Abstract
Free full text

Co-delivery of Doxorubicin and Bcl-2 siRNA by Mesoporous Silica Nanoparticles Enhances the Efficacy of Chemotherapy in Multidrug Resistant Cancer Cells**
Associated Data
Development of multidrug resistance in cancer cells and adverse side effects are the major obstacles for effective cancer chemotherapy.[1-3] Therapeutic strategies to overcome drug resistance and specific tumor targeting with minimal premature drug release should have a great impact on the treatment of cancer. The term multidrug resistance (MDR) is used to define a resistance phenotype where cancer cells become resistant simultaneously to multiple drugs with no obvious structural resemblance and with different molecular targets.[4, 5] The multidrug resistance can be divided into two distinct classes, pump and nonpump resistance.[3] The pump resistance is caused by certain proteins that form membrane-bound ATP-dependent active drug efflux pumps, which significantly decrease the intracellular concentration of the drug and thereby the efficacy of the treatment. Membrane proteins, P-glycoprotein (Pgp) and multidrug resistance-associated protein (MRP) have been shown to be the main players for pump resistance to a broad range of structurally and functionally distinct cytotoxic agents.[6] The main mechanism of nonpump resistance is an activation of cellular antiapoptotic defense, mainly by Bcl-2 protein. Most of the anticancer drugs trigger apoptosis and simultaneously activate both pump and nonpump cellular defense of multidrug resistance, which prevents cell death. Therefore, to effectively suppress the overall resistance to chemotherapy, it is essential to simultaneously inhibit both pump and nonpump mechanisms of cellular resistance by targeting all the intracellular molecular targets.[3, 7-9]
The field of RNA interference (RNAi) therapeutics has made significant progress since the first demonstration of gene knockdown in mammalian cells. [10, 11] Short interfering RNA (siRNA)-based formulations offer significant potential as therapeutic agents to induce potent, persistent, and specific silencing of a broad range of genetic targets.[12-14] Special sequences of siRNAs targeted against mRNA encoding major proteins responsible for pump and nonpump cellular defense have been developed and shown a substantial efficacy in vitro.[8, 9, 15-18]
However, reports on delivering such types of siRNA simultaneously with a traditional anticancer drug to cancer cells for enhanced chemotherapy efficacy have been scant, due to the lack of efficient co-delivery methods.[3, 15]
There is a surge of interest in using inorganic engineered nanoparticles (NPs) for medical and biological applications. They are expected to solve some difficult human health problems, due to their unique properties and their remarkably large surface area. Studies using inorganic engineered NPs modified with various functional groups have demonstrated enhancement in drug delivery, including DNA and siRNA condensation and delivery into mammalian cells.[19-27] Luo et al. also reported that DNA transfection by cationic transfection reagents can be dramatically enhanced by unmodified silica nanoparticles via a unique “enhanced concentration” mechanism.[28] However, simultaneous delivery of an anticancer drug as an apoptosis inducer and siRNAs as suppressors of pump resistance and cellular antiapoptotic defense to enhance the therapeutic effects has not been reported yet. Inspired by the finding that mesoporous silica nanoparticles (MSNs) can simultaneously deliver DNA and chemicals into plant cells reported by Lin's group,[29] we explored to utilize MSNs to co-deliver doxorubicin (Dox) (as a model apoptosis-inducing anticancer drug), and a siRNA (as a suppressor of cellular antiapoptotic defense) simultaneously into multidrug resistant cancer cells for efficient cancer therapy.
As illustrated in Scheme 1, the MSNs were modified to encapsulate Dox inside the pores to achieve minimal premature drug release. Then the Dox-loaded MSNs were modified with generation 2 (G2) amine-terminated polyamidoamine (PAMAM) dendrimers. The dendrimer-modified MSN can efficiently complex with siRNAs targeted against mRNA encoding Bcl-2 protein, which is the main player for nonpump resistance. We found thus-formed complex can be delivered into multidrug resistant A2780/AD human ovarian cancer cells to induce cell death. The anticancer efficacy of Dox increased 132 times compared to free Dox, mainly because the simultaneously delivered siRNA significantly suppressed the Bcl-2 mRNA, and efficiently overcome the nonpump resistance. Moreover, our data suggested that the delivered Dox by the MSNs are primarily localized in perinuclear region upon internalization, providing additional advantage in possibly bypassing pump resistance, therefore further enhancing the drug efficacy. This result is much different from some liposome co-delivery systems, in which antisense oligonucleotides targeted to pump and nonpump resistances must be simultaneously delivered in order to significantly increase drug efficacy.[3, 7] We attributed this difference to the different internalization pathway and drug release mechanism. We envisioned that this co-delivery system can be generalized to other anticancer drugs and other cancer cell lines.
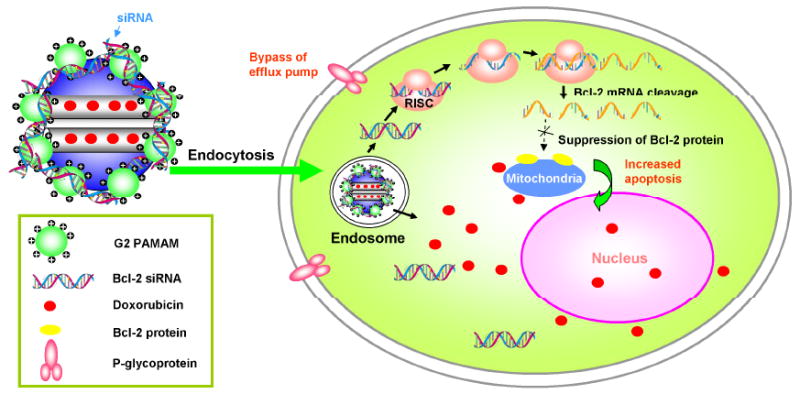
Schematic diagram of a co-delivery system based on MSNs to deliver Dox and Bcl-2-targeted siRNA simultaneously to A2780/AD human ovarian cancer cells for enhanced chemotherapy efficacy.
We synthesized the MCM-41 type of MSNs using a reported surfactant-templated, base-catalyzed condensation method for our study.[21, 30] The MSNs were imaged by transmission electron microscope (TEM) and shown in Figure 1a. The MSNs were then reacted with 3-iso-cyanatopropyltriethoxysilane to yield isocyanatopropyl (ICP)-modified surface both inside the pores and outside surface of the MSNs. The portions of MSNs with small diameters were isolated by dispersing the MSNs in H2O and collecting the supernatant after settling for 2 h. The supernatant was subsequently lyophilized to obtain MSN powders. Thus-obtained ICP-modified MSNs (ICP-MSN) showed a mean particle size of ~200 nm by transmission electron microscope (TEM) (Figure 1b) and dynamic light scattering (DLS), a mean pore size of 2.88 nm, surface area of 816 m2/g, and pore volume of 0.77 cm3/g (supporting information, Figures S-1 and 2).
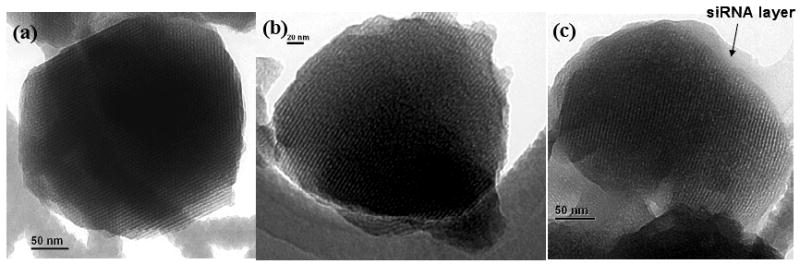
TEM image of an (a). MSN nanoparticle with no modification; (b). isocyanatopropyl-modified MSN nanoparticle; (c). MSN-Dox-G2 complex with siRNA. Bar indicates 50 nm in panels (a) and (c) and 20 nm in panel (b).
To encapsulate Dox into the pores of MSNs, 4.9 mg of lyophilized ICP-MSNs were suspended in 8.0 ml Dox solution in 3:5 (v:v) MeOH:H2O (4.5 mg/ml) for 24 h. After loading, the suspension was centrifuged and the supernatants and pellets were collected respectively. To confer the Dox loaded-MSN to complex with siRNA, the pellet collected was further redispersed into 2.0 ml of H2O and reacted with 34.4 mg of G2 PAMAM to form urea linkage between amines of G2 PAMAM and ICP groups of MSN. After 3-h reaction, the suspension was centrifuged to remove the supernatant. The pellet was then washed extensively with H2O to remove residual free Dox and G2 PAMAM by re-dispersing and centrifuging a few times. The encapsulation efficiency (weight ratio of Dox loaded into MSNs to MSNs, w/w %) was determined by UV-Vis spectroscopy to be approximately 40%. The successful loading of Dox into the MSN pores and modification of G2 PAMAM on the MSN surfaces were further confirmed by 13C solid state NMR (supporting information, Figure S-3).
The Dox-loaded and G2 PAMAM-modified MSNs (MSN-Dox-G2) were analyzed by fluorescence spectroscopy and the results showed that the fluorescence of Dox was completely quenched after encapsulated into the MSNs (supporting information, Figure S-4). The same phenomenon was observed when Dox was encapsulated in liposomes and micelles, which was attributed to a self-quenching effect of Dox.[3, 31, 32] This feature is extremely useful as it enables us to use fluorescence to directly monitor the Dox release from the MSN both outside and inside cells. We evaluated the release of Dox from MSN-Dox-G2 in PBS buffer at room temperature and found that only ~2.6% Dox was released up to 24 h. This is remarkable since this property of the loaded Dox in MSN can ensure our pursued intracellular triggered release to minimize side effects of Dox to normal tissues and cells. However, almost 100% of Dox was released in 4.9 mM glutathione solution (supporting information, Figure S-5). Considering the relatively high concentration of glutathione in drug resistant cancer cells,[33-35] we expect that 100% of Dox could be released upon internalization into cells. The detailed releasing mechanism study will be reported in a separate work.
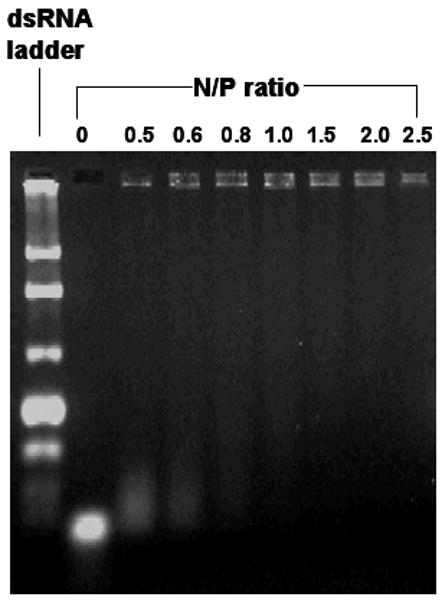
It is widely accepted that the main prerequisite for delivery of siRNA into cytoplasm where it can guide sequence specific mRNA degradation is their packaging into nanometer-sized complexes by their delivery vehicle, which is the Dox-loaded and G2 PAMAM-modified MSNs in our case. The complex formation is driven mainly by electrostatic interaction between negatively charged siRNA and positively charged dendrimers on the MSNs. Such binding led to the formation of positively charged complexes that can be retarded in gel electrophoresis when compared with free siRNA which can not. Therefore the sufficient amount of MSN-Dox-G2 needed for the complex formation can be determined from an agarose gel electrophoresis experiment. As shown in Figure 2, when the N/P (nitrogen/phosphate) ratio, which refers to the ratio of the positively charged primary amine groups of the G2 dendrimer on the MSNs to negatively charged phosphate groups from siRNAs, was around 1, the siRNA was completely retained in the sample wells with no electrophoretic shift corresponding to free siRNA. This indicated that all siRNAs formed stable complex with MSN-Dox-G2 at N/P=1. The morphology of MSN-Dox-G2 complex with siRNA was further visualized by TEM, showing that a ~20-nm thick layer of siRNAs was formed surrounding the surface of MSN-Dox-G2 (Figure 1c).
To investigate the cellular internalization and the intracellular release of Dox and siRNA, MSN-Dox-G2 was complexed with siGLO green siRNA transfection indicator (FAM-labeled) and then added to A2780/AD human ovarian cancer cells and incubated for 6 h at 37 °C. The cells were then washed with PBS buffer and fresh medium was added for fluorescence imaging. Red and green fluorescence imaging was performed to visualize the released Dox and siRNAs respectively. Since the fluorescence of Dox encapsulated inside the MSN pores was completely quenched, the presence of red fluorescence is a hallmark of the Dox released from the MSN pores. As shown in Figure 3c, the cells displayed strong red fluorescence both in perinuclear regions of cytoplasm and in nucleus, indicating efficient release of Dox after being delivered inside the cells. Strong green fluorescence was also shown in similar perinuclear regions of cytoplasm indicating efficient co-delivery of siRNA into cytoplasm. However, no significant green fluorescence signals were detected in nucleus, suggesting the absence of siRNAs inside nucleus (Figure 3b). The absence of siRNAs in nucleus is an advantage for RNA interference (RNAi) as RNAi occurs in cytoplasm and delivery of siRNA into nucleus may lower the siRNA activity.[36] Furthermore, for the case of Dox, it was noted that the red fluorescence in perinuclear regions was much stronger than that in nucleus, which was in contrast with that observed when incubating cells with free Dox, where Dox was primarily localized in nucleus as shown in Figure 3d, similar to previous reports.[31, 37] These observations are important for several reasons. First, the different distribution of Dox in the cells from the free Dox suggested the MSN delivery system was stable in the presence of cells and serum-containing cell medium, achieving minimal premature release of Dox, as the rapid release of Dox in the extracellular environment would be expected to result in an image similar to that observed for free Dox. In addition, the primary localization of both Dox and siRNA in perinuclear regions of cytoplasm with very weak signals in the regions close to cell membrane suggested our delivery system likely internalized through endocytosis and could bypass the efflux pump resistance as reported for several other delivery systems.[1, 38, 39] This is because P-glycoprotein (Pgp) and multidrug resistance-associated protein (MRP), which are the two main ATP binding transporter proteins responsible to reduce cellular drug accumulation, are mainly located in the plasma membrane. These proteins recognize and efflux drugs out of the cell only when the drug is internalized by passive diffusion, present in or close to the plasma membrane, and not when the drug is delivered directly into perinuclear region of cells, through endocytosis.[1, 2, 39-42]
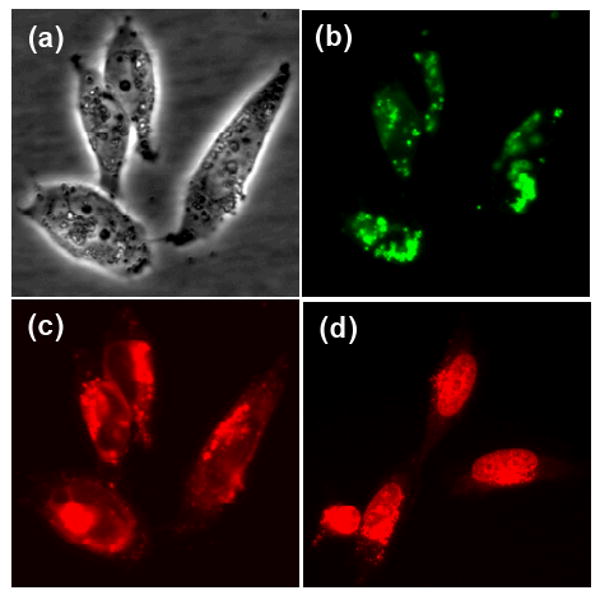
Fluorescence microscopy images of A2780/AD human ovarian cancer cells (a) light image. (b) green fluorescence (siRNA) and (c) red fluorescence (Dox) images, following incubation with MSN-Dox-G2 complex with siGLO green siRNA transfection indicator (FAM-labeled) for 6 h at 37 °C or (d) red fluorescence image following incubation with free Dox for 5 h at 37 °C. The red fluorescence in panel (c) indicated the released Dox from MSN pores.
We next studied the ability of the MSN-Dox-G2/siRNA complexes to silence the targeted mRNA expression with quantitative reverse transcriptase-polymerase chain reaction (RT-PCR). The Bcl-2-targeted siRNA was used for this study. Gene expression was calculated as the ratio of mean band density of analyzed RT-PCR product to that of the internal standard (β2-m) and then the ratio of each sample was normalized to that of sample without treatment. As shown in Figure 4, after the cells was incubated with MSN-Dox-G2/siRNA complexes at 37 °C for 24 h, the Bcl-2 mRNA level was effectively suppressed to ~20%, while cells incubated with MSN-Dox-G2 without Bcl-2 siRNA showed similar Bcl-2 mRNA level as the control cells without treatment. This data indicated that our co-delivery system can not only efficiently deliver the siRNA simultaneously with Dox into cytoplasm, thus-delivered siRNA can also be efficiently released and effectively silence the targeted mRNAs.
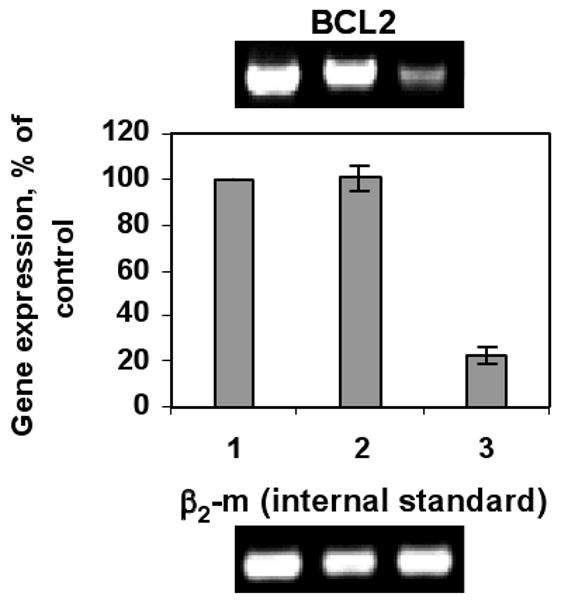
Effect of MSN-Dox-G2 complex with siRNA on silencing of Bcl-2 mRNA in A2780/AD human ovarian cancer cells. Lane 1. No treatment. Lane 2. MSN-Dox-G2. Lane 3. MSN-Dox-G2 with Bcl-2 siRNA. Gene expression was calculated as a ratio of the band intensity of Bcl-2 gene to that of the internal standard, β2-m and then the ratio of each sample was normalized to that of sample without treatment.
To further examine whether the efficient knockdown of Bcl-2 mRNA can effectively increase the chemotherapy efficacy of Dox, the cellular cytotoxicity of free Dox and MSN-Dox-G2 with or without Bcl-2 siRNA was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. The A2780/AD cells in the cell growth medium were separately incubated in 96-well plates with different concentrations of free Dox, MSN-Dox-G2, and MSN-Dox-G2/Bcl-2 siRNA, respectively, for 24 h at 37 °C. Based on these measurements, the IC50 dose (the dose that kills 50% of cells) of free Dox was determined to be 2.25 μM and that of MSN-Dox-G2 was determined to be 1.07 μM (Figure 5). The MSN-Dox-G2 showed significantly higher cytotoxicity than the free Dox. This increased cytotoxicity was not due to MSN-G2 as the MSN-G2 alone showed minimal toxicity at the concentration used (supporting information, Figure S-6). Combined with the fluorescence microscopy data, the higher cytotoxicity of MSN-Dox-G2 than free Dox can be explained by the possible higher accumulation of Dox inside the cell by bypassing the pump resistance.[1, 39, 41] Moreover, one can speculate that the Dox encapsulated inside the MSN pores can be protected from the drug degradation by intracellular enzymes and environment during delivery and therefore preserves its anticancer activity.[43] While one important mechanism of action of Dox involves DNA intercalation,[44] other mechanisms have also been proposed, including the inhibition of mitochondrial function.[45] Therefore, last but not least, the coexistence of Dox both in the nucleus and cytoplasm of cells may exert synergistic cytotoxicity effects to the cells, thus contributing to the higher toxicity of Dox by this delivery system. More detailed investigation is underway in order to further explore the mechanism of cell uptake and intracellular trafficking, and cytotoxic mechanism of Dox delivered by this system.
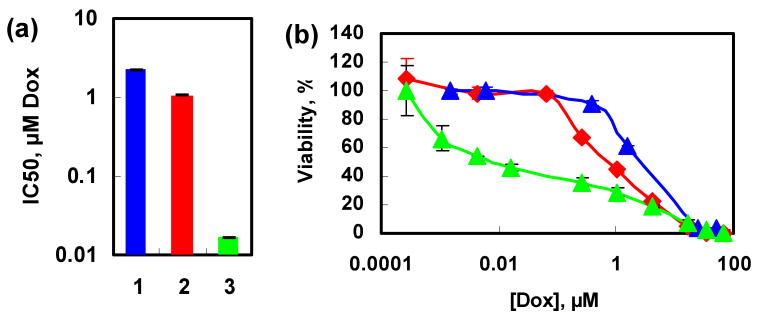
Viability of A2780/AD human ovarian cancer cells incubated for 24 h with the indicated formulations. (a). Cytotoxicity, represented by IC50 of the different formulations that contain Dox; (b). Actual dose-response curves of the different formulations that contain Dox. 1. Free Dox (blue); 2. MSN-Dox-G2 (red); 3. MSN-Dox-G2 and Bcl-2 siRNA(green).
The major goal of the present investigation is to confirm whether the simultaneous delivery of Bcl-2 siRNA and Dox can enhance the cytotoxicity of Dox and thus its efficacy of chemotherapy. As shown in Figure 5, indeed the MSN-Dox-G2/Bcl-2 siRNA complex significantly enhanced the cytotoxicity of Dox by decreasing the IC50 (the concentration of drugs that kills 50% of the cells) 64-fold to 0.017 μM from 1.07 μM for MSN- Dox-G2 alone. This represents a ~132-fold increase of cytotoxicity of Dox when compared to free Dox. This data confirmed that by co-delivering Dox and Bcl-2 siRNA using MSNs, the cytotoxicity of Dox can be significantly enhanced by efficiently knocking down the Bcl-2 mRNA and possibly bypassing the efflux pump resistance due to the different cell uptake pathway than free Dox.
To further confirm the enhanced cytotoxicity was indeed due to increased apoptosis not due to necrosis,[46] the cell apoptosis was analyzed based on the detection of single- and double-stranded DNA breaks (nicks) by an in situ cell death detection kit using terminal deoxynucleotidyl transferase mediated dUTP-fluorescein nick end labeling (TUNEL) method.[47] Briefly, after incubation with MSN-Dox-G2/Bcl-2 siRNA for 24 h, the cells were fixed, permeabilized and incubated with the TUNEL reaction mixture. The label incorporated at the damaged sites of the DNA was visualized by a fluorescence microscope. As shown in Figure 6, control cells without treatment showed almost no green fluorescence, indicating the absence of apoptosis. The cells incubated with MSN-Dox-G2 (0.13 μM Dox) showed some weak fluorescence, indicating the activity of apoptosis in some cells. In contrast, when incubated with MSN-Dox-G2 of the same concentration with an addition of only 0.95 nM of Bcl-2 siRNA, very strong fluorescence was detected in almost every cells, indicating significantly enhanced apoptosis.
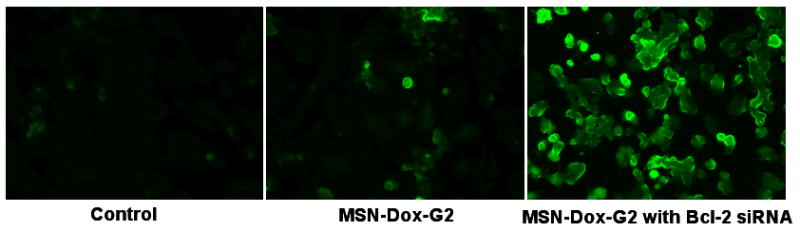
Typical fluorescence microscope images of TUNEL-labeled A2780/AD human ovarian cancer cells. Cells were incubated with medium (no treatment), with MSN-Dox-G2 and with MSN-Dox-G2 containing Bcl-2 siRNA respectively for 24 h.
In summary, we have reported the first effort of utilizing MSNs as a system to simultaneously deliver Dox and Bcl-2-targeted siRNA into multidrug resistant A2780/AD human ovarian cancer cells for enhanced efficacy of chemotherapy. Our results showed that by delivering Dox and Bcl-2 siRNA simultaneously into cancer cells, the Bcl-2 siRNA can effectively silence the Bcl-2 mRNA and significantly suppress the nonpump resistance and substantially enhance the anticancer action of Dox. Our data also suggested that the Dox delivered by MSNs has minimal premature release in the extracellular environment, which can greatly eliminate side effects of Dox. Furthermore, delivered by MSNs Dox was primarily localized in perinuclear region after internalization, possibly bypassing the pump resistance and further enhancing the cytotoxicity. In order to limit adverse side effects and further enhance antitumor efficiency of the complex, this delivery system may be conjugated with targeting moieties specific to tumor cell. Currently, MSNs with different sizes are under investigation on different cancer cell lines to achieve the long-term goal of in vivo targeted simultaneous delivery of chemical and genetic drugs specifically to cancer cells for efficient cancer therapy.
Footnotes
**The research was supported in part by Charles & Johanna Busch Biomedical and CA100098, CA111766 NIH grants. A. M. Chen acknowledges Merck & Co., Inc. for support.
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Dr. Alex M. Chen, Department of Chemistry, Rutgers, The State of New Jersey, Newark, NJ, 07102 (USA), Fax: (+1) 973-353-1264.
Min Zhang, Department of Pharmaceutics, Rutgers, The State of New Jersey, Piscataway, NJ 08854 (USA), Fax: (+1) 732-445-3134.
Dr. Dongguang Wei, Carl Zeiss SMT, Inc., Peabody, MA 01960 (USA)
Dr. Dirk Stueber, Pharmaceutical Research, Merck & Co., Inc., Rahway, NJ 07065.
Dr. Oleh Taratula, Department of Chemistry, Rutgers, The State of New Jersey, Newark, NJ, 07102 (USA), Fax: (+1) 973-353-1264.
Dr. Tamara Minko, Department of Pharmaceutics, Rutgers, The State of New Jersey, Piscataway, NJ 08854 (USA), Fax: (+1) 732-445-3134.
Dr. Huixin He, Department of Chemistry, Rutgers, The State of New Jersey, Newark, NJ, 07102 (USA), Fax: (+1) 973-353-1264.
References
Full text links
Read article at publisher's site: https://doi.org/10.1002/smll.200900621
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2833276?pdf=render
Citations & impact
Impact metrics
Article citations
Neurotoxicity of the antineoplastic drugs: "Doxorubicin" as an example.
J Mol Histol, 55(6):1023-1050, 01 Oct 2024
Cited by: 1 article | PMID: 39352546
Review
Enhanced Efficacy against Drug-Resistant Tumors Enabled by Redox-Responsive Mesoporous-Silica-Nanoparticle-Supported Lipid Bilayers as Targeted Delivery Vehicles.
Int J Mol Sci, 25(10):5553, 20 May 2024
Cited by: 0 articles | PMID: 38791591 | PMCID: PMC11122197
Personalized Versus Precision Nanomedicine for Treatment of Ovarian Cancer.
Small, 20(41):e2307462, 11 Feb 2024
Cited by: 2 articles | PMID: 38342698
Overcoming Chemotherapy Resistance in Metastatic Cancer: A Comprehensive Review.
Biomedicines, 12(1):183, 15 Jan 2024
Cited by: 4 articles | PMID: 38255288 | PMCID: PMC10812960
Review Free full text in Europe PMC
Oncofetal reprogramming in tumor development and progression: novel insights into cancer therapy.
MedComm (2020), 4(6):e427, 02 Dec 2023
Cited by: 4 articles | PMID: 38045829 | PMCID: PMC10693315
Review Free full text in Europe PMC
Go to all (284) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Codelivery of doxorubicin and MDR1-siRNA by mesoporous silica nanoparticles-polymerpolyethylenimine to improve oral squamous carcinoma treatment.
Int J Nanomedicine, 13:187-198, 28 Dec 2017
Cited by: 25 articles | PMID: 29343957 | PMCID: PMC5749394
Redox-responsive mesoporous silica nanoparticles: a physiologically sensitive codelivery vehicle for siRNA and doxorubicin.
Antioxid Redox Signal, 21(5):707-722, 28 Sep 2013
Cited by: 16 articles | PMID: 23931896
Quick synthesis of a novel combinatorial delivery system of siRNA and doxorubicin for a synergistic anticancer effect.
Int J Nanomedicine, 14:3557-3569, 15 May 2019
Cited by: 6 articles | PMID: 31190812 | PMCID: PMC6526930
Codelivery of anticancer drugs and siRNA by mesoporous silica nanoparticles.
Ther Deliv, 7(9):649-655, 01 Sep 2016
Cited by: 16 articles | PMID: 27582236
Review
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: R01 CA111766-04
Grant ID: CA100098
Grant ID: R01 CA100098
Grant ID: R01 CA100098-05A1
Grant ID: CA111766
Grant ID: R01 CA111766
 and
and