Abstract
Free full text

Coevolution between a Family of Parasite Virulence Effectors and a Class of LINE-1 Retrotransposons
Abstract
Parasites are able to evolve rapidly and overcome host defense mechanisms, but the molecular basis of this adaptation is poorly understood. Powdery mildew fungi (Erysiphales, Ascomycota) are obligate biotrophic parasites infecting nearly 10,000 plant genera. They obtain their nutrients from host plants through specialized feeding structures known as haustoria. We previously identified the AVR k1 powdery mildew-specific gene family encoding effectors that contribute to the successful establishment of haustoria. Here, we report the extensive proliferation of the AVR k1 gene family throughout the genome of B. graminis, with sequences diverging in formae speciales adapted to infect different hosts. Also, importantly, we have discovered that the effectors have coevolved with a particular family of LINE-1 retrotransposons, named TE1a. The coevolution of these two entities indicates a mutual benefit to the association, which could ultimately contribute to parasite adaptation and success. We propose that the association would benefit 1) the powdery mildew fungus, by providing a mechanism for amplifying and diversifying effectors and 2) the associated retrotransposons, by providing a basis for their maintenance through selection in the fungal genome.
Introduction
There is strong selection pressure on parasites to develop strategies to successfully infect whilst evading host detection and defense mechanisms [1]. Important components of the pathogenicity arsenal of parasites are effectors, usually secreted proteins that influence host metabolism or defense mechanisms to provide an environment for successful infection [2]. Resistance (R) genes are part of the plant defense system, and are widely used in agriculture to control parasites. Most of the known R genes encode nucleotide binding site leucine rich repeat (NBS-LRR) receptors [1]. When an NBS-LRR protein recognizes specific parasite avirulence (AVR) molecules, plant defense responses that prevent further infection are induced in accordance with the gene-for-gene (GFG) model [3]. Some bacterial and oomycete AVR proteins are known to be effectors, but little is known about the function of most fungal AVR molecules [2], [4]. Parasites may evolve to overcome host resistance by altering their AVR genes to avoid R-dependent recognition [1], [5], [6].
GFG resistance has been extensively investigated in the interaction between barley and barley powdery mildew (Blumeria graminis f. sp. hordei, Bgh), an obligate fungal parasite. More than 85 barley R genes, including 28 alleles at the Mla locus, have been described, each conferring resistance to Bgh isolates with matching AVR genes [7]. Mla proteins are nucleotide binding site leucine rich repeat (NBS-LRR) receptors. They share >90% amino acid sequence identity but recognise isolate-specific Bgh AVR gene products [8]. More than 25 independent AVR gene loci have been described in Bgh isolates [9], [10], and genetic crosses have shown that genes for up to eight linked AVR specificities are clustered at a complex set of loci [11], [12]. B. graminis exhibits a high level of host specialization and eight formae speciales (ff. spp.) infecting cereals and forage grasses are known [13], [14]. The genetic basis for such host specialization is as yet unknown, but several genes are likely to be involved [15].
We previously isolated AVR k1 (Q09QS2) and AVR a10 (Q09QS3) genes which, when present in Bgh isolates, induce resistance in barley lines containing Mlk1 and Mla10 genes, respectively [16]. We also provided the first evidence that these fungal AVR genes encode effectors that contribute to the establishment of haustoria, the essential feeding structures of Bgh [16]. The predicted amino acid sequences of AVR k1 and AVR a10 do not contain signal peptides, indicating that they are not secreted from the parasite in the same way as the majority of known fungal and oomycete AVR proteins [17], [18]. When expressed in barley cells, AVRa10 induces an association between Mla10 and a WRKY-2 transcription factor in the nucleus, which may initiate defense gene activation [19]. AVR k1 and AVR a10 belong to a family of closely-related paralogs (hereafter called AVR k1 family or AVR k1 paralogs) which encode proteins with a core domain of conserved amino acids [16].
Some parasite effector genes are found in the proximity of transposable elements (TEs), which have been postulated to provide a mechanism for their expansion and movement within and among genomes [5], [6]. Some transposon insertions into AVR gene loci have resulted in the loss of avirulence (i.e. gain of virulence on hosts with specific resistance genes) of bacterial and fungal parasites [16], [20]–[22]. We previously demonstrated that members of the AVR k1 family lie close to TE1a LINE-1 retrotransposons (RTs), and both sequences can be expressed as a single transcript [12], [16]. Here, we report the extensive proliferation of the AVR k1 gene family throughout the genome of B. graminis, with sequences diverging in ff. spp. adapted to infect different hosts. Furthermore we show that the AVRk 1 family has coevolved with the lineage of TE1a RTs, suggesting a mutual advantage from the association which may ultimately benefit parasite adaptation and success.
Results
The AVR k1 effector gene family is unique to powdery mildew fungi
An initial BLAST [23] of the draft Bgh genome sequence (http://www.blugen.org/), resulted in 1145 homologs to AVR k1 with Expect (E) values ranging from 7e−62 to 1e−5. To investigate the phylogenetic diversity of these paralogs, we created an nrdb90 database (non-redundant set of the predicted open reading frames with 90% sequence identity threshold). Proteins shorter than 100 residues were discarded. This search resulted in 260 sequences which were clearly paralogous to AVRk1 (including 94 paralogs of AVRa10) with Expect (E) values ranging from 1e−152 to 1e−10. Homologous sequences were also found in the genomes of the powdery mildew fungi Erysiphe (Golovinomyces) orontii (six homologs, 1e−3<E<1e−8), which infects Arabidopsis thaliana, and Erysiphe pisi (six homologs, 1e−4<E<1e−17), which infects pea. None of the Erysiphe sequences grouped in the clades containing AVR k1 or AVR a10 (Fig. 1). AVRk1 or AVR a10 homologs were not found in BLAST searches (E value <1e−5) against the EMBL/GenBank [24], COGEME phytopathogen EST database [25], Broad Institute (Fungal Genome Initiative fungi) and Uniprot [26] databases, indicating that this gene family is specific to powdery mildew fungi.
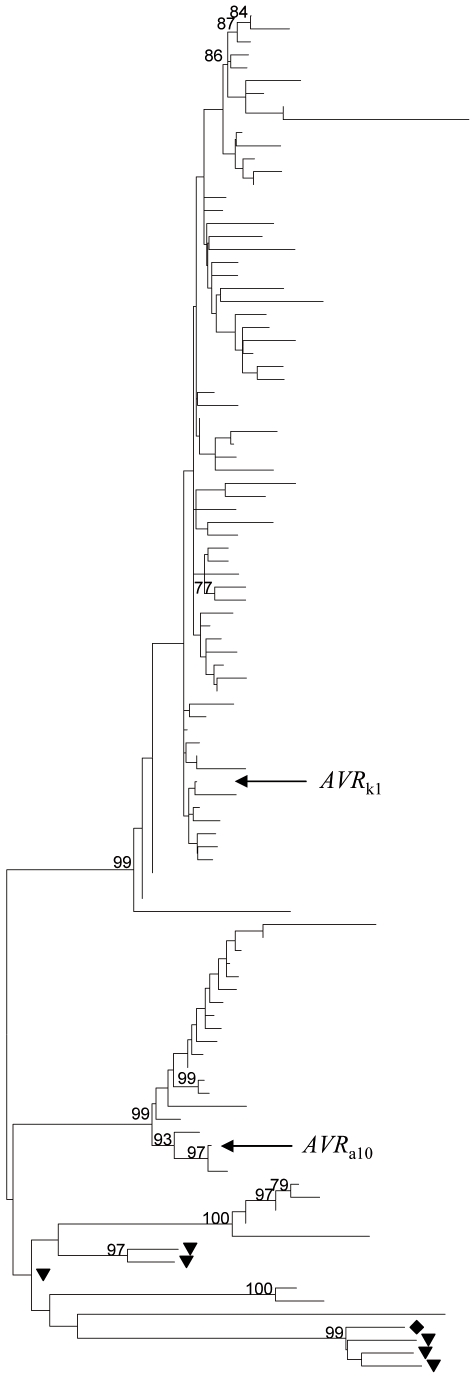
B. graminis sequences were retrieved from an nrdb90 database as described in the text; near-identical sequences were removed for clarity. The figure shows 105 amino acid sequences, including AVR k1, AVR a10 and 96 ORFs predicted from Bgh, six ORFs predicted from the Erysiphe pisi genome (marked with a triangle) and one ORF predicted from the Erysiphe (Golovinomyces) orontii genome (the closest homologue to AVRk1 of the six found, marked with a diamond). Bootstrap support (1,000 replicates) is shown if higher than 70%.
The AVR k1 gene family has diverged in accordance with B. graminis ff. spp. specialized on different hosts
On the basis of the known role of AVRk1 and AVRa10 proteins in pathogenicity, we predicted that sequences of AVR
k1 paralogs might have diverged from each other in B. graminis isolates adapted to infect different host genera. To test this hypothesis, degenerate PCR primers designed from the conserved core of the AVRk1 and AVRa10 protein sequences were used to amplify genomic DNA and clone the corresponding gene regions from ff. spp. infecting cereal crops and the grasses Elytrigia repens (synonym Agropyron repens) and Lolium perenne. The sequences obtained were classified into two subfamilies: the AVR
k1-like clade and the AVR
a10-like clade (Fig. 2A). Nucleotide identity within subfamilies was very high, around 80%. The number of sequences in the sub-family which grouped with AVR
a10 was four times higher than the number of AVR
k1-like sequences. Moreover, the relative number of sequences of each type differed significantly depending on the host of each f. sp. (χ2 =
= 34.1, P<10−3; Fig. 2B). None of the sequences amplified from powdery mildew isolates of oats (f. sp. avenae) or L. perenne grouped with the AVR
k1-like clade (Fig. 2A), indicating the absence or low abundance of this subfamily in these ff. spp.
34.1, P<10−3; Fig. 2B). None of the sequences amplified from powdery mildew isolates of oats (f. sp. avenae) or L. perenne grouped with the AVR
k1-like clade (Fig. 2A), indicating the absence or low abundance of this subfamily in these ff. spp.
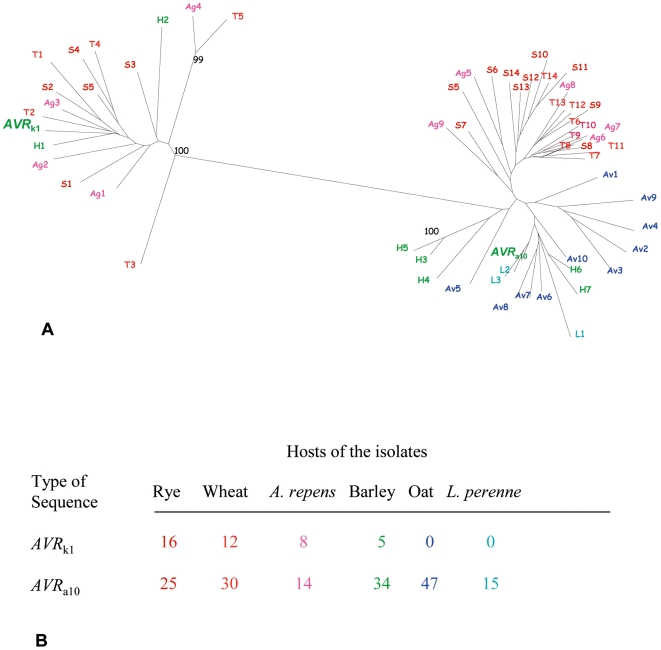
A. Neighbor Joining tree of the sequences obtained by degenerate primers from isolates of B. graminis from grass hosts: rye (f. sp. secalis, S, in red), wheat (f. sp. tritici, T, in orange), Agropyron spp. (f. sp. agropyri, Ag, in magenta), barley (f. sp. hordei, H, in green), oat (f. sp. avenae, Av, in blue) and Lolium perenne (L, in cyan). The sequences of the genes AVR k1 and AVR a10 are in a larger font. Bootstrap support (1,000 replicates) is shown if higher than 90%. Only sequences with a maximum identity to other sequences in the family less than 90% were used in the analysis. B. Number and type of sequences homologous to AVR k1 and AVR a10 obtained by degenerate PCR from B. graminis from different hosts.
The internal branches of both AVR k1-like and AVR a10-like clades were not supported statistically, possibly due to a phase of rapid divergence during expansion of the gene family [27]. Therefore, we used a likelihood mapping test [28] to examine if there was a relationship between the groups of sequences within each clade and the f.sp. from which they originated. There was no statistical support for any such grouping within the AVR k1-like clade. By contrast, an association between the AVR a10 sequences and ff.spp. was found: 91% of the quartets grouped the sequences from ff.spp. tritici, secalis and agropyri separately from the sequences from ff.spp. avenae, hordei and the isolate from L. perenne (Fig. 2A, Fig. S1). Therefore the AVR a10 sequences have diverged with the powdery mildew formae speciales infecting different Poaceae host genera.
AVR k1 paralogs contain conserved and diversified regions
The very large number of AVR k1 paralogs detected in the B. graminis genome may not reflect the actual number of expressed genes. Indeed, many gene duplications can be subject to gene inactivation through mutation or deletion/insertion events as well as DNA methylation. To study the expressed AVR k1 paralogs, we analyzed the B. graminis transcriptome amplified by 5′ and 3′RACE RT-PCR. In total, 49 5′ RACE sequences and 84 3′RACE sequences were obtained from four isolates of f. sp. hordei and one isolate of f. sp. tritici, revealing considerable divergence in their length and degree of homology with AVR k1 (Table 1). The 3′RACE sequences were significantly less conserved than those obtained by 5′RACE (t-test for comparison of average nucleotide identities with AVR k1, P<10−14).
Table 1
| RACE 5′ | RACE 3′ | ||||||
| Forma specialis | Isolate | No of different seq. | Length | % Identity to AVR k1 | No of different seq. | Length | % Identity to AVR k1 |
| hordei | A6 | 6 | 417±291 | 83±16 | 13 | 724±501 | 45±13 |
| CC52 | 7 | 560±34 | 87±1 | 14 | 205±70 | 73±13 | |
| CC148 | 11 | 422±178 | 82±5 | 18 | 695±527 | 58±16 | |
| DH14 | 12 | 417±204 | 78±17 | 19 | 578±354 | 53±14 | |
| tritici | JIW11 | 13 | 466±165 | 86±7 | 20 | 150±65 | 75±9 |
Several parasite effectors are under diversifying selection (DS), evolving rapidly to avoid immune detection systems within the host [2]. We tested for DS in a set of 113 AVR
k1 paralogs obtained by RACE RT-PCR. We used a maximum likelihood method to identify specific amino acid residues that are under positive selection (with a nonsynonymous/synonymous rate ratio higher than one, ω =
= dN/dS >1) [29]. Most analyzed residues in the core region of the expressed AVR
k1 paralogs are under purifying selection. This indicates a high level of sequence conservation, possibly due to protein functional or structural constraints. DS was evident in a region immediately 5′ to the core. This indicates that this region is evolving rapidly, so it could be involved in adaptation to avoid R gene recognition, as proposed for Phytophthora effectors [30] (Fig. S2A). By comparing complete cDNAs, breakpoints of nucleotide divergence could be identified shortly after the sequence homologous to the AVR
k1 protein (Fig. S2B and S3A, B). This suggests that AVR
k1 sequence proliferation has occurred through gene duplication and insertion at several distinct sites within the Bgh genome.
dN/dS >1) [29]. Most analyzed residues in the core region of the expressed AVR
k1 paralogs are under purifying selection. This indicates a high level of sequence conservation, possibly due to protein functional or structural constraints. DS was evident in a region immediately 5′ to the core. This indicates that this region is evolving rapidly, so it could be involved in adaptation to avoid R gene recognition, as proposed for Phytophthora effectors [30] (Fig. S2A). By comparing complete cDNAs, breakpoints of nucleotide divergence could be identified shortly after the sequence homologous to the AVR
k1 protein (Fig. S2B and S3A, B). This suggests that AVR
k1 sequence proliferation has occurred through gene duplication and insertion at several distinct sites within the Bgh genome.
AVR k1 paralogs are associated with TE1a retrotransposons
Of the 17 3′RACE sequences longer than 800 nucleotides, 65% had homology with retrotransposons (RTs) at their 3′end, increasing to 90% for sequences longer than 1200 nucleotides. Most (10/11) of the predicted homologies had an amino acid identity of 70–80% with the nucleic acid binding domain of Bgh TE1a RTs that we reported previously [12], [16]. Full-length sequences were also obtained by hybridization to a cDNA library, with similar results. Four of 22 full-length cDNA clones were natural antisense transcripts [NATs, 31] with a polyT tail at the 5′ end before the ATG translation start site. The genomic region containing the NATs was identified by BLAST with the draft Bgh genomic sequence. The presence of polyT at the 5′ end of the cDNA sequences confirms that the sequences are transcribed in the reverse orientation (Fig. S4).
We further investigated the association between the AVR k1 gene family and RTs, by testing the extent to which TE1a and AVR k1 predicted open reading frames occurred together in the draft Bgh genome sequence. Three categories of hits were identified: 1) ‘Common’ hits were those in which AVR k1 and TE1a sequences occurred in the same open reading frame. 2) ‘Adjacent’ hits were those in which AVR k1 and TE1a sequences occurred on the same contig but were separated by a stop codon. Pairs were not considered adjacent if one hit was on the complementary strand. Additionally, we specified that each member of a pair could only belong to a maximum of one pair. 3) ‘Unique’ hits matched a specific contig containing either AVR k1 or TE1a paralogs, but not both. We found that 57.8% of AVR k1 paralogs were either ‘common’ or ‘adjacent’ to TE1a homologs. This proportion is significantly higher than the proportion of TE1a homologs found common or adjacent to the two largest Bgh gene families other than AVR k1 (Table 2, χ2 test, P<10−4). Conversely, the proportion of TE1a homologs common or adjacent to AVR k1 paralogs was significantly higher than the proportion found with the four largest families of repetitive elements other than TE1a (Table 3, χ2 test, P<10−4). These two results demonstrate that there is a significant association between AVR k1 and TE1a sequences.
Table 2
| Gene family | Repetitive element | Number of common hits | Number of adjacent hits | Number of hits with no repeat neighbor | Total number of homologs |
| AVR k1 | TE1a | 532 | 130 | 483 | 1145 |
| EGH24 | 1 | 154 | 990 | ||
| A8U3S4 | TE1a | 0 | 18 | 405 | 423 |
| EGH24 | 0 | 57 | 366 | ||
| Q9HGU6 | TE1a | 0 | 1 | 57 | 58 |
| EGH24 | 0 | 0 | 58 |
Table 3
| Repetitive element | Number of common hits | Number of adjacent hits | Number of hits with no AVR k1 neighbor | Total number of homologs |
| TE1a | 532 | 130 | 1085 | 1747 |
| EGH24 | 1 | 154 | 4274 | 4429 |
| Q5BBQ3 | 0 | 50 | 2008 | 2058 |
| Q9ZT24 | 0 | 43 | 1927 | 1970 |
| Q2AA50 | 0 | 46 | 1747 | 1793 |
We examined which other sequences were found in the proximity of the 483 AVR k1 homologs that were not situated next to TE1a sequences (Table 2). We retrieved 10 kb-long contig sequences (5 kb either side of the hit), fragmented them into 2 kb segments (each overlapping by 1 kb) and searched for sequence homology of each fragment establishing a cut-off of E≤10−5. A total of 59 different proteins were found. Fifty three of them had homologs that appeared 10 times or less (31 appeared only once, which means that no homolog was found for these particular genes). The sequences most commonly found close to these AVR k1 sequences were TE1a sequences (284 hits), followed by another retrotransposon family, TE1b (192 hits, Table 4). Therefore, no other type of sequence is associated with the AVR k1 family.
Table 4
| Gene familya | Number of homologs | Putative function/homology with other sequences |
| A8U3R5 | 12 | Hypothetical protein |
| Q8S7A3 | 32 | Putative retroelement |
| A8U3S6 | 38 | TE3 retrotransposon |
| A8U3R2 | 192 | TE1b retrotransposon |
| A8U3R0 | 284 | TE1a retrotransposon |
We investigated if associations between retrotransposable elements and gene families are common events in the Bgh genome. We searched for cases where the most frequent repetitive element found in Bgh genome (EGH24) occurred close to other gene families. We did not find any case with a proportion of common or adjacent hits equivalent to that found with TE1a and AVR k1 paralogs (Table 2). To further test if other types of sequence could be associated with TE1a homologs, we examined the 1085 TE1a hits that were neither common nor adjacent to AVR k1 paralogs (Table 3) with the same procedure used for AVR k1 explained above. A total of 112 different proteins were found, of which 101 had homologs that appeared 10 times or less. The family that was most commonly found close to TE1a sequences was a reverse transcriptase (1415 hits). The other most frequent families were Gag-like or reverse transcriptases, typical of retrotransposons (Table 5). Therefore apart from the AVR k1 family, only retrotransposable elements are frequently found in the proximity of TE1a sequences.
Table 5
| Gene familya | Number of homologs | Putative function/homology with other sequences |
| Q2GV21 | 20 | Hypothetical protein |
| A8U3R5 | 39 | Hypothetical protein |
| Q9C436 | 51 | Gag protein |
| A4QX15 | 74 | Hypothetical protein |
| Q2HI73 | 105 | Hypothetical protein |
| Q7XUD9 | 132 | Retrotransposable element |
| Q2PWB3 | 459 | Gag-like protein |
| Q2PWB2 | 1000 | Reverse transcriptase |
AVR k1 paralogs have coevolved with TE1a retrotransposons
The strong linkage between AVR k1 paralogs and the retroelement TE1a suggests a benefit to this association and, as a consequence, coevolution of the two genetic structures in the genome of Bgh. If two associated lineages coevolve, each lineage is expected to track the other over evolutionary time, which will be reflected in congruence between their phylogenies. Congruence between phylogenies of organisms is commonly ascribed to cospeciation in host-parasite systems [32], whereas incongruence is generally explained by events such as duplications, host-switch and parasite extinction. The equivalent processes for this genome analysis can be interpreted as codivergence instead of cospeciation, gene transfer within the genome instead of host-switch and gene loss instead of parasite extinction [33].
To explore the coevolutionary history of AVR k1 paralogs and TE1a sequences, we compared the phylogeny of these two groups by using the adjacent hits identified above. We used a mathematical model, Jungle [34], which contains all the combinations of associations between the two trees considering the events of codivergence, duplication, gene transfer and gene loss. We initially analyzed the 49 sequences that contained the entire conserved AVR k1 core sequence as previously defined [16], i.e. sequences that aligned to the central region of AVR k1, and were adjacent to a TE1a element. We applied cophylogenetic analysis to these 49 pairs of elements (Fig. S5) and then reduced the dataset to a more manageable subtree of 29 sequences that were selected because they form a large single clade in the larger tree (Fig. 3A). Two sub-clades of this group of AVR k1 sequences matched with similar clades in the TE1a phylogeny (subclades 1 and 4, Fig. S5). Since the computational complexity of the reconstruction problem is prohibitive when the number of gene transfers is large [34], we limited the Jungles reconciliation analysis to a maximum number of three gene transfers. Four potentially optimum solutions were identified: all four reconstructions postulated 32 codivergence events (equivalent to 16 instances of cospeciation) (Table 6, Fig 3B). The number of codivergence events was highly significant (P<0.01, the null hypothesis being the two phylogenies are randomly related) for scenarios with 0, 1 or 2 gene transfers, giving a good indication that AVR k1 and TE1a sequences have coevolved. However, the use of strong constraints (gene transfer ≤3) signifies a possible overestimation of the number of codivergence events and a probable underestimation of gene transfers.
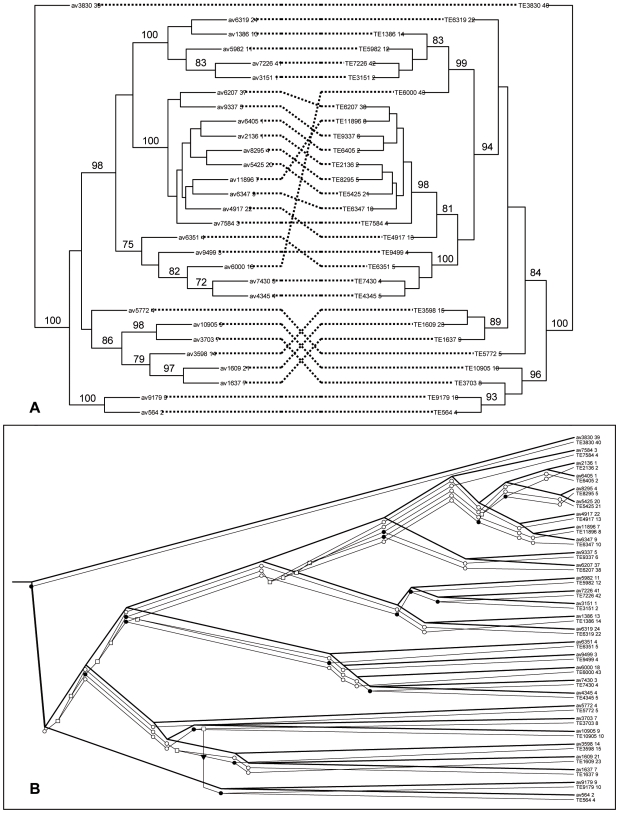
A. Tanglegram for AVR k1 (left) and TE1a (right) sequences, based on predicted ORFs from Bgh genome. Lines connecting sequences indicate associations. Bootstrap support (1,000 replicates) is shown below the branch if higher than 70%. B. One of the four potentially optimal reconciled trees between AVR k1 and TE1a trees. The two trees are superimposed. Hypothetical evolutionary events are represented as black circles for codivergence events, white squares for duplication events, white circles for gene losses and arrows for gene transfers.
Table 6
| Solution | Codivergences | Duplications | Gene transfers | Gene losses | Cost | P |
| 1 | 32 | 24 | 0 | 55 | 79 | <0.01 |
| 2 | 32 | 24 | 1 | 49 | 74 | <0.01 |
| 3 | 32 | 24 | 2 | 44 | 70 | <0.01 |
| 4 | 32 | 24 | 3 | 38 | 65 | n/a |
We also used an event-based parsimony approach [35] to test the fit between the AVR
k1 and TE1a phylogenies. This method finds the most likely explanation of observed data by minimizing the cost of implied events. We tested different reconstructions by preventing particular events from happening by applying a very high cost. We assigned a high cost to all four events in turn (codivergence, duplication, gene transfer and gene loss), and found a significant global fit between the two trees (P<0.001, the null hypothesis being the two phylogenies are randomly related) in all analyses, except when codivergence was prevented (P =
= 1), indicating that the similarity of AVR
k1 and TE1 phylogenies is due to the number of codivergence events [36]. Using the same default values as in our first approach, we found that 10 to 12 codivergence events and 16 to 18 gene transfers maximize the likelihood of the model (P<0.001). These results indicate 1) a moderate fit between both phylogenies, and 2) that incongruences in the cophylogeny have most likely arisen by gene transfers from one genomic location to another. This means that the AVR
k1 paralogs have coevolved with the TE1a sequences adjacent to them, although there have also been AVR
k1 sequences that, in being transferred in the genome, have become close to TE1 retrotransposons with which they have not coevolved.
1), indicating that the similarity of AVR
k1 and TE1 phylogenies is due to the number of codivergence events [36]. Using the same default values as in our first approach, we found that 10 to 12 codivergence events and 16 to 18 gene transfers maximize the likelihood of the model (P<0.001). These results indicate 1) a moderate fit between both phylogenies, and 2) that incongruences in the cophylogeny have most likely arisen by gene transfers from one genomic location to another. This means that the AVR
k1 paralogs have coevolved with the TE1a sequences adjacent to them, although there have also been AVR
k1 sequences that, in being transferred in the genome, have become close to TE1 retrotransposons with which they have not coevolved.
Discussion
This work reveals that the AVR k1 family has extensively colonized the Bgh genome, representing the largest family of effector paralogs discovered so far in a fungal genome. A similar example of an extended number of related sequences within a given genome is the RXLR-containing effector family in oomycetes [30]. Functional redundancy of AVR genes within the genome may facilitate rapid evolution of the parasite to overcome host resistance by allowing elicitor genes to become inactivated without compromising parasite fitness [5], [37], [38]. The exceptionally high number of AVR genes described in Bgh [7] supports the idea of such an evolutionary history of this parasite.
Blumeria was the first genus that split from the rest of the Erysiphales 76 million years ago [39]. We found AVR k1 homologs in two Erysiphe species, so the gene family must predate the split. However, the Erysiphe sequences lie in the base of the phylogeny, not in the two large clades formed by AVR k1 or AVR a10 paralogs, so these subfamilies may have differentiated and proliferated extensively only in Blumeria. AVR k1 paralogs have evolved differentially in B. graminis ff.spp. from different grass hosts. The AVR a10-like sequences from ff. spp. tritici, secalis and agropyri group separately from those in ff. spp. avenae, hordei and the isolate from Lolium perenne. This corresponds with the phylogeny of other genes [40], in which isolates from ff. spp. tritici, secalis and agropyri form a distinct clade, with f.sp. hordei as a sister clade and ff. spp. avenae and isolates from Lolium sp. in more distantly related clades. Differential selection for a battery of effectors that are not recognized by the host could be the basis of host specialization of B. graminis [41]. Thus, it is possible that AVR k1 paralogs may be involved in the extreme host specialization encountered in this strictly biotrophic pathogen.
The selection pressure exerted on crops during the development of agriculture could have played an important role in promoting the proliferation and diversification of the AVR k1 family in B. graminis. After early cultivation of domesticated wheat, new powdery mildew resistance genes arose [42]. In the GFG system, mutation of the AVR genes would allow new, virulent isolates to escape recognition by these new resistance specificities. The greater abundance of AVR k1-like sequences in the ff. spp. from wheat, rye and barley, compared to those from oats, suggests that the proliferation of these genes could be related to the specialization of the parasite during the evolution of cereal crops in agriculture. Wheat, rye and barley originated in the near East during the 11th–9th millennia BP [43]. Oats originated much later as a crop in Northern Europe [4th–3rd millennia BP, 44], and have been subject to less intensive breeding than wheat and barley.
These data provide the first direct evidence that a parasite effector gene family and a particular retrotransposon lineage are consistently associated and have coevolved. The frequency with which members of the AVRk 1 and TE1a retrotransposon lineages occur together in the genome is highly significant, and two independent analyses show that their phylogenies are congruent. The coevolution between these two entities indicates that they move and evolve together, so their occurrence close to each other is not merely due to a retrotransposon insertion site bias. An association with transposable elements has been postulated as a mechanism for the expansion and movement of effector genes within genomes [5], [6]. The coevolution of these two entities implies a mutual benefit to the association, which could ultimately contribute to parasite adaptation and success. The association would benefit 1) the powdery mildew fungi, by providing a mechanism for amplifying and diversifying effectors, which would increase the pathogen's mean fitness in the presence of diverse plant resistance genes and 2) the associated RTs, by providing a basis for their maintenance in the fungal genome through natural selection for genomes which contain numerous effector genes and thus contribute to increased fitness.
In addition to a role in gene mutation, RTs play an important role in genome evolution [45]–[47]. There is also considerable evidence that eukaryotic organisms have co-opted functions from RTs, including the epigenetic regulation of associated genes required for adaptation [48]. Such mechanisms could also apply to effectors, and be related to host adaptation [49]. We have found AVR k1 paralogs expressed as natural antisense transcripts (NATs) which can be a mechanism for epigenetic control of neighboring genes [31]. With an increasing number of genomes sequenced [50], it will be possible to establish whether coevolution between families of effectors and RTs occurs more widely, and how the association may contribute to parasite adaptation and host specialization.
In conclusion, we show that an effector gene family required for virulence in the powdery mildew fungus has coevolved with TE1a, a class of LINE-1 retrotransposon. To our knowledge, this is the first demonstration of the coevolution between parasite effectors and retrotransposons. An association between effectors and retrotransposons had already been postulated in many cases, but this is the first work that shows that this association is significant and has an evolutionary basis. Our discovery that effectors and retrotransposons have coevolved leads to a much deeper understanding of pathogenicity and specialization in parasites.
Materials and Methods
Fungal isolates and samples
Isolates of Blumeria graminis from different cultivated and wild grasses were obtained from the laboratory collection at the John Innes Centre. The Bgh isolate Race I [51] was used for making a cDNA library.
RACE-PCR reactions
RNA was extracted with an RNAeasy kit (Qiagen) from leaves of barley cultivar Golden Promise, three days after inoculation with Bgh isolates A6, CC52, CC148, DH14 and from leaves of wheat cultivar Cerco, three days after inoculation with B. graminis f. sp. tritici (Bgt) isolate JIW11. Amplification of the 5′ and 3′ cDNA was performed with the SMART™ RACE kit (BD Biosciences). Twenty genomic sequences from a Bgh BAC library [16] were first obtained by hybridization to AVR k1. Primers were then designed to amplify expressed AVR k1 paralogs from four different Bgh isolates and a Bgt isolate. Following initial screening of primers to achieve the highest diversity in lengths for all the isolates, the primers used were: RACEK15′2 (5′AATGGCGGCGCGTAGGTAGACTCT3′) for the 5′end, nested with NESTEDK15′2 (5′CCCGTTGGTCAAAGGAAGAAGGGT3′) and RACE13′2 (5′TCGATGAGAGTCTACCTACGCGCC3′) for the 3′end, nested with NESTED15′2 (5′ATTGCGCAATACATGGCCACGGTG3′). Amplification products were cloned in the pGEM®-T Easy vector (Promega) and a random set of 24 clones per isolate were sequenced. The sequences have been deposited in the EMBL/GenBank [24], and accession numbers are GQ470737 to GQ470866.
Sequencing of paralogs from different ff. spp
DNA was extracted as described previously [16] from conidia of B. graminis f. sp. hordei isolates DH14 and CC148; tritici isolates JIW11 and FEL09; secalis isolates RyeRMasBlue and RyeRmas6W; avenae isolates MO892 and MOH15; agropyri isolate CF3a. B. graminis and isolate LSSB1 from L. perenne. PCR was performed using AmpliTaq (Applied Biosystems) and degenerate PCR primers: AVRDEGF (5′GTCGARGCMRCCCTTCWWCC3′, where R =
= A+G, M
A+G, M =
= A+C, W
A+C, W =
= A+T) and AVRDEGR (5′GTGGCMCSWGTGCTTYTGAG3′, where Y
A+T) and AVRDEGR (5′GTGGCMCSWGTGCTTYTGAG3′, where Y =
= C+T, S
C+T, S =
= G+C). Sixteen to twenty six clones per isolate were sequenced. Only sequences with identities lower than 99% to any other sequence were considered as unisequences. The sequences have been deposited in the EMBL/GenBank [24], and accession numbers are GQ470682 to GQ470736.
G+C). Sixteen to twenty six clones per isolate were sequenced. Only sequences with identities lower than 99% to any other sequence were considered as unisequences. The sequences have been deposited in the EMBL/GenBank [24], and accession numbers are GQ470682 to GQ470736.
Isolation of cDNA clones
Full-length cDNA clones were isolated from a Lambda ZAP Express cDNA library [52], made from epidermal strips of barley leaves, cultivar Manchuria, 14–16 h after inoculation with Bgh isolate Race I [51]. The library was screened according to the ZAP Express manual (Stratagene) with a probe made from the conserved region of the AVR k1 gene family using the primers R1 and R3 [16] and 192 positive plaques were initially picked. From these, 22 clones were purified, in vivo excised and the inserts of the plasmids were sequenced. The sequences have been deposited in the EMBL/GenBank [24], and accession numbers are GQ470867 to GQ470888.
Sequence analyses
Nucleotide sequence analysis and contig assembly were done with the STADEN package [53]. Protein sequences were aligned with MUSCLE [54] and edited with Genedoc (distributed by Nicholas KB, Nicholas HB and Deerfield DW, http://www.psc.edu/biomed/genedoc/gdfeedb.htm). Protein sequences were converted back to coding DNA sequences to conserve the codons position in the alignment using RevTrans [55]. Homologies were detected using the BLAST program [23] against the EMBL/GenBank [24], COGEME phytopathogen EST database [25], Broad Institute (http://www.broad.mit.edu/) and Uniprot [26] databases. Open reading frames were predicted from the draft genomes of Bgh (www.blugen.org), Erysiphe (Golovinomyces) orontii and Erysiphe pisi using the program getorf from the EMBOSS package [56].
Neighbor-Joining (NJ) and Maximum Likelihood trees were generated using the PHYLIP 3.6 package [57] and MEGA version 4 [58]. Distance matrices of the NJ trees were calculated under the Jones-Taylor-Thornton and the Jukes Cantor models of evolution for Figure 1 and Figure 2A respectively. Bootstrapping (100 or 1,000 replicates) was used to determine the strength of support for individual nodes. Likelihood mapping analyses [28] were done using the program TREE-PUZZLE 5.3 [59]. The dataset of sequences was classified in four groups under different hypotheses: a) depending on the host of origin (all possible combinations) and b) randomly. The posterior weights of the possible topologies of each quartet under each hypothesis were analyzed using the quartet puzzling algorithm.
The diversifying selection analyses were done using codeml from PAML 3.15 [60] with alignments of N-terminal and C-terminal regions. Two pairs of codon substitution models (M1a/M2a and M7/M8) were used to study ω variation among amino acid sites [61]. M1a and M7 assumes no site with ω >1 (no positive selection, null hypothesis) while M2a and M8 assumes the presence of positively selected sites. To test for positive selection, the likelihood ratio test (LRT) between the models in each pair was compared with a χ2 distribution. Whenever the LRT suggested the presence of positively selected sites, an empirical Bayes approach was used to calculate the conditional (posterior) probability distribution of ω for each site enabling the identification of positively selected residue in the alignment. Both Naive Empirical Bayes (NEB) and Bayes Empirical Bayes (BEB) methods were used [62].
In the cophylogenetic analysis, we compared AVR k1 and TE1a trees, using reconciliation analysis with Jungles [34] as implemented in the program TreeMap 2.0β. The analysis was performed with a maximum number of three host switches (or gene transfers). We used the default values for event costs: 0 for codivergence and 1 for duplication, loss and gene transfer (host switch) events. The significance of the codivergence events was determined by generating 99 random TE1a trees and determining how many of those supported solutions had as many codivergence events as the observed AVR k1 tree [63]. TreeFitter 1.0 [35] was used for parsimony-based tree fitting. The significance of the results was tested by performing 1,000 random permutations of the TE1a tree terminals.
Sequences of E. pisi and E. orontii
E. pisi (Birmingham isolate, kindly provided by Dr. Timothy Carver from The Welcome Trust Sanger Institute, Hinxton, Cambridge, CB10 1SA, UK) and E. orontii (isolate MPIZ) genomic DNA was extracted from vacuum-harvested conidia and purified on a CsCl gradient. DNA sequencing by pyrosequencing (454 Technology) was performed by imaGenes, formerly RZPD German Resource Center for Genome Research in Berlin, Germany (http://www.imagenes-bio.de/) using GS-20 and FLX sequencer systems and automatically assembled on site. The available sequence corresponds to 400–450 Megabases each for E. orontii and E. pisi genomes.
Supporting Information
Figure S1
Grouped likelihood mapping diagrams produced from the AVRa10 clade (Fig. 2A). A. The dataset was grouped in two clusters, a: agropyri - tritici - secalis and b: hordei - avenae - L. perenne. 91% of the quartets are (a,a) - (b,b), supporting the clusters defined. B. Sequences were randomly distributed in two clusters, a and b; any topology is favored. The analysis is consistent with the hypothesis that sequences from ff.spp. agropyri, tritici and secalis form a distinct clade in the phylogeny shown in Fig. 2A.
(0.99 MB TIF)
Figure S2
A. Diversifying selection at amino acid residues in AVRk1 homologs. Consensus representation of DS analysis on an alignment of RACE3′ or RACE5′ sequences. Sites were defined as diversified (in black) whenever the probability exceeds 90%. Otherwise, sites were defined as non-diversified (in grey). A residue with undefined adaptation (dotted) signifies discrepancy of results between the alignments of RACE3′ and RACE5′ sequences. Positions that were not analyzed are shown in white. The core sequence as defined in ref 16 is marked by dots above the sequence. Arrows show boundaries for 5′ and 3′ analysis. B. Breakpoints of divergence in expressed AVRk1 homologs. Representation of three full-length cDNA sequences obtained by hybridization to AVRk1, selected to illustrate how the sequence diverges after the conserved core region of AVRk1 (horizontal dotted line above the degree of homology to AVRk1). Sudden sequence divergence typically occurs in the break point region (shaded). Length of homology obtained by BLASTN against EMBL nucleotide database is shown by an horizontal line. Homologies identified by TBLASTX to expressed sequence tag (EST) of unknown function: * EST clone SL011D12–5, accession AU250405 from B. graminis-infected Lolium multiflorum.
(0.08 MB TIF)
Figure S3
A. Alignment of full-length cDNA sequences of AVRk1 paralogs from Fig. S2B showing sequence divergence breakpoint at arrow. B. Alignment of the other full-length cDNA sequences from Fig. S2B showing sequence divergence breakpoint at arrow.
(1.92 MB TIF)
Figure S4
Alignment of a natural antisense transcript (NAT) from two cDNA clones against the genomic sequence containing the AVRk1 sequence. Start of the AVRk1 coding sequence is highlighted in red. Conserved DNA sequence bases are indicated by an asterisk. The presence of poly dT at the 5′ end of the cDNA indicates polyadenylation of the transcript in the reverse orientation to that expected when compared to the AVRk1 sequence.
(1.06 MB TIF)
Figure S5
Tanglegram for AVRk1 (left) and TE1a (right) sequences, based on predicted ORFs from the Bgh genome. Lines connecting sequences indicate associations. Bootstrap support (100 replicates) is shown below the branch if higher than 70%. The groups of associated sequences selected for further analysis are numbered 1 to 4.
(0.93 MB TIF)
Acknowledgments
We thank Sandra Noir, Mariam Benjdia, Ralph Panstruga and Paul Schulze-Lefert for the sequences of E. pisi and G. orontii and Michael Charleston for the help with interpreting TreeMap results.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grant reference BB/C506299/1, European Union Framework VI programme (BIOEXPLOIT), the Max Planck Society, a Marie Curie Intra-European Fellowship award to S. Sacristan, a Hellenic Republic Studentships Foundation (I.K.Y.) award and a Leverhulme Trust Early Career Research Fellowship to P. Skamnioti, a Villum Kann Rasmussen Foundation grant to C. Pedersen and funding from the Alexander von Humboldt Foundation for C. Micali. The Blumeria graminis genome sequencing project (http://www.blugen.org/) was funded by BBSRC grant reference: BBE0009831. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0007463
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0007463&type=printable
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pone.0007463
Article citations
Deciphering the Genomic Landscape and Virulence Mechanisms of the Wheat Powdery Mildew Pathogen Blumeria graminis f. sp. tritici Wtn1: Insights from Integrated Genome Assembly and Conidial Transcriptomics.
J Fungi (Basel), 10(4):267, 03 Apr 2024
Cited by: 0 articles | PMID: 38667938 | PMCID: PMC11051031
The Venturia inaequalis effector repertoire is dominated by expanded families with predicted structural similarity, but unrelated sequence, to avirulence proteins from other plant-pathogenic fungi.
BMC Biol, 20(1):246, 03 Nov 2022
Cited by: 15 articles | PMID: 36329441 | PMCID: PMC9632046
Transcriptional activity and epigenetic regulation of transposable elements in the symbiotic fungus Rhizophagus irregularis.
Genome Res, 31(12):2290-2302, 12 Nov 2021
Cited by: 11 articles | PMID: 34772700 | PMCID: PMC8647823
Variable genome evolution in fungi after transposon-mediated amplification of a housekeeping gene.
Mob DNA, 10:37, 27 Aug 2019
Cited by: 1 article | PMID: 31462936 | PMCID: PMC6710886
Novel jack-in-the-box effector of the barley powdery mildew pathogen?
J Exp Bot, 69(15):3511-3514, 01 Jun 2018
Cited by: 2 articles | PMID: 29947808 | PMCID: PMC6022647
Review Free full text in Europe PMC
Go to all (43) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Genes & Proteins (Showing 15 of 15)
- (2 citations) UniProt - A8U3R5
- (1 citation) UniProt - Q9ZT24
- (1 citation) UniProt - Q2HI73
- (1 citation) UniProt - Q2PWB2
- (1 citation) UniProt - Q9C436
- (1 citation) UniProt - Q2PWB3
- (1 citation) UniProt - Q8S7A3
- (1 citation) UniProt - Q2AA50
- (1 citation) UniProt - Q7XUD9
- (1 citation) UniProt - A8U3S4
- (1 citation) UniProt - A8U3R2
- (1 citation) UniProt - A8U3R0
- (1 citation) UniProt - Q9HGU6
- (1 citation) UniProt - Q2GV21
- (1 citation) UniProt - A8U3S6
Show less
Nucleotide Sequences (Showing 7 of 7)
- (1 citation) ENA - GQ470866
- (1 citation) ENA - GQ470888
- (1 citation) ENA - AU250405
- (1 citation) ENA - GQ470736
- (1 citation) ENA - GQ470867
- (1 citation) ENA - GQ470682
- (1 citation) ENA - GQ470737
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance.
Plant Cell, 18(9):2402-2414, 11 Aug 2006
Cited by: 152 articles | PMID: 16905653
Evolution of the EKA family of powdery mildew avirulence-effector genes from the ORF 1 of a LINE retrotransposon.
BMC Genomics, 16:917, 10 Nov 2015
Cited by: 16 articles | PMID: 26556056 | PMCID: PMC4641428
Rapid turnover of effectors in grass powdery mildew (Blumeria graminis).
BMC Evol Biol, 17(1):223, 31 Oct 2017
Cited by: 24 articles | PMID: 29089018 | PMCID: PMC5664452
Long-term and rapid evolution in powdery mildew fungi.
Mol Ecol, 33(10):e16909, 20 Mar 2023
Cited by: 13 articles | PMID: 36862075
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (4)
Sequencing the genome of the powdery mildew fungus Blumeria graminis
Dr Spanu, Imperial College London
Grant ID: BB/E000983/1
Sequencing the genome of the powdery mildew fungus Blumeria graminis
Dr Spanu, Imperial College London
Grant ID: BBE0009831
Pathogenicity function of an avirulence gene family in barley powdery mildew
Professor James Brown, John Innes Centre
Grant ID: BB/C506299/1
Exploitation of natural plant biodiversity for the pesticide-free production of food
Professor James Brown, John Innes Centre
Grant ID: BBS/E/J/0000A252




