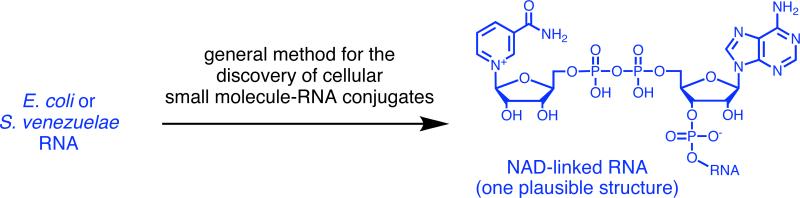
Abstract
Free full text

LC/MS analysis of cellular RNA reveals NAD-linked RNA
Abstract
We developed a general method to detect cellular small molecule-RNA conjugates that does not rely on the reactivity of the small molecule, revealing NAD-linked RNA in E. coli and S. venezuelae. Subsequent characterization shows NAD is a 5’ modification of RNA, cannot be installed in vitro through aberrant transcriptional initiation, is only found among smaller cellular RNAs, and is present at a surprisingly high abundance of ~3000 copies per cell.
The known chemical diversity of natural RNA has remained limited primarily to canonical polyribonucleotides, 3’-aminoacylated tRNAs, and modified nucleobases in tRNA and rRNA1 in contrast with a growing number of elucidated biological roles for RNA that include catalysis, gene regulation, and defense against viral infection2,3. Researchers have previously speculated that early biotic systems carried out biochemical reactions using small molecule-RNA conjugates as participants in RNA-templated chemistries4-6. Recent studies of non-natural DNA-templated chemistries7,8 also highlight the unusual functional capabilities of small molecule-nucleic acid conjugates. These observations collectively led us to speculate that small molecule-RNA conjugates beyond those previously described may exist in modern cells9.
In the six decades since their initial discovery10, researchers have identified over 100 modified nucleosides, almost all in RNA11,12. Virtually all past efforts to characterize modified nucleotides, however, have examined modifications in a targeted manner by studying a specific cellular RNA or class of RNAs13,14. We recently described the development of a broad approach to the discovery of biological small molecule-RNA conjugates that uses size-exclusion chromatography and mass spectrometry to detect base- or nucleophile-labile small molecules that are cleaved from any cellular RNA9. While fruitful, we sought to develop a more general method to detect small molecule-RNA conjugates that can be applied to any such conjugate regardless of its chemical reactivity.
Here we report the development and application of a method that in principle enables the detection of any small molecule-RNA conjugate independent of its chemical structure (Fig. 1a and Supplementary Results). Whole cellular RNA is subjected to size-exclusion chromatography and the macromolecular fraction (> ~2,500 Da) is divided into two halves (Supplementary Methods). One half is treated with nuclease P115, while the second half is treated with heat-inactivated nuclease P1 under otherwise identical conditions. Both samples are subjected to size-exclusion chromatography again and the small-molecule fraction from each is subjected to comparative high-resolution LC/MS9. Non-canonical species more abundant in the active nuclease-treated sample compared with the inactive nuclease-treated are considered possible novel small molecule-RNA conjugates (Supplementary Fig. 1). After validating this method by observing 16 of the 20 major 3’-aminoacyl adenosine monophosphates as enriched ≥ 2-fold in samples treated with active nuclease compared with heat-inactivated nuclease (Supplementary Figs. 2 and 3), we applied the general approach to small molecule-RNA discovery to E. coli and S. venezuelae RNA, revealing 24 and 28 unknown species, respectively, that were enriched ≥ 2-fold (Supplementary Fig. 4).

Discovery of a small molecule-linked nucleotide of [M-H]- m/z = 540.0533 from E. coli and S. venezuelae RNA. (a) The general method for biological small molecule-RNA conjugate discovery developed in this work. Paired samples for comparative LC/MS analysis are generated by treatment of cellular RNA with active nuclease P1 versus treatment with heat-inactivated nuclease P1 under otherwise identical conditions. (b) The extracted ion chromatograms (EICs) for [M-H]- m/z = 540.0533 from E. coli RNA exposed to active nuclease P1 (cleavage conditions) or to heat-inactivated nuclease P1 (control conditions). (c) Same as (b), but using S. venezuelae RNA. (d) Isotope-labeled S. venezuelae total RNA reveals that the [M-H]- m/z = 540.0533 ion contains five nitrogen atoms and 15 carbon atoms. (e) The EICs for [M-H]- m/z = 662.1018 from E. coli RNA or (f) from S. venezuelae RNA digested with nuclease P1 or control conditions (heat-inactivated nuclease incubation). (g) One possible structure of NAD-linked cellular RNA. We note that our data is also consistent with RNA attachment to the 2’ hydroxyl of NAD, or to either of the nicotinamide-linked ribose hydroxyl groups.
One unknown species from both E. coli and S. venezuelae that was enriched 8-fold in the nuclease versus heat-inactivated nuclease samples was [M-H]- m/z = 540.0533 (Figs. 1b and 1c). Culturing S. venezuelae in media containing 13C-glucose (1) as the sole carbon source, or containing 15N-ammonium sulfate as the sole nitrogen source, resulted in mass increases of this species of +15 Da and +5 Da, respectively (Fig. 1d). These results enabled us to deduce a molecular formula of C15H20N5O13P2 (expected [M-H]- m/z = 540.0538). The MS/MS spectrum of this ion revealed ADP (expected [M-H]- m/z = 426.0221) as a major fragment (Supplementary Fig. 5). We therefore reasoned that the 540.0533 Da species likely consists of a 115.0395 Da group (C5H7O3) attached to ADP.
An elimination reaction on an adenosine-containing cofactor could generate an unsaturated ribose group. We therefore speculated that the [M-H]- m/z = 540.0533 ion is a breakdown product of a larger small molecule-RNA conjugate that undergoes elimination during MS ionization. Indeed, when MS analysis was performed using milder ionization conditions, a [M-H]- m/z = 662.1032 species appeared at the same retention time as the [M-H]- m/z = 540.0533 species (Figs. 1e and 1f). Collectively, these observations led us to propose that the [M-H]- m/z = 540.0533 species is a fragment of nicotinamide adenine dinucleotide (NAD) (2) (expected [M-H]- m/z = 662.1018). This hypothesis was confirmed by LC/MS and MS/MS comparisons of the cellular species with authentic NAD (Supplementary Fig. 5 and Supplementary Fig. 6). The analysis of whole E. coli RNA using the milder ionization conditions revealed that [M-H]- m/z = 662.1032 was also significantly enriched (8-fold) in the nuclease versus heat-inactivated nuclease samples, consistent with these conclusions.
Since NAD is prevalent as a cellular metabolite and cofactor, we sought to confirm that the detected NAD species was a bona fide small molecule-RNA conjugate such as the structure shown in Fig. 1g rather than an intracellular contaminant that was unexpectedly carried through RNA purification and size exclusion. We spiked varying quantities of authentic NAD or NADH into E. coli and S. venezuelae cell lysates, and repeated the RNA isolation, nuclease P1 digestion, and LC/MS analysis. The addition of up to 10,000-fold more NAD and NADH than was observed in the unspiked samples did not significantly change the abundance of the corresponding NAD species (Fig. 2a). These results demonstrate that the NAD species observed in our experiments on E. coli and S. venezuelae RNA cannot be accounted for by endogenous NAD (or by the oxidation of endogenous NADH), and further support the conclusion that these species arise from cellular small molecule-RNA conjugates.
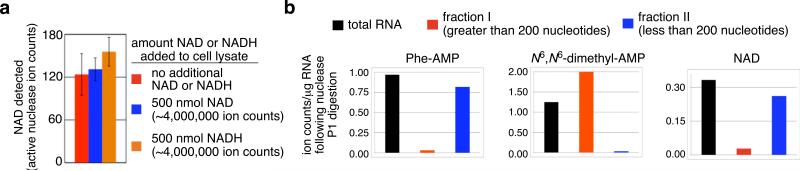
Characterization of NAD-RNA. (a) Spiking large quantities of NAD or NADH into E. coli cell lysate before RNA isolation and treatment with active nuclease P1 does not change the observed ion counts of these species. Error bars represent the standard deviation of three independent trials. (b) Total E. coli RNA was separated into RNAs of length greater than 200 nucleotides (fraction 1) and RNAs of length less than 200 nucleotides (fraction II) using a silica column (Qiagen RNeasy). Each fraction was subjected to nuclease P1 digestion and analyzed by LC/MS. The presence of NAD in fraction II, similar to that of Phe-AMP, suggests that the NAD-linked RNA(s) are primarily less than 200 nucleotides in length.
The structure of NAD led us to hypothesize that NAD is a 5’ RNA modification. Because nuclease P1 catalyzes the attack of a water molecule on RNA to generate 5’-phosphonucleotides15, all nuclease P1 digestion products other than the nucleotides at the 5’ termini undergo a mass shift when the digestion is performed in the presence of isotopically labeled water, compared with digestion in unlabeled water. We carried out nuclease P1 digestion of total RNA from both E. coli and S. venezuelae in 18O-enriched water. As expected, Phe-AMP (3) exhibited a +2 Da mass shift when the digestion was carried out in 18O water compared to in 16O water (Supplementary Fig. 7a). In contrast, there was no mass shift for NAD in both isotopically-labeled and unlabeled water (Supplementary Fig. 7b). This finding is consistent with a model in which NAD is present at the 5’ termini of RNA (Fig. 1g).
We quantified the amount of NAD-linked RNA in E. coli cells using two methods. First, we generated a standard curve that relates known quantities of authentic NAD to observed ion counts under both the original (harsher) and the milder ionization conditions. Ion counts from E. coli RNA when plotted on the resulting curve result in the estimate of ~3000 copies of NAD-RNA per E. coli cell for both ionization conditions (Supplementary Figs. 8a and b). In the second approach, we added a known quantity of authentic NAD to the nuclease P1-digested cellular RNA sample before LC/MS analysis, and used the resulting increase in the NAD ion count to relate added NAD concentration with observed ion counts. This second method resulted in a similar estimate of ~3300 copies of NAD-RNA per E. coli cell (Supplementary Fig. 8b). This abundance level is comparable to that of Phe-linked tRNA in E. coli16, ~4-fold higher than that of E. coli tmRNA17, and ~300-fold higher than that of short RNAs generated by DNA primase during DNA synthesis18.
Structural similarities between NAD and ATP led us to speculate that NAD might be incorporated into RNA at the 5’-terminus through aberrant transcriptional initiation with NAD or NADH instead of ATP. Indeed, NAD has been incorporated into the 5’-terminus of RNA transcripts in vitro using T7 RNA polymerase19. To test if transcriptional initiation was responsible for incorporating NAD into RNA transcripts, we used E. coli RNA polymerase to carry out in vitro transcription in the presence of high concentrations of NADH using a modified pUC19 plasmid or E. coli genomic DNA as templates. When the RNA was purified, digested with nuclease P1, and analyzed by LC/MS, no NAD or NADH was detected (Supplementary Fig. 9). In contrast, when an authentic 5’-NAD-linked transcript (generated using T7 RNA polymerase19) was spiked into an in vitro transcription reaction and processed in the same way, NAD-linked RNA was readily detected (Supplementary Fig. 9). If one assumes that the inability of E. coli RNA polymerase to incorporate these levels of NAD in vitro parallels an inability to do so in cells, these results suggest that NAD groups are installed following transcriptional initiation.
Prior to nuclease P1 digestion, size-exclusion chromatography retains molecules of molecular weight greater than ~2,500 Da. To narrow the size range of possible NAD-linked RNAs, we used silica-based RNA purification columns (Qiagen RNeasy columns) to further fractionate RNA molecules into two fractions that are less than or greater than ~200 nucleotides (Supplementary Fig. 10). Each of the two fractions was then subjected to nuclease P1 digestion and LC/MS analysis. The rRNA nucleoside modification N6,N6’-dimethyladenine (4) (conjugated to 1.5 kB-2.9 kB rRNAs) was detected in the > 200 base fraction and 3’-aminoacyl adenosine monophosphates conjugated to tRNAs (~76 nucleotides) were present predominantly in the <200 nucleotide flow-through fraction, as expected (Fig. 2b)20. The NAD-linked nucleotides were predominantly detected in the flow-through fraction. This result suggests that the NAD-linked RNA(s) from E. coli and S. venezuelae are not widely distributed in size but instead are predominantly below ~200 nucleotides in length, as was also observed in the case of CoA-linked RNA9. In addition, this finding further supports the hypothesis that the NAD modifications arise through a mechanism other than non-specific transcriptional initiation, which would be expected to generate a broader size distribution of NAD-linked RNAs.
The method developed in this work enables the detection, in principle, of any small molecule-RNA conjugate. The application of this method resulted in the discovery of NAD-RNA in E coli and S. venezuelae, as well as in the detection of a variety of additional unknown, non-canonical small molecule-RNA conjugates. Following the recent discovery of CoA-linked RNA, these results further indicate that the chemical diversity of biological RNA is greater than previously understood.
The biological role, if any, of the NAD-RNA conjugates remains unknown at this point. Perhaps the simplest such possible role is one in which the NAD group serves to localize NAD-linked RNA(s) to NAD-binding proteins. It is also tempting to speculate that NAD-linked RNA (or NADH-linked RNA) might mediate redox reactions. In such a scenario, the RNA component of the NAD-linked RNA might assist in localizing or positioning the NAD for reaction with the appropriate substrates. Indeed, laboratory-evolved ribozymes that use cofactors including NAD and CoA have been previously reported although no natural ribozyme has yet been discovered that requires either NAD or CoA21,22. Alternatively, based on previous examples of RNAs that are either conjugated to small molecules23 or that bind small molecules24, it is also possible that the NAD-linked RNAs might play a role in RNA stability or even in gene regulation. Indeed, the state of the 5’ terminus of RNA can serve as determinant of mRNA half-life25, and it is possible that NAD (or CoA) at the 5’ terminus of a cellular RNA protects an RNA from degradation.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and the NIH/NIGMS (R01GM065865). Y.G.C and W.E.K gratefully acknowledge NSF Graduate Research Fellowships.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nchembio.235
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2842606?pdf=render
Citations & impact
Impact metrics
Article citations
N1-Methylpseudouridine and pseudouridine modifications modulate mRNA decoding during translation.
Nat Commun, 15(1):8119, 16 Sep 2024
Cited by: 0 articles | PMID: 39284850 | PMCID: PMC11405884
Identification and in vitro characterization of UDP-GlcNAc-RNA cap-modifying and decapping enzymes.
Nucleic Acids Res, 52(10):5438-5450, 01 Jun 2024
Cited by: 3 articles | PMID: 38716860
Plasmodium RNA triphosphatase validation as antimalarial target.
Int J Parasitol Drugs Drug Resist, 25:100537, 26 Apr 2024
Cited by: 0 articles | PMID: 38810336
HIV-1 Infection Reduces NAD Capping of Host Cell snRNA and snoRNA.
ACS Chem Biol, 19(6):1243-1249, 15 May 2024
Cited by: 4 articles | PMID: 38747804 | PMCID: PMC11197007
Toll/interleukin-1 receptor (TIR) domain-containing proteins have NAD-RNA decapping activity.
Nat Commun, 15(1):2261, 13 Mar 2024
Cited by: 2 articles | PMID: 38480720
Go to all (111) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA.
Proc Natl Acad Sci U S A, 106(19):7768-7773, 28 Apr 2009
Cited by: 69 articles | PMID: 19416889 | PMCID: PMC2674394
Substrate recognition by a yeast 2'-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog.
Biochemistry, 40(46):14098-14105, 01 Nov 2001
Cited by: 22 articles | PMID: 11705403
LC-MS based quantification of 2'-ribosylated nucleosides Ar(p) and Gr(p) in tRNA.
Chem Commun (Camb), 47(18):5196-5198, 29 Mar 2011
Cited by: 7 articles | PMID: 21448475
Modification of tRNA as a regulatory device.
Mol Microbiol, 8(6):1011-1016, 01 Jun 1993
Cited by: 52 articles | PMID: 7689685
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIGMS NIH HHS (5)
Grant ID: R01 GM065865-06S1
Grant ID: R01GM065865
Grant ID: R01 GM065865-05A1
Grant ID: R01 GM065865
Grant ID: R01 GM065865-06