Abstract
Free full text

JAK-STAT Signal Inhibition Regulates Competition in the Drosophila Testis Stem Cell Niche
Abstract
Adult stem cells often reside in local microenvironments, or niches. Although niches can contain multiple types of stem cells, the coordinate regulation of stem cell behavior is poorly understood. In the Drosophila testis, Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling is directly required for maintenance of the resident germline and somatic stem cells. We found that the JAK-STAT signaling target and inhibitor Suppressor of cytokine signaling 36E (SOCS36E) is required for germline stem cell maintenance. SOCS36E suppresses JAK-STAT signaling specifically in the somatic stem cells, preventing them from displacing neighboring germline stem cells in a manner that depends on the adhesion protein integrin. Thus, in niches housing multiple stem cell types, negative feedback loops can modulate signaling, thereby preventing one stem cell population from outcompeting the other.
Adult stem cells reside in local microenvironments, or niches, in which signals from surrounding stromal cells inhibit differentiation (1). Niches are often structurally and molecularly complex, with diverse signaling pathways operating in a given niche (2). In niches containing multiple types of stem cells, it is unclear how behavior is coordinated to produce an appropriate ratio of differentiated cell types (3). Among the best-characterized niches are those in the Drosophila gonads, in which germline and somatic stem cells cooperate during gametogenesis (3, 4). In the ovary, germline stem cells (GSCs) and somatic stem cells (called escort stem cells) share a niche. However, separate signals appear to regulate escort stem cell and GSC maintenance (5). In the testis, germline and somatic stem cells (cyst progenitor cells, or CPCs) reside in a single niche created by a small cluster of stromal cells (the hub) (Fig. S1A), and both require Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling for their maintenance (6-8). Thus, the Drosophila testis provides a tractable paradigm for understanding the coordinate regulation of multiple stem cell types within a single niche.
Because JAK-STAT signaling is directly required for GSC and CPC maintenance in the Drosophila testis (Table S1, Fig. S2) (6-8), studying STAT targets should reveal regulatory mechanisms in one or both stem cell lineages. Transcriptional profiling identified putative STAT targets in this niche, including the JAK-STAT inhibitor Suppressor of cytokine signaling 36E (SOCS36E) (9). socs36E is expressed specifically at high levels in the hub and at lower levels in CPCs (Fig. S1B) (9), making it an excellent candidate regulator within the testis niche.
SOCS proteins, first identified in vertebrates, are highly conserved antagonists of JAK-STAT signaling that act in a classic negative feedback loop: in response to signal activation, SOCS proteins bind and inhibit JAK kinases or their associated receptors so as to down-regulate the pathway intracellularly (10-13). By analyzing socs36E expression in testes with altered JAK-STAT signaling, we found that socs36E behaves as an induced antagonist within the Drosophila testis niche (fig. S1, C and D) (14).
To determine the role of socs36E in the niche, we identified a P-element insertion in the coding region of the socs36E gene. This insertion mutation, designated socs36EPZ1647, creates a strong loss-of-function allele that interacts with JAK-STAT pathway components as a pathway antagonist (fig. S3) (14). socs36EPZ1647 flies are viable and fertile. Analysis by confocal microscopy, however, showed that the stem cell niche in socs36EPZ1647 testes from newly eclosed males is abnormal: It contains only three to four GSCs as compared with the wild-type average of 10 (Fig. 1A-C). This phenotype is due to disruption of socs36E by the P-element because introducing a wild-type socs36E cDNA transgene into the socs36EPZ1647 mutant background rescues the phenotype, and precise excision of the P-element restores the number of GSCs to wild-type. Moreover, the socs36EPZ1647 phenotype is due to loss, not gain, of socs36E function: socs36E heterozygotes have wild-type numbers of GSCs, whereas testes containing socs36EPZ1647 in trans to a deletion of the socs36E locus phenocopy socs36EPZ1647. Thus, socs36E is required for GSC maintenance in the testis.
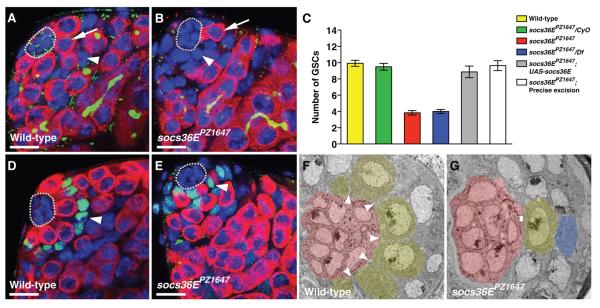
(A to E) Confocal section through the testis apex immunostained with [(A) and (B)] antibody to Vasa (germline, red), antibody to Fasciclin III (hub, green), antibody to Hts (1B1 fusome, green), and 4′,6′-diamidino-2-phenylindole (DAPI) (DNA, blue) or with [(D) and (E)] antibody to Vasa (red), antibody to ZFH-1 (CPCs and their immediate daughters, green), and DAPI (blue). The hub is outlined. (A) In wild-type testes, a rosette of GSCs (one indicated, arrow) surrounds the hub; Vasa-negative CPCs (one indicated, arrowhead) make minimal contact with the hub. (B) socs36EPZ1647 testes have fewer GSCs (arrow); instead, somatic cells (arrowhead) surround the hub. (C) Bar graph showing number of GSCs per testis. Homozygous mutant socs36EPZ1647 testes contain significantly fewer GSCs than do wild-type (red, 3.8 ± 0.2, n = 12 testes, versus yellow, 9.9 ± 0.4, n = 14 testes, P < 0.0001) or socs36E heterozygotes (green, 9.5 ± 0.4, n = 14 testes). Testes from socs36EPZ1647 in trans to a deletion of socs36E (Df(2L)M36F-S5) phenocopy socs36EPZ1647 (blue, 4.0 ± 0.2, n = 12 testes). Leaky expression from a socs36E cDNA transgene rescues the phenotype (gray, 8.9 ± 0.7 GSCs, n = 15 testes). Precise excision of the P-element restores GSC number to wild-type (white, 9.6 ± 0.6, n = 11 testes). (D) In wild-type testes, GSCs surround the hub; nuclei of ZFH-1–positive CPCs (arrowhead) are displaced from the hub. (E) In socs36EPZ1647, CPCs (arrowhead) occupy positions at the hub that usually contain GSCs. (F and G) Ultrastructural analysis of the testis niche. Hub cells (red), GSCs (yellow), and a gonialblast (blue), identified by differences in morphology and staining density (29), are indicated. (F) In wild-type testes, GSCs make broad contact with the hub; CPCs extend thin projections (arrowheads) between GSCs. (G) In socs36EPZ1647, CPCs broadly contact the hub, whereas GSCs make small areas of contact with the hub (rectangle). Scale bars, 10 μm.
Although the pool of GSCs is depleted in testes lacking socs36E, the positions typically occupied by these cells are filled with somatic cells (Fig. 1B). To identify these somatic cells we examined the expression of the zinc finger homeodomain-1 (ZFH-1) protein, which marks CPCs and their immediate cyst cell daughters (8). ZFH-1-positive nuclei are farther from the hub than GSC nuclei in wild-type testes but are often adjacent to the hub in socs36EPZ1647 testes (Fig. 1D-E). Ultrastructural analysis confirmed that CPCs in wild-type testes display a small region of contact with the hub whereas GSCs maintain large regions of contact (15), but in socs36E testes the situation is reversed: CPCs form aberrantly broad contacts with the hub (Fig. 1F-G). We considered that the decreased pool of GSCs in socs36EPZ1647 testes could be accompanied by a corresponding increase in the number of CPCs. However, the number of ZFH-1-positive cells was the same in socs36EPZ1647 and wild-type testes (34.8 ± 1.6, n = 17 versus 35.7 ± 1.6, n = 17, P > 0.05) and these cells displayed similar rates of proliferation (measured by mitotic ZFH-1–positive cells per testis 0.4 ± 0.1, n = 22 versus 0.5 ± 0.1, n = 50 testes, P > 0.05). Thus, the GSC loss and concomitant expansion of CPC/hub cell contacts in socs36EPZ1647 testes is not due to CPC overproliferation.
Because both GSCs and CPCs respond directly to JAK-STAT signaling, socs36E could be directly or indirectly required to prevent GSC loss. To distinguish between these possibilities we created socs36E loss-of-function clones (14) and analyzed the effects on each lineage. This analysis revealed that GSCs do not directly require socs36E for their maintenance (Fig. 2A-B, Table S2). In contrast, we uncovered a role for socs36E in CPCs. Testes initially containing only a few mutant CPCs gain significantly more mutant CPCs over time than those of controls, indicating that socs36E prevents CPCs from accumulating within the niche (Fig. 2C-D, Table S2). Over the timecourse of this experiment, the total number of CPCs remains constant, but the number of mutant CPCS increases, indicating that CPCs lacking socs36E outcompete wild-type CPCs (Table S2).
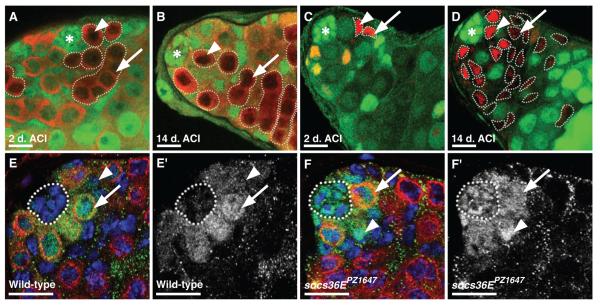
(A to F) Confocal sections through the testis apex. [(A) to (D)] Hubs denoted by asterisks. [(A) and (B)] socs36EPZ1647 GSC clones (outlined, arrowhead) and their differentiating progeny (outlined, arrow) identified by the absence of green fluorescent protein (GFP) (green) and the presence of Vasa (red) at (A) 2 days and (B) 14 days after clonal induction (ACI). GSCs at the hub and their progeny are maintained. [(C) and (D)] socs36EPZ1647 CPC clones (outlined, arrowhead) and their differentiating daughters (outlined, arrow) identified by the absence of GFP (green) and the presence of ZFH-1 (red) at (C) 2 days and (D) 14 days ACI. CPCs lacking socs36E accumulate around the hub, competing for niche occupancy and leading to a loss of GSCs over time. [(E) and (F)] Testes immunostained with antibody to Stat92E (green), antibody to Vasa (red), and DAPI (blue); GSCs (arrows), CPCs (arrowheads), and hubs (outlined) are indicated. [(E′) and (F′)] Stat92E channel. [(E) and (E′)] In wild-type testes, Stat92E is detectable at higher levels in GSCs than in CPCs or the hub. [(F) and (F′)] In socs36EPZ1647 testes, Stat92E levels in hub cells and CPCs are increased relative to wild-type controls. Scale bars, 10 μm.
The advantage for niche occupancy that mutant CPCs display over their wild-type counterparts even extends to GSCs: As the number of mutant CPCs increases, there is a corresponding decline in the number of GSCs (Table S2). Even small clones of CPCs lacking socs36E recapitulate the socs36EPZ1647 phenotype. Thus, our clonal analysis indicates that socs36E is not needed in GSCs to prevent them from ‘losing their grip’ and leaving the niche. Instead, socs36E acts directly in CPCs to prevent them from displacing GSCs.
These data suggest that inhibition of JAK-STAT signaling via socs36E within CPCs maintains the proper ratio of germline and somatic stem cells within the niche. We assessed the distribution of Stat92E protein in testes (16), which reflects JAK-STAT signaling activity and thus likely reflects the limits of the niche (17-19). In wild-type testes, Stat92E is highly enriched in GSCs but rapidly declines in their differentiating daughters. CPCs and hub cells contain relatively little Stat92E, suggesting that GSCs normally have higher levels of pathway activation than CPCs (Fig. 2E) (20). In contrast, hub cells and CPCs contain elevated levels of Stat92E in socs36EPZ1647 testes (Fig. 2F). We conclude that socs36E normally attenuates JAK-STAT signaling in hub cells and CPCs, thereby maintaining the appropriate balance of the two stem cell lineages in this niche.
We considered that socs36E could regulate competition between GSCs and CPCs by regulating adhesion and/or signaling within the niche. Although cadherin-mediated adhesion sustains stem cell maintenance in this and other niches (20, 21), we found no discernible differences in the expression patterns of DE- or N-cadherin between wild-type and socs36E mutant testes (Fig. S4). In contrast, socs36EPZ1647 testes have elevated levels of the common beta subunit of position-specific integrins (βPS-integrin), particularly at the CPC-hub cell interface (Fig. 3A-B), suggesting that integrin-mediated adhesion plays a role in maintaining the correct GSC-to-CPC ratio. To provide genetic evidence for this hypothesis, we asked if reducing integrin function reverses the GSC loss phenotype in socs36EPZ1647 testes. Removing one copy of rhea, which encodes Drosophila talin, an integrin-binding cytoskeletal linker essential for integrin-mediated adhesion (14, 22), did not cause GSC loss. However, this level of reduction in integrin function was sufficient to rescue the socs36EPZ1647 phenotype (Fig. 3C), indicating that SOCS36E normally prevents CPCs from outcompeting GSCs from the niche via an integrin-mediated mechanism.
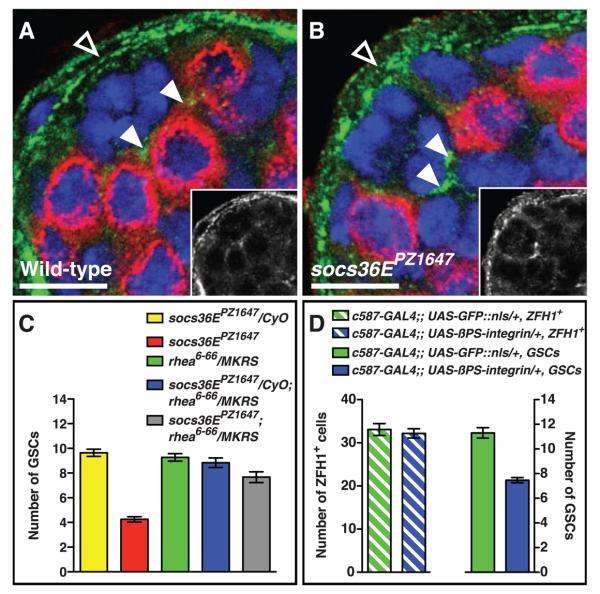
(A and B) Confocal section through the testis apex immunostained with antibody to Vasa (red), antibody to βPS-integrin (green, and in insets), and DAPI (blue). (A) In wild-type testes, βPS-integrin is expressed strongly in the sheath (open arrowhead) and at moderate levels throughout the apex; within the niche, it is enriched at points of CPC-hub cell contact (arrowheads). (B) In socs36EPZ1647 testes, expression of βPS-integrin at the CPC-hub interface (arrowheads) is increased relative to wild-type controls. (C) socs36EPZ1647 GSC loss is partially rescued by introducing one copy of a talin loss-of-function allele (rhea6-66) (red, 4.2 ± 0.2, n = 24 testes, versus gray, 7.7 ± 0.4, n = 18 testes, P < 0.0001). Testes from control flies (socs36EPZ1647 or rhea6-66 heterozygotes and socs36EPZ1647; rhea6-66 transheterozygotes) contain wild-type numbers of GSCs (yellow, 9.6 ± 0.3, n = 14 testes; green, 9.3 ± 0.2, n = 22 testes; and blue, 8.8 ± 0.3, n = 24 testes, respectively). CyO and MKRS denote balancer chromosomes containing wild-type alleles of socs36E and rhea. (D) Expression of ectopic βPS-integrin in CPCs and cyst cells significantly reduces the number of GSCs per testis as compared with UAS-GFP controls. Scale bars, 10 μm.
These data suggest that CPCs with aberrantly high levels of JAK-STAT signaling have increased integrin-mediated adhesion to the hub and therefore preferentially gain niche occupancy. Thus, increasing the level of βPS-integrin in CPCs may enable them to outcompete neighboring wild-type GSCs. Consistent with this hypothesis, overexpression of βPS-integrin in CPCs and cyst cells reduced the number of GSCs per testis significantly (11.3 ± 0.4, n = 10 testes, versus 7.5 ± 0.2, n = 30 testes, P < 0.0001) (Fig. 3D). Overexpression of βPS-integrin in CPCs and cyst cells did not significantly alter their numbers (Fig. 3D) or rate of proliferation (measured by mitotic ZFH-1–positive cells per testis 0.4 ± 0.1, n = 24 testes, versus 0.7 ± 0.2, n = 33 testes, P > 0.05). Thus, as in socs36EPZ1647 testes, CPCs with ectopic βPS-integrin gain an advantage for niche occupancy that is not driven by overproliferation. Together these data indicate that increased expression of integrin in CPCs leads to their enhanced adhesion to the hub, enabling them to outcompete GSCs.
We propose that socs36E acts in a negative feedback loop to inhibit JAK-STAT signaling in CPCs, thereby ensuring that the proper balance of germline and somatic stem cells is maintained in the niche. Although JAK-STAT is directly activated in both stem cell lineages, socs36E expression in CPCs prevents them from displacing GSCs, most likely by limiting integrin-mediated adhesion of CPCs to the hub. Although our work highlights the importance of adhesion in competition between these two distinct stem cell lineages, it is also possible that changes in cell signaling occur when socs36E is disrupted. For example, because the epidermal growth factor receptor (EGFR) pathway acts within the CPC lineage to promote GSC differentiation (23, 24), and socs36E can inhibit both EGFR and JAK-STAT signaling (11), socs36E might interact with additional signaling pathways within the niche.
Our work exemplifies how one signaling pathway coordinately maintains two types of stem cells in a single niche. Furthermore, it underscores the importance of feedback inhibition at the cellular and molecular levels within stem cell niches. Both these features may emerge as common themes of mammalian niches as they become better characterized. Finally, both aberrant JAK-STAT signaling (including the loss of SOCS gene expression) and disrupted integrin function can lead to tumorigenesis in mammals (25-27). Since mis-regulation of stem cells is implicated in many types of cancer (28), it will be important to determine if altered JAK-STAT signaling and integrin-mediated adhesion enhance the ability of additional types of cells to outcompete normal stem cells to establish occupancy of stem cell niches.
Acknowledgments
We thank the Bloomington Stock Center, Developmental Studies Hybridoma Bank, and members of the fly community for reagents; M. Sepanski for Electron Microscopy; M. Gold for inverse PCR; B. Ohlstein, M. Wawersik, and M. Matunis for helpful comments; and J. Leatherman and S. DiNardo for sharing unpublished data. Supported by NIH grant RO1HD40307 (E.M.) and Basil O'Connor/March of Dimes grant 5-FY02-285 (M.C.).
References and notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1176817
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3073347?pdf=render
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/content/full/326/5949/153
Free to read at www.sciencemag.org
http://www.sciencemag.org/cgi/content/abstract/326/5949/153
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/reprint/326/5949/153.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Mechanisms of Germline Stem Cell Competition across Species.
Life (Basel), 14(10):1251, 01 Oct 2024
Cited by: 0 articles | PMID: 39459551 | PMCID: PMC11509876
Review Free full text in Europe PMC
A regulatory loop of JAK/STAT signalling and its downstream targets represses cell fate conversion and maintains male germline stem cell niche homeostasis.
Cell Prolif, 57(10):e13648, 10 Jul 2024
Cited by: 1 article | PMID: 38987866 | PMCID: PMC11471429
The Drosophila histone methyltransferase SET1 coordinates multiple signaling pathways in regulating male germline stem cell maintenance and differentiation.
Development, 151(15):dev202729, 09 Aug 2024
Cited by: 0 articles | PMID: 39007366 | PMCID: PMC11369688
Integrin restriction by miR-34 protects germline progenitors from cell death during aging.
Aging Cell, 23(6):e14131, 07 Mar 2024
Cited by: 1 article | PMID: 38450871
Computational simulation of JAK/STAT signaling in somatic versus germline stem cells.
Dev Dyn, 253(7):648-658, 21 Dec 2023
Cited by: 0 articles | PMID: 38126664
Go to all (115) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling.
J Cell Physiol, 223(2):500-510, 01 May 2010
Cited by: 46 articles | PMID: 20143337 | PMCID: PMC2894562
Histone demethylase dUTX antagonizes JAK-STAT signaling to maintain proper gene expression and architecture of the Drosophila testis niche.
Development, 140(5):1014-1023, 30 Jan 2013
Cited by: 19 articles | PMID: 23364332 | PMCID: PMC3583039
A regulatory loop of JAK/STAT signalling and its downstream targets represses cell fate conversion and maintains male germline stem cell niche homeostasis.
Cell Prolif, 57(10):e13648, 10 Jul 2024
Cited by: 1 article | PMID: 38987866 | PMCID: PMC11471429
Local signaling within stem cell niches: insights from Drosophila.
Curr Opin Cell Biol, 24(2):225-231, 30 Jan 2012
Cited by: 16 articles | PMID: 22296770 | PMCID: PMC4821423
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NICHD NIH HHS (5)
Grant ID: R01 HD052937-01A1
Grant ID: R01HD40307
Grant ID: R01 HD040307
Grant ID: R01 HD052937
Grant ID: R01 HD040307-05A1





