possibility, we sequenced the patients for mutations in RET. The 3 F1 sibs were homozygous for the common intronic hypomorphic T allele; both parents were heterozygous. In F2, only the 2 BBS+HSCR, but not the other 3 BBS-only patients were homozygous for the same RET allele. Last, the sporadic SB25 case was WT for the intron 1 allele, but had a novel, maternally inherited heterozygous 11-bp deletion (del AAGCAACTGCC). This deletion lies 106-bp downstream of the known intron 1 hypomorphic allele, maps within the highly conserved enhancer region, and was not found in over 800 control chromosomes (Fig. 1; Fig. S1).
Functional Assessment of BBS and RET Mutations.
Our genetic data provide strong evidence that the BBS5 and BBS7 alleles in families F1 and F2 are sufficient to cause BBS. By contrast, the pathogenic potential of the BBS4 S457I allele cannot be established unequivocally, despite its absence from ethnically matched controls. Therefore, we investigated the (non)neutrality of this allele using an established zebrafish BBS model. We have reported previously that suppression of Bbs proteins results in Wnt-dependent gastrulation movement defects in zebrafish (15, 16), which can be rescued by coinjection of WT human mRNA. This system has enabled us establish the pathogenic potential of several novel BBS-causing alleles (12).
Therefore, we suppressed endogenous bbs4 and evaluated the ability of a 457I-encoding BBS4 message to rescue the phenotype. Approximately 63% of embryos injected with bbs4 morpholino (MO) had broadened somites, kinked notochords, and shortened body axes (Fig. 2 A and B). The observed phenotypes not only fully recapitulated our previous observations (15), but were also shown to be specific to the MO, because coinjection of mRNA encoding WT human BBS4 efficiently rescued the morphant phenotypes (Fig. 2B). By contrast, embryos injected with 457I were indistinguishable from embryos injected with bbs4 MO alone (n = 96 embryos, scored blind to injection mixture), suggesting that the introduction of the 457I residue results in functionally null protein (Fig. 2 A and B); expression of 457I-encoding mRNA alone yielded modest phenotypes, indicating that this allele does not act in a dominant-negative fashion (Fig. 2B). To validate the pathogenic nature of this mutation further, we investigated the cellular localization of the 457I protein in ciliated mammalian cells. Previous studies have established the reproducible localization of BBS4 near the 2 centrioles by overexpression of tagged protein in transformed cell lines (17, 18). Upon transient transfection of a BBS4-MYC construct, we observed a similar localization pattern (Fig. 2C). However, expression of a BBS4 S457I mutant protein resulted in complete mislocalization of the protein (Fig. 2C), with the mutant protein forming apparent intracellular aggregates away from the pericentriolar region (visualized by γ-tubulin staining), providing further evidence that the mutation is a likely null.

In vivo assessment of the pathogenic potential of BBS4 S457I variant. (A) Zebrafish embryos coinjected with a MO against bbs4 and mRNA expressing the S457I variant exhibited defects, including short body axes, broad somites, wider and kinked notochords, and detachment of cells along the dorsal axis. (B) Compared to rescue with WT BBS4 mRNA, rescue with the mutated mRNA produced an increase in the proportion of both moderately (Class I) and severely (Class II) affected embryos, but injection of S457I mRNA alone does not produce severe defects, suggesting that the variant is functionally null. (C) HeLa cells transfected with either WT or S457I mutant BBS4 protein (red). WT protein localizes to the basal body (γ-tubulin, green), but mutant protein completely mislocalizes to form cytoplasmic aggregates.
Next, we turned our attention to the RET alleles in our patients. Although the functional significance of the intronic HSCR predisposing allele in RET is known, the pathogenic potential of the 11-bp deletion within intron 1 is highly suggestive, but not conclusive. We have shown previously that the multispecies conserved sequence (MCS) within intron 1 (MCS + 9.7) acts as an enhancer element, and contains the previously described hypomorphic allele (5); the 11-bp deletion is 106 bp downstream of this SNP. To investigate the function of the deletion, we constructed pDSma-RET vectors with luciferase under the control of an SV40 promoter and the MCS + 9.7 enhancer plus a 1215-bp encompassing region either with (WT) or without (Δ11bp) the 11-bp sequence, and assayed relative luciferase activity in SK-N-SH neuroblastoma cells or HeLa cells. The WT RET enhancer sequence significantly decreased reporter gene expression (P = 0.001) compared with the promoter alone, and also as compared with MCS + 9.7, suggesting that it contains a putative transcription repressor-binding site. However, the vector harboring the 11-bp deletion significantly derepresses reporter gene expression compared with WT (P = 0.05) to a level statistically indistinguishable from the promoter alone (NS), suggesting specific disruption of the repressor element binding motif (Fig. 3). Reporter assays conducted in HeLa cells produced similar results. These findings suggest that deletion of 11 bp within the intronic enhancer results in defects in transcriptional regulation in vitro, although they do not abolish the effect of this genomic segment, raising the possibility that Δ11bp represents a mild allele. Together, these results suggest that the individual BBS and RET mutations identified in the sporadic case of BBS and HSCR are likely detrimental to gene function. Among putative consensus binding sites encompassing the 11-bp deletion, only the zinc finger transcription factor SNAIL is expressed in hNCC. However, ChIP experiments in SK-N-SH cells did not detect an interaction with this protein, suggesting that potentially novel components are associated with the observed suppression regulatory element.

Deletion in the RET regulatory sequence perturbs reporter gene expression in vitro. (A) Schematic representation of vectors used to assay the effect of the 11-bp deletion on reporter gene expression in vitro. WT or mutated (Δ11bp) RET enhancer sequence (green) was placed upstream of an SV40 promoter (gray) driving firefly luciferase expression (yellow). The pDSma-RET MCS + 9.7, described previously (5), was used as a positive control. (B) Relative luciferase activity of pDSma-RET vectors (depicted in A) in SK-N-SH neuroblastoma cells. Cells were lysed 48 h posttransfection, and relative luciferase activity was normalized to values obtained from the promoter alone (pDSma-Promoter). The WT RET enhancer sequence significantly decreases reporter gene expression (P = 0.001) compared with promoter alone, suggesting that it contains a putative transcription repressor-binding site. However, the vector harboring the 11-bp deletion significantly derepresses reporter gene expression compared with WT (P = 0.05) to a level statistically indistinguishable (NS) from the promoter alone, suggesting specific disruption of the repressor element binding motif. Assays were performed in triplicate wells and repeated thrice.
Genetic Interaction of BBS and RET in Vivo.
Our data are consistent with previous studies, in which a higher-than-expected frequency of RET hypomorphic alleles were found in BBS-HSCR patients, and suggest that this property is not restricted to a specific BBS gene (9), not least because BBS4, BBS5, and BBS7 are 3 of the least commonly mutated BBS genes (13). To test this possibility in vivo, we assayed the effects of suppression of each of Ret and Bbs proteins on enteric neuron migration in the developing zebrafish gut. As shown previously, suppression of ret by MO injection results in a reduction of enteric neurons populating the proximal and distal hindgut, as labeled by in situ hybridization with a phox2b riboprobe (Fig. 4 B and I) (19). Likewise, and consistent with recent data for bbs8, suppression of either bbs4, bbs5, or bbs7 also results in a reduction of enteric neuron migration, although not as severe (Fig. 4 C, E, G, and I). However, coinjection of ret MO with any of the 3 bbs genes tested resulted in a significantly greater decrease in the enteric neurons in the hindgut, with phenotypic synthesis particularly prominent in the distal hindgut (Fig. 4 D, F, H, and I). The increased severity is greater than a mere additive effect, suggesting an epistatic interaction between ret and bbs function. These findings indicate that suppression of ret in addition to loss of bbs function results in more severe defects in ENS development than with loss of these genes independently.

Genetic interaction of Ret and Bbs in zebrafish. (A) Migration of enteric neurons in 4-day-old embryos normally proceeds along the developing hindgut to reach the anus (asterisks). Embryos injected with MO against either (B) ret or (C) bbs4, (E) bbs5, or (G) bbs7 exhibit premature termination of enteric neuron migration (arrowhead). This defect is exacerbated in embryos injected with a combination of ret MO and either (D) bbs4, (F) bbs5, or (H) bbs7 MO. (I) The extent of the defect was quantified by assessment of the presence of phox2b expression in the proximal hindgut (somites 4–9) or the distal hindgut (somites 10 and beyond) as previously described (31).
NCCs as Likely Sites of Epistasis.
The finding that ret and bbs genes interact genetically to produce enteric neuron defects suggests the possibility that these genes function in overlapping pathways to regulate ENS development. Several lines of evidence support this conjecture: the role of RET signaling in NCC migration is well established (20, 21), whereas suppression of bbs8 has been shown to result in similar defects, potentially in a Shh-dependent fashion (14). Also, transcriptional profiling of NCC cells demonstrated the presence of mRNA message for several BBS proteins, as well as mRNA for genes encoding numerous ciliary and basal body proteins (Table S1; Fig. S2 A and B). Last, immunocytochemistry using antiacetylated α-tubulin revealed a clear presence of monocilia and basal bodies, respectively, in these cells (Fig. S2 C and D). To confirm that the ciliated NCCs are, in fact, those that will contribute to enteric innervation, we performed immunofluorescence using either anti-RET or anti-SOX9 and observed expression of both proteins in NCCs positive for antiacetylated tubulin staining (Fig. S2D).
Given these findings, one possibility is that RET signaling functions through the cilium, because several other paracrine receptors are known to localize there or to indirectly require the primary cilium for signal transduction (22). However, RET immunofluorescence has never detected the protein in the ciliary axoneme, suggesting that the observed genetic interaction between RET and the BBS proteins might occur at a higher level of systems organization. To test this possibility we attempted to rescue the enteric neuron phenotype of RET knock-down by coinjecting BBS4 mRNA in embryos injected with ret MO; we did not observe any amelioration of the gut innervation defect (Fig. 4I), suggesting that BBS4 is not downstream of RET and supporting the notion that the 2 pathways likely converge at the same endpoint.
Discussion
Here, we have described 3 families segregating BBS in concert with HSCR. Mutation detection in affected individuals revealed mutations in BBS4, BBS5, and BBS7, consistent with their phenotypic presentation. Also, each affected was also homozygous for the common RET intronic hypomorphic allele or, in one instance, harbored a novel heterozygous 11-bp deletion that was absent from control chromosomes. We verified the functional potential of all novel alleles identified, integrating in vitro and in vivo approaches to determine their impact. These data led us to posit that interactions between RET and either BBS4, 5, or 7 in each instance would explain the coexpression of BBS and HSCR phenotypes. We have used nonallelic, noncomplementation through MO-based approaches in zebrafish to test these observations.
We have demonstrated that pairwise compound reduction in RET and each independent BBS protein product (BBS4, 5, or 7), which independently yield mild intestinal aganglionosis, severely exacerbates the observed enteric defects, with particular prominence in the distal hindgut. Interestingly this synergy in mutational effect appears to be tissue dependent, not resulting in an overt worsening of other associated phenotypes (e.g., convergence extension) suggesting that the pleiotropic activities of these genes may only be amenable to interaction with each other in a cell type dependent manner. As with other systems in the vertebrate organism, nervous system development relies on a delicate balance of interactions. Importantly, our data also illuminate interrelated roles for these BBS loci and RET in buffering the effects of genetic variation during ENS development, and between their mutant alleles in the copresentation of BBS and HSCR phenotypes. Interestingly, HSCR has been recurrently reported in association with other ciliopathies, namely Jeune and Joubert syndromes (23, 24).
Complex inheritance is underlined by a requirement for the combined influence of alleles at >1 locus in the transmission and expression of a phenotype; homeostatic genetic networks minimize the consequence of variation. Both BBS and HSCR are models of such traits, requiring the interaction of multiple genes for disease expression.
Just as interactions between different BBS loci explain the risk of BBS in subsets of families, interactions between RET and other loci can also explain disease risk in subsets of isolated and syndromic HSCR cases/families (1–3, 7, 12, 25, 26). Many of the genes implicated in these disorders display markedly pleiotropic effects when evaluated independently and occasionally these effects can coincide.
The interaction of genes involved in the same biochemical system and the observation of their failure to complement each other (intrinsic noncomplementation) is well established (3, 27, 28). However, there is no evidence to suggest that RET-mediated receptor tyrosine kinase signaling, and its role in neuronal precursor proliferation (21), directly intersects the role of BBS proteins in the development and function of ciliated cells and their proposed role in cell migration (13). Thus, their observed synergy is unlikely to arise from biochemical interaction, but rather from extrinsic noncomplementation raising predisposition to a common phenotype. Importantly, intrinsic noncomplementation is the commonly recognized basis of both HSCR and BBS. Although it cannot at this stage be distinguished definitively, we speculate that the syndromic copresentation of BBS and HSCR observed here, as well as similar interactions between HSCR and WS, support a model of extrinsic noncomplementation, where perturbations in unrelated biochemical signaling cascades converge at the same biological process (namely, neuronal migration and specification).
Materials and Methods
Patient Genotyping and Sequencing.
Blood samples were obtained with informed consent, and DNA was extracted according to standard protocols. Genotyping the BBS1-12 loci was performed by using fluorescent microsatellite markers (available on request). Mutation screening of RET and BBS genes have been described elsewhere (29), and intronic primers designed for BBS genes are available on request. Direct sequencing on both strands was performed using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and was analyzed on an ABI 3100 automated sequencer (Applied Biosystems). Also, patients were genotyped for SNP rs2435357 lying in the intron 1 of the RET gene. Of note, variable allelic frequencies are observed according to ethnicity and range from <5% in South Africa to ≈50% in Asia. The European Caucasian population has been thoroughly investigated in France, Italy, Spain, the Netherlands, and Germany, allowing approximation of the frequency of the T HSCR predisposing allele to ≈25% in the Caucasian population (30).
Luciferase Reporter Assay.
SK-N-SH or HeLa cells were plated in 24-well plates at a density of 105 cells/well, and were transfected at ≈60% confluence with FuGENE6 transfection reagent (Roche) according to manufacturer's instructions, using a 2:1 ratio of pDSma vector and pRLSV40 Renilla luciferase control vector (Promega), respectively. At 48 h posttransfection, cells were lysed and assayed using the Dual-Luciferase Reporter Assay System (Promega), and a Tecan microplate reader equipped with the XFluor4GENiosPro macro. Assays were performed on triplicate wells, and were repeated at least twice. Relative luciferase activity was normalized to values obtained from the promoter alone (pDSma-Promoter).
Zebrafish Embryo Manipulation and MO Injection.
Embryos were injected with a previously described and validated oligonucleotide MO against bbs4 to suppress translation of the endogenous protein, resulting in phenotypes consistent with PCP defects including shortened body axes, broad notochords, and widened somites (15, 16). They were subsequently coinjected with mRNA encoding WT human BBS4 or the S457I variant of BBS4. Embryos were classified as normal, moderately (Class I), and severely (Class II) affected according to phenotype.
In Situ Hybridization.
Embryos were cultured for 4 days in embryo media at 28 °C, fixed in 4% Paraformaldehyde overnight at 4 °C, and stored at −20 °C in 100% MeOH until in situ hybridization, which was carried out according to previously described protocols (15). Enteric neurons were labeled using a phox2b digoxigenin ripoprobe (31).
Human NC Isolation and Culture.
Human embryos were collected from pregnancies legally terminated using the mefiprestone protocol, in concordance with French bioethics law 2004-800 and with the approval of the Necker hospital ethics committee. Human NCCs were isolated from posterior hindbrain and thoracic-level neural tube and cultured as previously described (32) to 80% confluency on 8-well collagen I-coated slides (BD BioCoat).
Immunocytochemistry.
Twenty-four hours after seeding, hNCC were starved in DMEM/F12 with 0.4 ng/mL FGF2 for a further 72 h. Cells were fixed for 20 min in 4% paraformaldehyde, then treated in 50 mM NH4Cl 10 min, 0.3% Triton X-100 in PBS 15 min, followed by 1 h in a solution of 1% BSA, 0.1% Tween-20, 10% goat serum in PBS. Cells were then incubated with antiacetylated α-tubulin (TUBA4A; Sigma T6793), anti-RET (SC13104, 1:50; Santa Cruz), or anti-SOX9 (ab5535, 1:200; Chemicon) for 2 h at room temperature, and rinsed. SOX9 and RET were revealed using an Alexa Fluor 488 goat anti-rabbit IgG and TUBA4A with an Alexa Fluor 555 donkey anti-mouse IgG (1:200 each; Invitrogen). The slides were mounted with Prolong Gold antifade reagent with DAPI (Invitrogen).
HeLa cells were grown on glass coverslips in DMEM supplemented with 10% FBS at 37 °C in 5% CO2. Transient transfections of WT or mutant BBS4 constructs were carried out using calcium phosphate (Invitrogen), and cells processed 24 h later with anti-MYC mouse monoclonal and anti-γ-tubulin rabbit polyclonal antibodies (T3559; Sigma).
Microarray and RT-PCR.
Total RNA from 5 human trunk NCCs were extracted using Rneasy Mini kit (Qiagen). Gene profiling was done using the Affymetrix Human Genome U133 Plus 2.0 GeneChip array. Hybridization data were analyzed with dChip software. Heat map was constructed using a list of 75 experimentally confirmed ciliary and basal body/centrosomal proteins defined in ref. 33. Reverse transcription was performed using GeneAmp RNA PCR core kit (Applied Biosystems), and PCR amplifications were performed using 50 ng of cDNA and 35 amplification cycles. Primers are available on request. Affymetrix data are available from the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE 14340.
Acknowledgments.
We thank the families and their referring clinical teams for participation, the Association Françcaise de la Maladie de Hirschsprung for its support, Sophie Audollent for technical assistance, and the Johns Hopkins University Finz Center for husbandry and technical support with the zebrafish experiments. This work was supported by grants from the Association Nationale pour la Recherche, the Fondation pour la Recherche Médicale, and the National Institute of General Medical Sciences (A.S.M.), by National Institute of Child Health and Development Grant R01HD04260, by National Institute of Diabetes, Digestive, and Kidney Disorders Grants R01DK072301 and R01DK075972, by the Macular Vision Research Foundation (N.K.), by a Visual Neuroscience Training Program fellowship (N.A.Z.), by and National Institute of Diabetes, Digestive, and Kidney Disorders National Research Service Award Fellowship F32 DK079541 (to E.E.D.). A.S.M. and D.M. were supported by award GM071648 (to A.S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901219106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0901219106
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2728996?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/33/13921
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/33/13921
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/33/13921.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Integrative genomic analyses identify neuroblastoma risk genes involved in neuronal differentiation.
Hum Genet, 143(11):1293-1309, 27 Aug 2024
Cited by: 0 articles | PMID: 39192051 | PMCID: PMC11522082
Unsaturated Fatty Acid Synthesis Is Associated with Worse Survival and Is Differentially Regulated by MYCN and Tumor Suppressor microRNAs in Neuroblastoma.
Cancers (Basel), 16(8):1590, 21 Apr 2024
Cited by: 0 articles | PMID: 38672672 | PMCID: PMC11048984
Comprehensive characterization of the genetic landscape of familial Hirschsprung's disease.
World J Pediatr, 19(7):644-651, 01 Mar 2023
Cited by: 2 articles | PMID: 36857021 | PMCID: PMC10258170
Review Free full text in Europe PMC
Size matters: Large copy number losses in Hirschsprung disease patients reveal genes involved in enteric nervous system development.
PLoS Genet, 17(8):e1009698, 06 Aug 2021
Cited by: 13 articles | PMID: 34358225 | PMCID: PMC8372947
Genetic Characterization of a Model Ciliopathy: Bardet-Biedl Syndrome.
J Pediatr Genet, 10(2):126-130, 31 Mar 2020
Cited by: 1 article | PMID: 33996183 | PMCID: PMC8110344
Go to all (39) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases
- (1 citation) OMIM - 164761
SNPs
- (1 citation) dbSNP - rs2435357
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Epistatic interactions with a common hypomorphic RET allele in syndromic Hirschsprung disease.
Hum Mutat, 28(8):790-796, 01 Aug 2007
Cited by: 52 articles | PMID: 17397038
Mutations of the RET gene in isolated and syndromic Hirschsprung's disease in human disclose major and modifier alleles at a single locus.
J Med Genet, 43(5):419-423, 27 Jan 2006
Cited by: 34 articles | PMID: 16443855 | PMCID: PMC2649010
Germline mutations of the RET ligand GDNF are not sufficient to cause Hirschsprung disease.
Nat Genet, 14(3):345-347, 01 Nov 1996
Cited by: 122 articles | PMID: 8896569
Comprehensive characterization of the genetic landscape of familial Hirschsprung's disease.
World J Pediatr, 19(7):644-651, 01 Mar 2023
Cited by: 2 articles | PMID: 36857021 | PMCID: PMC10258170
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NICHD NIH HHS (1)
Grant ID: R01HD04260
NIDDK NIH HHS (6)
Grant ID: R01 DK072301
Grant ID: R01DK075972
Grant ID: R01 DK075972
Grant ID: R01DK072301
Grant ID: F32 DK079541
Grant ID: P30 DK079637
NIGMS NIH HHS (2)
Grant ID: GM071648
Grant ID: R01 GM071648




