Abstract
Free full text

Hepatic Autophagy Is Suppressed in the Presence of Insulin Resistance and Hyperinsulinemia

Abstract
Autophagy is essential for maintaining both survival and health of cells. Autophagy is normally suppressed by amino acids and insulin. It is unclear what happens to the autophagy activity in the presence of insulin resistance and hyperinsulinemia. In this study, we examined the autophagy activity in the presence of insulin resistance and hyperinsulinemia and the associated mechanism. Insulin resistance and hyperinsulinemia were induced in mice by a high fat diet, followed by measurements of autophagy markers. Our results show that autophagy was suppressed in the livers of mice with insulin resistance and hyperinsulinemia. Transcript levels of some key autophagy genes were also suppressed in the presence of insulin resistance and hyperinsulinemia. Conversely, autophagy activity was increased in the livers of mice with streptozotocin-induced insulin deficiency. Levels of vps34, atg12, and gabarapl1 transcripts were elevated in the livers of mice with insulin deficiency. To study the mechanism, autophagy was induced by nutrient deprivation or glucagon in cultured hepatocytes in the presence or absence of insulin. Autophagy activity and transcript levels of vps34, atg12, and gabarapl1 genes were reduced by insulin. The effect of insulin was largely prevented by overexpression of the constitutive nuclear form of FoxO1. Importantly, autophagy of mitochondria (mitophagy) in cultured cells was suppressed by insulin in the presence of insulin resistance. Together, our results show that autophagy activity and expression of some key autophagy genes were suppressed in the presence of insulin resistance and hyperinsulinemia. Insulin suppression of autophagy involves FoxO1-mediated transcription of key autophagy genes.
Introduction
Macroautophagy (autophagy) is a catabolic process whereby long lived large molecules and cellular organelles, such as mitochondria and endoplasmic reticulum (ER),3 are degraded by lysosomes for an alternative energy source during starvation (1, 2). Autophagy is normally activated by glucagon or deprivation of amino acids during starvation (1) but inhibited by amino acids and/or insulin through the mTOR- or/and Akt-dependent pathways after food intake (3, 4). Thus, autophagy activity fluctuates with food intakes and fasts. Importantly, autophagy is also essential for maintaining cellular health by removing misfolded large molecules and aged/dysfunctional cellular organelles, such as mitochondria and ER (1, 5). In other words, decreased autophagy will inevitably slow the removal of misfolded large molecules and aged/dysfunctional cellular organelles. Accumulation of these molecules and dysfunctional cellular organelles may not only contribute to the development of cancers (1) but also contribute to the development of metabolic diseases, such as insulin resistance. For example, the accumulation of dysfunctional mitochondria will most likely cause increased mitochondrion-derived oxidative stress, which is known to contribute to the development of insulin resistance (6,–10).
Insulin resistance is either a precursor or a key component of numerous diseases including obesity, metabolic syndrome, T2DM, cardiovascular disorders (including strokes), Alzheimer disease, depression, asthma, chronic inflammatory diseases, cancers, and aging (11,–19). Insulin resistance is primarily caused by the positive energy imbalance between the intake and expenditure of calories. The mechanisms of insulin resistance have been under intense investigation but remain unestablished.
However, it is known that the induction of insulin resistance in mice by obesity or high fat diet (HFD) is prevented or reversed when production of the mitochondrion-derived reactive oxygen species (mtROS) is blocked (6,–8). Induction of insulin resistance in cultured cells is also blocked when mtROS is scavenged (6). It has been clearly shown that oxidative stress is a precursor of insulin resistance (9). Therefore, mtROS plays a critical/necessary role in the development of insulin resistance.
The level of mtROS production may be influenced by many factors, such as mitochondrial mass and integrity. Much attention has been paid to the production of mitochondria via biogenesis. Numerous studies have shown that mitochondrial mass may be decreased in the subjects with insulin resistance/hyperinsulinemia due to reduced mitochondrial biogenesis. It is well known that decreased mitochondrial number is a cardinal feature of insulin resistance (20,–22). The ratio between mitochondrion-rich (type I) muscle fibers and glycolytic (type II) muscle fibers is decreased in subjects with insulin resistance (23, 24). Mitochondrial DNA (mtDNA) copy number is decreased in subjects with insulin resistance (25). Suppression of mitochondrial biogenesis by antiretroviral nucleoside analogues is associated with the development of insulin resistance in patients with AIDS (26). We have recently shown that mitochondrial production is decreased by prolonged exposure to insulin, which resembles insulin resistance and hyperinsulinemia (10, 27, 28). However, little attention has been paid to the removal of aged/dysfunctional mitochondria in the presence of insulin resistance and hyperinsulinemia, probably due to the current perception that insulin is not functional in the presence of insulin resistance. Obviously, the removal of aged/dysfunctional mitochondria can affect both the mass and integrity of mitochondria and, hence, influence the production of mtROS. There is no doubt that the removal of mitochondria and other cellular organelles, such as ER, is autophagy-dependent (1, 2). Thus, in this study, we have examined the level of hepatic autophagy in the presence of insulin resistance/hyperinsulinemia and the associated mechanism.
MATERIALS AND METHODS
Antibodies and Reagents
Antibodies against LC3 and p62 were from MBL International (Woburn, MA). The monoclonal antibody against β-actin, the antibody against ubiquitin, the recombinant human insulin, and glucagon were from Sigma. Antibodies against total and phospho-Akt (serine 473/308) or phospho-FoxO1 were from Cell Signaling Technology (Danvers, MA). Antibodies against IRS1(Tyr(P)416) was from Abcam (catalog number ab66153). Antibodies against PARP1 were from Santa Cruz Biotechnology (catalog number SC-7150). LY294002 was from Calbiochem. Blood glucose concentrations were measured using a Breese® 2 glucose meter (Bayer HealthCare). Plasma insulin levels were measured with a Linco insulin enzyme-linked immunosorbent assay kit. Protein assay kits were from Bio-Rad. The plasmid LC3-GFP and the recombinant adenoviruses encoding a constitutively nuclear FoxO1 mutant (ADA-FoxO1) were kind gifts from Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan) and Dr. Domenico Accili (Columbia University, New York, NY), respectively. Other materials were all obtained commercially and are of analytical quality.
Animal Experiments
All animal studies were approved by the Institutional Animal Care and Use Committee of The Hamner Institutes for Health Sciences and fully complied with the guidelines from the National Institutes for Health. Animals were housed under the usual day (12-h daylight) and night (12-h darkness) circadian and fed ad libitum. The following animal experiments were included: 1) C57BL/6 (B6) mice were fed with either the HFD (Research Diets catalog number D12330: 58.0 kcal % fat, 16.0 kcal % protein, and 26 kcal % carbohydrate) or the normal rodent chow diet (ND) for 8 weeks (28); 2) B6 mice were treated with streptozotocin (STZ; 60 mg/kg/day) for 7 days via intraperitoneal injection to induce T1DM as previously described (28); 3) to study the effect of insulin signaling blockade, B6 mice were similarly fed with either ND or HFD as described above for 4 weeks. During the last week of the ND or HFD, some mice were administered the phosphatidylinositol 3-kinase inhibitor LY294002 (10 mg/kg body weight, intraperitoneal injection, once a day at 9 a.m., 7 days) during day time, when mice normally sleep and do not eat.
Cells
Mouse primary hepatocytes were isolated from B6 mice that were fed with the ND and cultured in Williams' medium E supplemented with 10% fetal bovine serum as previously described (10, 29,–35). Mouse hepatoma 1c1c7 cells were maintained in high glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Calculation of Insulin Sensitivity
Insulin sensitivity was evaluated by measuring levels of blood glucose and plasma insulin and calculated as previously described (36): quantitative insulin sensitivity check index (QUICKI) = 1/log(I0 + log(G0)) and homeostasis model assessment (HOMA) = (G0 × I0)/22.5), where I0 represents fasting insulin (microunits/ml) and G0 represents fasting glucose (mg/dl) for QUICKI, and for HOMA, G0 is in mmol/liter.
DNA Transfection and Adenoviral Infection
DNA plasmids were introduced into the indicated cells by Lipofectamine 2000 transfection agents (Invitrogen). Adenoviruses encoding the nuclear form of FoxO1 (ADA-FoxO1) or empty adenovirus, amplified in HEK-293 cells, were applied to infect primary hepatocytes at 108 plaque-forming units/well in 6-well plates as described (35).
Immunoblotting
Cells were lysed in Nonidet P-40 lysis buffer (1% Nonidet P-40, 150 mm NaCl, 10% glycerol, 2 mm EDTA, 20 mm Tris (pH 8.0), 1 mm dithiothreitol, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, and 10 μg/ml aprotinin). Cell lysates (10–15 μg/ lane) were resolved in 4–20% Tris-glycine gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad). Target proteins were detected by immunoblotting with primary antibodies as indicated and alkaline phosphatase-conjugated secondary antisera. Fluorescent bands were visualized with a Typhoon 9410 variable mode Imager from GE Healthcare and then quantified by densitometry analysis using ImageQuant version 5.2 software from GE Healthcare.
RNA Extraction and Real-time PCR
Total RNAs were extracted from cells or tissues with an RNeasy minikit (Qiagen) and reverse transcribed into cDNAs, which were quantified by TaqMan® real-time PCR with specific primers listed in Table 1 and normalized to levels of 36B4 transcripts.
TABLE 1
Full names and sequences of primers for real-time PCR used in this study
| Gene name | DNA sequence |
|---|---|
| beclin1 | |
    Forward Forward | 5′-ggccaataagatgggtctga-3′ |
    Reverse Reverse | 5′-cactgcctccagtgtcttca-3′ |
| vps34 | |
    Forward Forward | 5′-tgtcagatgaggaggctgtg-3′ |
    Reverse Reverse | 5′-ccaggcacgacgtaacttct-3′ |
| ulk2 | |
    Forward Forward | 5′-cagccctggatgagatgttt-3′ |
    Reverse Reverse | 5′-ggatgggtgacagaaccaag-3′ |
| atg12 | |
    Forward Forward | 5′-ggcctcggaacagttgttta-3′ |
    Reverse Reverse | 5′-cagcaccgaaatgtctctga-3′ |
| gabarapl1 (GEC1) | |
    Forward Forward | 5′-catcgtggagaaggctccta-3′ |
    Reverse Reverse | 5′-atacagctggcccatggtag-3′ |
| 36B4 | |
    Forward Forward | 5′-gcagacaacgtgggctccaagcagat-3′ |
    Reverse Reverse | 5′-ggtcctccttggtgaacacgaagccc-3′ |
Confocal Microscopy
Hepatoma 1c1c7 cells were plated onto glass bottom cell culture chamber (MatTek, Ashland, MA). The next day, LC3-GFP or GFP was introduced into cells by Lipofectamine 2000 transfection reagent (Invitrogen). Some cells were then cultured in regular Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37 °C. Cells were next pretreated with 1 nm insulin for 30 min, followed by incubation in the nutrient starvation medium (Earle's balanced salt solution (EBSS)) (Invitrogen) (37) or regular medium plus 100 nm glucagon for 8 h. To test whether insulin suppresses the formation of mitochondrial autophagy (mitophagy) in the presence of insulin resistance, some cells were pretreated with insulin (10 nm) for 24 h to induce insulin resistance as described (27). Mitochondria were labeled with the mitochondrion-specific dye (tetramethylrhodamine methyl ester, 25 nm) (catalog number T668, Invitrogen) at room temperature for 30 min. Cells were subsequently imaged live at room temperature in Dulbecco's modified Eagle's medium without phenol (Invitrogen) using a Zeiss LSM 510Meta confocal imaging system with a 30-milliwatt argon laser and a ×63 1.4 numerical aperture oil immersion objective at ×2 zoom. GFP was detected using a 488-nm argon laser and detected with a 515–540-nm band pass filter. Tetramethylrhodamine methyl ester fluorescence was excited with a 563-nm helium-neon laser attenuated and detected at >570 nm. The images were manipulated with Zeiss LSM software.
Statistical Analyses
Data are presented as mean ± S.E. Data were compared by Student's t test with GraphPad Prism version 4.0 for Windows (San Diego, CA). Differences at values of p < 0.05 were considered significant.
RESULTS
Autophagy Is Suppressed in the Livers of Mice with Insulin Resistance and Hyperinsulinemia Induced by the HFD
It is known that autophagy is normally suppressed by insulin (1, 2). However, it is unknown whether autophagy is increased or decreased in the presence of hyperinsulinemia and insulin resistance. The current general perception is that insulin is not properly functional in the presence of insulin resistance due to the fact that the response to acute insulin challenge is blunted in the presence of insulin resistance. Thus, some have speculated that autophagy may be increased in the presence of insulin resistance (38). To address this issue, mice were fed with the HFD for 8 weeks, followed by measuring markers of autophagy. As shown in Fig. 1, A and B, fasting plasma glucose levels were not yet altered by the 8-week HFD, whereas plasma insulin levels were significantly increased. As a result, insulin sensitivity evaluated by HOMA and QUICKI was significantly decreased, suggesting the presence of insulin resistance. The basal level of Akt phosphorylation in the liver after a 12-h fast was increased by the HFD (Fig. 1C). To examine the level of autophagy in the liver, three autophagy markers were measured. First, the ratio of LC3-II over LC3-I is normally increased when autophagy is activated, and vice versa (1, 2). As shown in Fig. 1D, the hepatic ratio of LC3-II/LC3-I was decreased by the HFD, suggesting that hepatic autophagy was suppressed by the HFD. Second, the level of p62, another autophagy marker (long lived protein), is normally increased when autophagy is suppressed (39). Our results clearly show that the hepatic level of p62 protein was indeed increased by the HFD (Fig. 1E), also suggesting that hepatic autophagy was decreased by the HFD. Third, levels of polyubiquitin are normally elevated when autophagy is suppressed (40). Results in Fig. 1F showed that levels of polyubiquitin in the liver were increased by the HFD. These results from multiple aspects all show that the autophagy activity in the liver was decreased in the presence of insulin resistance and hyperinsulinemia induced by the HFD.
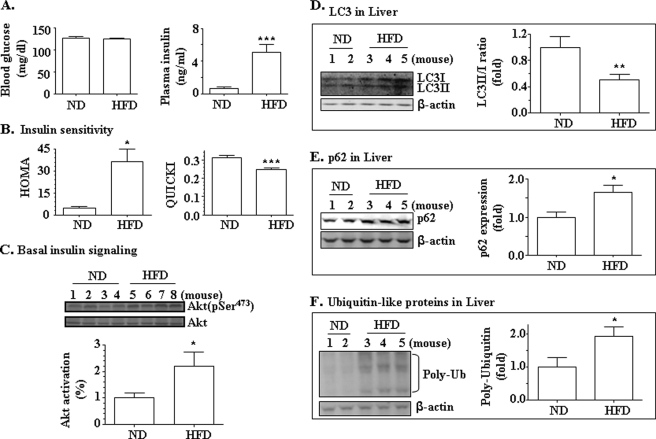
Autophagy was suppressed in the livers of mice on the HFD. B6 mice were fed with either the ND (n = 10) or HFD (n = 11) for 8 weeks. A, levels of blood glucose and plasma insulin were measured. B, insulin sensitivity was analyzed by HOMA and QUICKI. C, levels of basal phospho-Akt and total Akt in the livers were evaluated by immunoblotting. D, levels of LC3I and LC3II in the liver were determined by immunoblotting and normalized to levels of β-actin. E, levels of p62 in the liver were determined by immunoblotting and normalized to levels of β-actin. F, levels of polyubiquitin (Poly-Ub) in the liver were measured and normalized to levels of β-actin. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Hepatic Expression of Key Genes Involved in Activation of Autophagy Is Suppressed in the Presence of the HFD-induced Insulin Resistance and Hyperinsulinemia
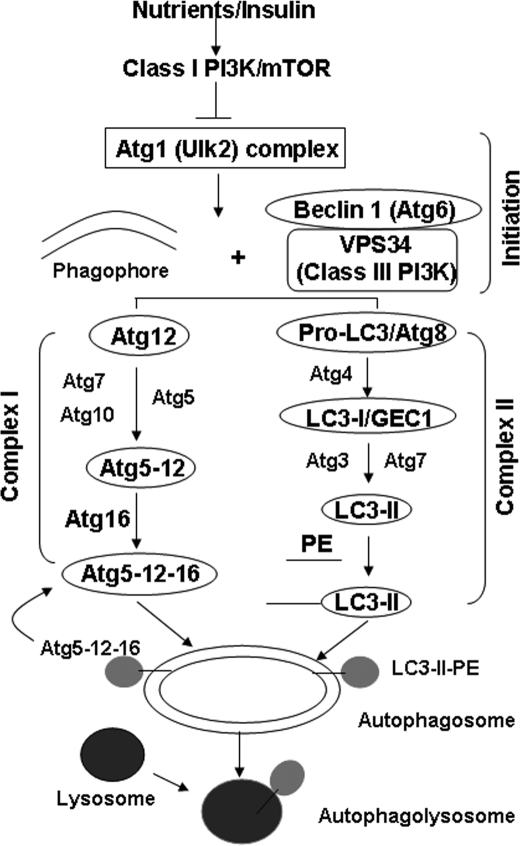
The autophagy machinery includes two main ubiquitin-like complexes (1, 2). In complex I (Fig. 2), Atg12 is activated by Atg7 and then transferred by Atg10 to Atg5. The Atg12-Atg5 dimer will be joined by Atg16 to form an Atg12-Atg5-Atg16 multimer complex, which is localized to membranes of early autophagosomes and will be subsequently joined by the complex II. The formation of autophagy complex II is initiated when LC3-I (a mammalian homologue of Atg8) is cleaved by Atg4 to produce LC3-II. LC3-II will then be conjugated to phosphatidylethanolamine by Atg7 and Atg3. The unconjugated LC3-I stays in the cytosol, but the conjugated LC3-II binds to the autophagosomal membrane, following the formation of the complex I. Complex I will eventually detach from mature autophagosomes, whereas LC3-II stays on the membrane until it is degraded by the lysosomes. That is why LC3-II has been widely used as a marker for monitoring activation of autophagy. Additionally, several other proteins are required for the initiation of autophagy. These proteins include at least Ulks (a mammalian homologue of Atg6), Beclin1 (another homologue of Atg6), and Vps34 (a class III phosphatidylinositol 3-kinase).
To further examine the autophagy process in the presence of insulin resistance/hyperinsulinemia, mRNA levels of some of the above described autophagy genes were examined in the livers of the mice detailed in Fig. 1. As shown in Fig. 3, the mRNA levels of vps34, atg12, and gabarapl1 (protein name GEC, another homologue of Atg8 (41)) genes were significantly decreased by the HFD, although Beclin1 and Ulk2 gene transcripts were not affected. Because vps34, atg12, and gabarapl1 genes are involved in the initiation and formation of complexes I and II of autophagy, respectively, these results further demonstrate that hepatic autophagy level is decreased in the presence of insulin resistance/hyperinsulinemia induced by the HFD.
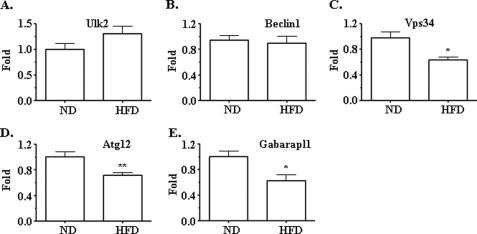
Expression levels of some key autophagy genes in the livers of mice were suppressed by the HFD. mRNAs from the liver samples of mice described in Fig. 1 were extracted, converted into cDNA, and used for quantifications of target genes. Transcripts of ulk2 (A), beclin1 (B), vps34 (C), atg12 (D), and gabarapl1 (E) were measured by using real-time PCR and normalized to the level of 36B4 transcript. *, p < 0.05; **, p < 0.01; ND (n = 10) versus HFD (n = 11).
To explore whether or not insulin signaling was responsible for the suppressed autophagy in the presence of insulin resistance/hyperinsulinemia, insulin resistance in mice was induced by the HFD (4 weeks) as we previously described (28). Some mice were administered the phosphatidylinositol 3-kinase inhibitor LY294002 during day time (once a day for the last week), when mice normally sleep and do not eat. The basal level of insulin signaling was evaluated by measurements of Akt phosphorylation. Autophagy markers were detected as detailed above. Results showed that the basal level of Akt phosphorylation was increased by the HFD as we previously described (28) but reversed by the treatment with LY294002 (supplemental Fig. 1, A and B). The autophagy marker p62 in the liver was increased by the HFD, and the increase was largely prevented by LY294002 (supplemental Fig. 1C). Transcript levels of key autophagy genes (vps34, atg12, and gebarapl1) in the liver were decreased by the HFD but reversed by the treatment with LY294002 (supplemental Fig. 1, D–F). These results imply that insulin is responsible for the suppressed autophagy in the presence of insulin resistance and hyperinsulinemia.
Autophagy and Expression of Key Autophagy Genes Are Increased in the Livers of Mice with Insulin Deficiency Induced by STZ
It has previously been shown that the autophagy level is increased in STZ-induced insulin deficiency, which resembles but is not an equivalent of T1DM (42). However, the associated mechanism has not been studied. Because the basal Akt-dependent insulin signaling is increased in the presence of insulin resistance/hyperinsulinemia, as shown in Fig. 1 and supplemental Fig. 1, and expression of some key autophagy genes was decreased by the HFD (Fig. 3), it is possible that insulin suppresses autophagy through inhibiting expression of key autophagy genes. To further examine the effect of insulin on expression of key autophagy genes, insulin secretion in mice was deleted by STZ, followed by measurements of levels of autophagy and transcripts of some key autophagy genes. As shown in Fig. 4, A and B, application of STZ significantly damaged the pancreatic insulin secretion, leading to overt hyperglycemia. As predicted, the LC3-II/LC3-I ratio in the liver was increased by the treatment with STZ (Fig. 4C), whereas levels of both p62 proteins and polyubiquitin were decreased (Fig. 4, D and E). Together, these results demonstrate that autophagy was activated in the livers of mice when insulin secretion was crippled by STZ.
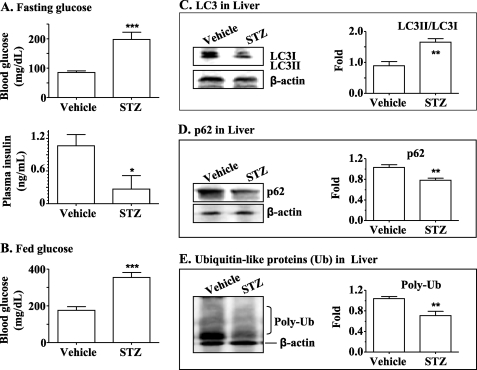
Autophagy was increased in the livers of mice with T1DM. T1DM was induced in B6 mice by using STZ (60 mg/kg body weight, once a day, intraperitoneally, 7 days). Levels of fasting blood glucose (A, top), plasma insulin (A, bottom), and blood glucose during the fed state (B) were measured. Levels of LC3-I and -II (C), p62 (D), and polyubiquitin (Poly-Ub) (E) in the liver were measured by immunoblotting and normalized to the level of β-actin. **, p < 0.01; ***, p < 0.001; STZ group (n = 5) versus vehicle group (n = 5).
Next, transcript levels of vps34, atg12, and gabarapl1 genes were quantified in the livers of mice with STZ-induced insulin deficiency. As shown in Fig. 5, expression of all of these genes was significantly increased in the livers of mice with insulin deficiency. These results further support the notion that insulin suppresses autophagy by inhibiting expression of key autophagy genes directly or indirectly.
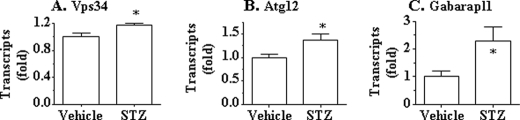
Expression of key autophagy genes in the livers of mice with T1DM was increased. mRNAs in the liver samples from the mice described in the legend to Fig. 4 were extracted, converted into cDNA, and used for quantifications of target genes. Transcripts of vps34 (A), atg12 (B), and gabarapl1 (C) were determined by using real-time PCR and normalized to the level of 36B4 transcript. *, p < 0.05; **, p < 0.01; STZ group (n = 5) versus vehicle group (n = 5).
Insulin Inhibits Autophagy and Expression of Key Autophagy Genes in Cultured Hepatocytes
To investigate the mechanism of insulin suppression of autophagy, we used a cellular system (primary hepatocytes). Autophagy was activated by either nutrient starvation or glucagon, followed by evaluating the expression levels of key autophagy genes. As shown in Fig. 6, A and B, the LC3-II/LC3-I ratio was significantly increased by the nutrient starvation, indicating increased autophagy. The increased LC3-II/LC3-I ratio was reversed by the treatment with insulin. The effect of insulin was largely prevented by the pretreatment with the phosphatidylinositol 3-kinase inhibitor LY294002. Akt phosphorylation was significantly elevated by the treatment with insulin but largely prevented by LY294002. Similarly, in Hepa1c1c7 hepatoma cells stained by immunofluorescence, both nutrient starvation and treatment with glucagon stimulated LC3-II aggregation as intense immunofluorescence dots, and the aggregation of these dots was reduced by insulin (Fig. 6, C–I). These results demonstrate the induction and inhibition of autophagy in our culture system.
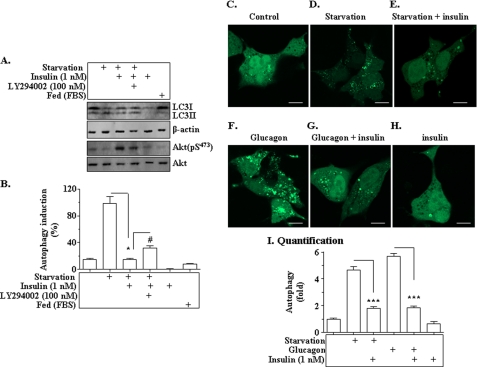
Autophagy is suppressed by insulin in cultured hepatocytes. A and B, primary hepatocytes isolated from mice were incubated in either the regular Dulbecco's modified Eagle's medium or nutrient-deprived starvation medium (EBSS) for 8 h. Insulin (1 nm) was added into some cells for 8 h as noted. The phosphatidylinositol 3-kinase inhibitor LY294002 (100 nm) was added to some cells prior to insulin as noted. Levels of LC3-I and II, phospho-Akt and total Akt, and β-actin were measured by immunoblotting, quantified, and normalized. * and #, p < 0.05. C–H, Hepa1c1c7 cells were plated onto glass slips in culture chambers for 12 h, followed by the introduction of GFP-labeled LC3 (LC3-GFP) into the cells via transfection. Cells were incubated in either the starvation medium (EBSS) or regular medium with glucagon (100 nm) in the presence or absence of 1 nm insulin for 8 h. LC3-GFP was visualized with confocal microscopy as detailed under “Materials and Methods.” Scale bar, 10 μm. I, quantification analysis of autophagy dots in hepatocytes. Data represent mean ± S.E. from three independent experiments of C–H. ***, p < 0.001.
Next, we examined the effect of insulin on the expression level of key autophagy genes. As shown in Fig. 7, mRNA levels of vps34, atp12, and gabarapl1 were elevated by nutrient starvation, but the elevations were significantly reversed by insulin. The effect of insulin was largely prevented by the phosphatidylinositol 3-kinase inhibitor LY294002. Together, these results show that insulin suppresses expression of key autophagy genes in cultured hepatocytes.
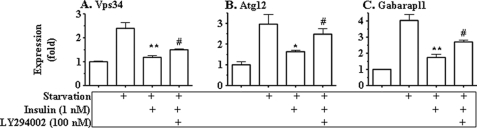
Expression of key autophagy genes (vps34 (A), atg12 (B), and gabarapl1 (C)) was suppressed by insulin in cultured cells. Primary hepatocytes were treated as detailed in the legend to Fig. 6A. Transcript levels of target genes were determined by real-time PCR and normalized to the level of 36B4 transcript. Results represent mean ± S.E. from three independent experiments. *, p < 0.05 insulin plus starvation versus starvation only. #, p < 0.05 LY294002-treated cells versus cells treated with insulin plus starvation.
Insulin Inhibits Autophagy and Expression of Key Autophagy Genes in a FoxO1-dependent Manner
To examine the mechanism by which insulin inhibits expression of autophagy genes, the constitutive nuclear form of FoxO1 was introduced into Hepa1c1c7 cells prior to the treatment with insulin as noted (34), followed by measurements of LC3-II and mRNA levels of key autophagy genes. As shown in Fig. 8A, nutrient starvation increased the autophagy marker, LC3-II, but the increase was reversed by insulin, which stimulated Akt phosphorylation. However, in the presence of the overexpressed constitutive nuclear form of FoxO1, the effect of insulin was prevented, suggesting that insulin suppression involved FoxO1-mediated gene transcription. To determine the role of FoxO1 in insulin-mediated suppression of autophagy genes, the effect of the constitutive nuclear form of Foxo1 was examined. As shown in Fig. 8B, nutrient starvation-induced expression of vps34, atg12, and gabarapl1 was blocked by insulin. Nevertheless, in the presence of the overexpressed constitutive nuclear form of FoxO1, insulin failed to suppress expression of vps34, atg12, and gabarapl1 genes. Together, these results demonstrate that the FoxO1-mediated transcription of key autophagy genes is an intricate part of insulin suppression of autophagy.
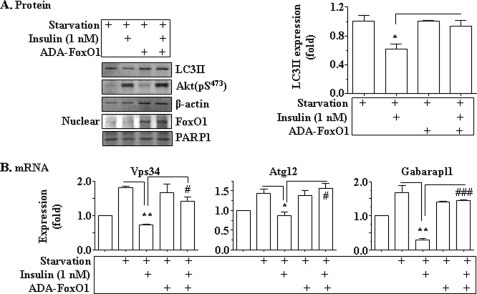
Insulin-suppressed expression of key autophagy genes was mediated by FoxO1. The constitutive nuclear form of FoxO1 (ADA-FoxO1) encoded by the recombinant adenovirus or the empty adenoviruses was introduced into Hepa1c1c7 cells via infection. Thirty-six hours later, cells were cultured in the starvation medium (EBSS) in the presence or absence of insulin (1 nm) for an additional 8h. A, levels of LC3, phosphorylated Akt, and β-actin were measured by immunoblotting. Levels of FoxO1 and PARP1 in nuclear protein extracts were measured by immunoblotting with the antibodies as noted. *, p < 0.05. B, transcripts of vps34, atg12, and gabarapl1 genes were determined by using real-time PCR and normalized to the level of 36B4 transcript. Results represent mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01 insulin plus starvation versus starvation only. #, p < 0.05; ###, p < 0.001 ADA-FoxO1 infection plus insulin plus starvation group versus starvation only group.
Autophagy of Mitochondria (Mitophagy) Was Suppressed in the Presence of Insulin Resistance in Cultured Cells
To examine the level of autophagy in cultured cells in the presence of insulin resistance, insulin resistance was induced in some hepatoma cells by the prolonged exposure to insulin as described (10), followed by evaluation of mitophagy induced either by starvation or glucagon. As shown in Fig. 9A, prolonged exposure to insulin blunted the response of cells to the acute challenge of insulin in terms of IRS1 tyrosine phosphorylation and Akt phosphorylation, indicating the presence of insulin resistance. Under the same conditions, autophagy of mitochondria (mitophagy) was examined by confocal microscopy. As shown in Fig. 9, D–E and H, nutrient starvation significantly induced mitophagy formation (yellow dots from merging of green GFP-LC3-II and red mitochondria), which was significantly attenuated by chronic exposure to insulin. Similarly, glucagon stimulated mitophagy, but the stimulation was significantly inhibited by the chronic presence of insulin (Fig. 9, E, G, and H). Together, these results show that mitophagy in cultured cells is decreased in the presence of insulin and insulin resistance.
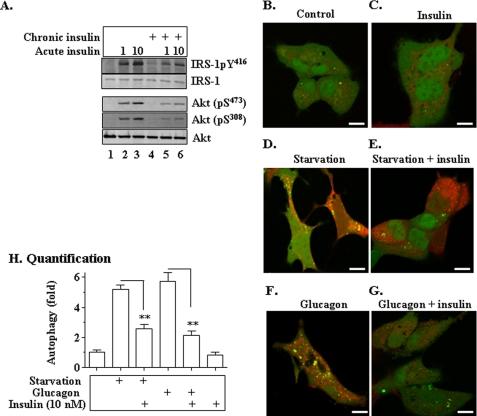
Mitophagy is suppressed in the presence of insulin and insulin resistance in cultured cells. A–G, LC3-GFP or GFP was introduced into hepa1c1c7 cells via transfection. After a 24-h culture in the presence of 10 nm insulin (chronic insulin treatment), cells were then incubated in either the starvation medium (EBSS) or regular medium with glucagon (100 nm) in the presence or absence of 10 nm insulin for 8 h. Cells were labeled with the mitochondria-specific dye tetramethylrhodamine methyl ester (25 nm) at room temperature for 30 min. LC3-GFP co-localization with mitochondria (mitophagy) was immediately analyzed with confocal microscopy as detailed under “Materials and Methods.” Scale bar, 10 μm. H, quantification of mitophagy (yellow dots). Data represent mean ± S.E. from three independent experiments of A–G. **, p < 0.01.
DISCUSSION
Autophagy is not only important for cell survival by providing fuel through the self-digestion of large molecules and cellular organelles but also essential for maintaining cellular health by removing and cleaning up the misfolded molecules and aged/dysfunctional cellular organelles, such as mitochondria and ER (1, 2). The role of autophagy in prevention of cancers and many other diseases has been extensively investigated (1, 2). Although autophagy activity is closely connected with nutrients and the central metabolic hormones (insulin and glucagon), the potential role of autophagy in the development of metabolic diseases, such as insulin resistance and its many associated diseases, has hardly been investigated.
It is well established that autophagy is suppressed by insulin and nutrients when autophagy is not needed for providing fuel during the fed state. In contrast, autophagy is activated during starvation, when plasma levels of nutrients and insulin are low and the plasma glucagon level is high. Thus, the autophagy activity fluctuates with activities of food intake and fast every day. This fluctuation has been established in humans through the evolution for millions of years and become a necessity for cell survival and health. In the presence of insulin resistance/hyperinsulinemia primarily caused by the positive energy imbalance due to overeating and/or physical inactivity, this fluctuation may be lost due to the continuous presence of insulin in the blood. The continuous increase in plasma insulin level can presumably inhibit autophagy continuously. However, the general perception now is that insulin is not functional in the presence of insulin resistance. As a result, some have speculated that autophagy level may be increased in the presence of insulin resistance and hyperinsulinemia (42). Results from this study clearly show that the autophagy level (including mitophagy) in the liver is actually decreased by continuous exposure to insulin in animals. The suppressed autophagy will evidently lead to various problems that are associated with the retention of misfolded large molecules and aged/dysfunctional cellular organelles.
The retention of large molecules due to suppressed autophagy may contribute to the development of many health problems. It is well known that the decreased autophagy is closely associated with the development of various cancers due to the retention of large molecules (2). Retention of some large molecules, such as amyloid and Tau proteins, may contribute to the development of Alzheimer disease, which is characterized by intracellular neurofibrillary tangles and extracellular deposits in the form of senile plaques (43,–46). That may be why hyperinsulinemia and insulin resistance is inseparable from Alzheimer disease (12, 13). Retention of large molecules may contribute to the hypertrophy of tissues/organs. For example, retention of myoglobins and other large molecules may lead to the hypertrophy of organs like the heart and smooth muscles. That may be why subjects with insulin resistance/hyperinsulinemia usually have cardiac hypertrophy (47). The thickened vascular wall due to the retention of large molecules may contribute to hypertension, which is a frequent companion of insulin resistance and hyperinsulinemia. Retention of large molecules in the airway may contribute to the thickened airway smooth muscles, render the airway hyper-responsive, and lead to asthma. That may be why subjects with obesity have increased prevalence and severity of asthma (16, 48).
Retention or delayed removal of cellular organelles due to suppressed autophagy may contribute to the development of metabolic diseases. For example, delayed removal of mitochondria (mitophagy) may cause accumulation of aged/dysfunctional mitochondria. It is conceivable that the aged/dysfunctional mitochondria are less efficient in converting the proton gradient into ATP without producing much reactive oxygen species. In other words, the accumulation of aged/dysfunctional mitochondria may contribute to or aggravate the increased oxidative stress in subjects with insulin resistance and hyperinsulinemia. Similarly, the accumulation of aged/dysfunctional ER may cause increased production of misfolded large proteins, such as Tau protein, seen in Alzheimer disease (43) and cause the so-called ER stress. ER stress is known to be associated with insulin resistance and hyperinsulinemia (49).
It is known that insulin can suppress autophagy through mTOR (1). mTOR inhibits autophagy via Akt/S6K (50). It has recently been shown that insulin can also inhibit autophagy through the Akt/FoxO3 pathway in skeletal muscles (41, 50). Specifically, insulin can inhibit expression of several autophagy genes, which are mediated by FoxO3 (41, 50). In this study, we show that insulin can inhibit autophagy by blocking the expression of vps34, atg12, and gabarapl1 genes. VPS34 is involved in the initiation of autophagy (1). ATG12 is required for the formation of Complex I (see Fig. 2). GEC1 is a part of Complex II (Fig. 2). Our results show that the expression of these genes is FoxO1-dependent in hepatocytes. Thus, our results provide new insight into the mechanism by which insulin suppresses autophagy.
In summary, results from this study demonstrate that in the presence of insulin resistance and hyperinsulinemia, autophagy is suppressed. The reduced autophagy will lead to delayed removal of misfolded large molecules and aged/dysfunctional cellular organelles. Thus, the reduced autophagy level may be a major contributor to the development of various major diseases that are associated with modern lifestyles characterized by overeating and/or physical inactivity. Our results also shed new light on the mechanism of insulin regulation of autophagy. Therefore, results from this study may provide new approaches for the prevention and treatment of many modern major diseases, such as metabolic syndrome (including insulin resistance), Alzheimer disease, asthma, cancers, and aging.
*This work was supported, in whole or in part, by National Institutes of Health Grant R01DK076039 (to W. C.). This work was also supported by the Investigator Development Fund from The Hamner Institutes for Health Sciences (to W. C.), American Heart Association Grant SDG-0530244N (to W. C.), and American Diabetes Association Grant 7-09-BS-27 (to W. C.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
3The abbreviations used are:
- ER
- endoplasmic reticulum
- HFD
- high fat diet
- mtROS
- mitochondrion-derived reactive oxygen species
- mtDNA
- mitochondrial DNA
- ND
- normal rodent chow diet
- STZ
- streptozotocin
- GFP
- green fluorescent protein
- EBSS
- Earle's balanced salt solution
- QUICKI
- quantitative insulin sensitivity check index
- HOMA
- homeostasis model assessment
- T1DM
- type1 diabetes mellitus.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m109.033936
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925820380479/pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/284/45/31484.pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/284/45/31484
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/284/45/31484
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Thyroid Hormone-Mediated Selective Autophagy and Its Implications in Countering Metabolic Dysfunction-Associated Steatotic Liver Disease.
Endocrinol Metab (Seoul), 39(5):686-692, 14 Oct 2024
Cited by: 0 articles | PMID: 39397515 | PMCID: PMC11525703
Review Free full text in Europe PMC
Autophagy and hepatic lipid metabolism: mechanistic insight and therapeutic potential for MASLD.
NPJ Metab Health Dis, 2(1):19, 02 Aug 2024
Cited by: 0 articles | PMID: 39100919 | PMCID: PMC11296953
Review Free full text in Europe PMC
Cinnamaldehyde ameliorates STZ-induced diabetes through modulation of autophagic process in adipocyte and hepatic tissues on rats.
Sci Rep, 14(1):10053, 02 May 2024
Cited by: 0 articles | PMID: 38698047 | PMCID: PMC11066029
Identification of relevant differential genes to the divergent development of pectoral muscle in ducks by transcriptomic analysis.
Anim Biosci, 37(8):1345-1354, 01 Apr 2024
Cited by: 0 articles | PMID: 38575126 | PMCID: PMC11222850
Dihydromyricetin ameliorates hepatic steatosis and insulin resistance via AMPK/PGC-1α and PPARα-mediated autophagy pathway.
J Transl Med, 22(1):309, 26 Mar 2024
Cited by: 1 article | PMID: 38532480 | PMCID: PMC10964712
Go to all (256) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1.
Nat Med, 18(3):388-395, 19 Feb 2012
Cited by: 231 articles | PMID: 22344295 | PMCID: PMC3296881
Pancreatic-derived factor promotes lipogenesis in the mouse liver: role of the Forkhead box 1 signaling pathway.
Hepatology, 53(6):1906-1916, 02 May 2011
Cited by: 38 articles | PMID: 21412813
FoxO1 links hepatic insulin action to endoplasmic reticulum stress.
Endocrinology, 151(8):3521-3535, 25 May 2010
Cited by: 22 articles | PMID: 20501674 | PMCID: PMC2940535
Skeletal Muscle Vascular Function: A Counterbalance of Insulin Action.
Microcirculation, 22(5):327-347, 01 Jul 2015
Cited by: 11 articles | PMID: 25904196
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (2)
Grant ID: R01 DK076039
Grant ID: R01DK076039