Abstract
Free full text

ROADTRIPS: Case-Control Association Testing with Partially or Completely Unknown Population and Pedigree Structure
Abstract
Genome-wide association studies are routinely conducted to identify genetic variants that influence complex disorders. It is well known that failure to properly account for population or pedigree structure can lead to spurious association as well as reduced power. We propose a method, ROADTRIPS, for case-control association testing in samples with partially or completely unknown population and pedigree structure. ROADTRIPS uses a covariance matrix estimated from genome-screen data to correct for unknown population and pedigree structure while maintaining high power by taking advantage of known pedigree information when it is available. ROADTRIPS can incorporate data on arbitrary combinations of related and unrelated individuals and is computationally feasible for the analysis of genetic studies with millions of markers. In simulations with related individuals and population structure, including admixture, we demonstrate that ROADTRIPS provides a substantial improvement over existing methods in terms of power and type 1 error. The ROADTRIPS method can be used across a variety of study designs, ranging from studies that have a combination of unrelated individuals and small pedigrees to studies of isolated founder populations with partially known or completely unknown pedigrees. We apply the method to analyze two data sets: a study of rheumatoid arthritis in small UK pedigrees, from Genetic Analysis Workshop 15, and data from the Collaborative Study of the Genetics of Alcoholism on alcohol dependence in a sample of moderate-size pedigrees of European descent, from Genetic Analysis Workshop 14. We detect genome-wide significant association, after Bonferroni correction, in both studies.
Introduction
It is well known that problems can arise in case-control genetic association studies when there is population structure.1 At its most basic, case-control association testing can be thought of as a comparison of the allele (or genotype) frequency distribution between cases and controls, and markers that are not directly associated with the trait of interest can be spuriously associated with the trait if ancestry differences between cases and controls are not properly accounted for. Similarly, failure to account for population structure can also reduce power. To correct for population structure in case-control studies with samples of unrelated individuals, a number of methods have been proposed, including genomic control (GC),2 structured association,3 spectral analysis,4–8 and other approaches.9–13 However, many genetic studies include related individuals. Several methods have been proposed for case-control association testing in related samples from a single population with known pedigrees14–16 or with unknown or partially known pedigrees.17 However, these methods might not be valid in the presence of population heterogeneity. For certain types of study designs, family-based association tests such as the TDT18 and FBAT19 have been used to protect against potential problems of unknown population substructure. Family-based tests, however, are generally less powerful than case-control association methods20,21 and are more restrictive because they typically require genotype data for family members of an affected individual. In contrast, case-control designs can allow, but do not require, genotype data for relatives of affected individuals.
We address the general problem of case-control association testing in samples with related individuals from structured populations. We do not put constraints on how the individuals might be related, and we allow for the possibility that the pedigree information could be partially or completely missing. We propose a new method, ROADTRIPS, where this name is inspired by the description of the method as a robust association-detection test for related individuals with population substructure. ROADTRIPS uses a covariance matrix estimated from genome-screen data to simultaneously correct for both population and pedigree structure. The method does not require the pedigree structure of the sampled individuals to be known, but when pedigree information is available, the method can improve power by incorporating this information into the analysis. ROADTRIPS is computationally feasible for genetic studies with millions of markers. Other features of ROADTRIPS include (1) appropriate handling of missing data and (2) the ability to incorporate both unaffected controls and controls of unknown phenotype (i.e., general population controls) in the analysis.
In order to compare ROADTRIPS to other methods, on the basis of type 1 error and power, we simulate case-control samples containing both related and unrelated individuals with various types of population structure, including admixture. We also apply ROADTRIPS to identification of SNPs associated with rheumatoid arthritis (RA [MIM 180300]) in small UK pedigrees22 from Genetic Analysis Workshop (GAW) 15, and we apply it to identification of SNPs associated with alcohol dependence (MIM 103780) in a sample of moderate-size pedigrees of European descent from the Collaborative Study of the Genetics of Alcoholism (COGA) data23 of GAW 14.
Material and Methods
We first describe a class of testing procedures suitable for known structure. Then we describe the ROADTRIPS method for extending these tests to the contexts of unknown and partially known structure.
Overview of Association Testing with Known Structure
Consider the problem of testing for association of a genetic marker with a particular phenotype in a sample of n genotyped individuals. For simplicity, we assume that the marker to be tested is a SNP, with alleles labeled “0” and “1.” (The extension to multiallelic markers can be obtained as in previous work.16) Let Y = (Y1, … Yn)T where Yi = ½ × (the number of alleles of type 1 in individual i), so the value of Yi is 0, ½, or 1. We treat the genotype data on the n individuals as random and the available phenotype information as fixed in the analysis, an approach that is appropriate, for example, with either random or phenotype-based ascertainment. Under the null hypothesis of no association and no linkage between marker and trait, we assume that the expected value of Y is E0(Y) = p1, where 1 is a column vector of 1s of length n and p is a parameter representing the frequency of the type 1 allele. In models incorporating population structure, p would typically be interpreted as an “ancestral” allele frequency or some kind of average allele frequency across subpopulations. We denote by Var0(Y) = Σ the n × n covariance matrix of Y under the null hypothesis of no association. It is often convenient to write Σ = σ2Ψ, where σ2 is defined to be the variance of Y for an outbred individual in the absence of population structure, and Ψ accounts for relatedness, inbreeding, and population structure. We use the term “known structure” to refer to the case when the matrix Ψ is known. We always take σ2 to be unknown and estimate it from the sample. Denote by
where V is a fixed, nonzero column vector of length n such that VT1 = 0. Note that Var0(VTY) = σ2VTΨV, so the denominator in Equation 1 can be viewed as an estimator of Var0(VTY). In a test for association, V would naturally include phenotype information and could also include pedigree information. One could include covariate information in V as well, although we do not treat that situation in the present work. There are a number of case-control association test statistics that have the general form in Equation 1, including the Pearson χ2 test, the Armitage trend test,24 the corrected χ2 test,15 the WQLS test,15 and the MQLS test16 (details on how these tests can be written in the form of Equation 1 are given in subsection Examples of Association Tests with Known Structure). Under standard regularity conditions, the test statistic given in Equation 1 has an asymptotic χ12 distribution under the null hypothesis of no association and no linkage.
Estimation of σ2 when Structure Is Known
In the context of Equation 1, when structure is known, we have two general approaches for estimating σ2 under the null hypothesis. If we assume that, for an outbred individual in the absence of population structure, HWE holds at the marker, then σ2 = ½p(1 − p), where p is the frequency of allele 1 at the SNP being tested, and a reasonable estimator of σ2 under this assumption is
and (3) a Bayesian estimator26 such as
Alternatively, an approach to estimation of σ2 that does not assume σ2 = 0.5p(1 − p) could be used. When Ψ is known, a reasonable estimator is
which is RSS/(n − 1) for generalized regression of Y on 1, where RSS is the residual sum of squares. Note that when Ψ = I, the n × n identity matrix, e.g., with unrelated individuals in the absence of population structure, then
Examples of Association Tests with Known Structure
Corrected Pearson χ2 and Armitage Trend Tests
In the standard Pearson χ2 and Armitage trend tests for allelic association, one assumes that the individuals are unrelated with no population structure, so that Ψ = I. A corrected version of the Pearson χ2 test has previously been described15 for the situation when sampled individuals are related with all relationships known, in which case Ψ = Φ, where Φ is the kinship matrix, which is obtained as a function of the known pedigree information and is given by
where hi is the inbreeding coefficient of individual i, and ϕij is the kinship coefficient between individuals i and j, 1 ≤ i, j ≤ n. We propose to use this same choice of Ψ in the corrected Armitage test. (More generally, for either test, one might consider known structure Ψ that does not necessarily equal Φ.) In both tests, one further assumes that every individual in the sample can be classified as either case or control. In that context, let 1c be the case indicator, i.e., the vector of length n whose ith entry is 1 if individual i is a case and 0 if individual i is a control. Then both the corrected Pearson χ2 and corrected Armitage test statistics are obtained as special cases of Equation 1 with the choice
where nc is the number of case individuals among the n total individuals. (In the most general specification of the Armitage test for genetic association, mean-zero, nonlinear functions of Y are allowed in place of VTY, but in practice, the test is almost always performed with the V given in Equation 5.) The difference between the two tests is in the estimation of σ2. The corrected Pearson χ2 test uses the estimator
WQLS Test
The WQLS test15 was proposed in the context of related individuals without additional population structure, in which case Ψ = Φ given in Equation 4. The WQLS test statistic is formed from Equation 1 by taking
This choice of V can be motivated by generalized least-squares regression, because VTY is proportional to the estimated regression coefficient for 1c in the generalized least-squares regression of Y on 1c with intercept. The WQLS test uses the estimator
In the context of trait mapping in samples of related individuals with known pedigrees, the WQLS test generally has lower power16 than the corrected χ2 and MQLS tests. Nonetheless, we include it in the present work because the ROADTRIPS extension of the WQLS (described in subsection Association Tests when Structure Is Partially or Completely Unknown) is equivalent to the method, recently proposed by Rakovski and Stram,13 for association testing in the presence of hidden population structure and hidden relatedness. Thus, we include the ROADTRIPS extension of the WQLS in our simulation studies to compare its power and type 1 error to those of our proposed methods.
MQLS Test
In contrast to the preceding tests, the MQLS test16 allows three possible values for an individual's phenotype: “affected,” “unaffected,” and “unknown,” where the label “unknown” is used to represent unphenotyped individuals, e.g., general population controls, or individuals who are deemed too young to have developed an age-related trait such as Alzheimer's, whereas the label “unaffected” is reserved for true unaffecteds. As they have different expected frequencies of predisposing alleles, the two types of controls are treated differently in the analysis. Furthermore, whereas the preceding tests use the phenotype information only for individuals who have genotype data at the marker being tested, the MQLS also uses the phenotype information for individuals with missing genotype data at the marker being tested, provided that those individuals have a sampled relative who is genotyped at the marker.
As a result of these considerations, instead of using the phenotype vector 1c that is used in the preceding four tests, the MQLS uses the vector A = (ANT, AMT)T, which contains more information than 1c. Here, AN is the phenotype vector for the n individuals with nonmissing genotype data at the marker being tested, and AM is the phenotype vector for the m individuals with missing genotype data at the marker being tested, where individual i's phenotype is coded as Ai = 1 if i is affected, −k/(1 – k) if i is unaffected, and 0 if i is of unknown phenotype, where 0 < k < 1 is a constant that represents an external estimate of the population prevalence of the trait for a suitable reference population. (The prevalence estimate is permitted to be very rough; the MQLS test is valid for arbitrary fixed k.)
The MQLS test was proposed in the context of related individuals without additional population structure, in which case Ψ = Φ, the n × n matrix given in Equation 4. In order to incorporate the information of AM into the MQLS test, one also needs the n × m matrix, ΦN,M, whose (i, j)th entry is 2ϕij, where ϕij is the kinship coefficient between the ith nonmissing and jth missing individuals. The MQLS test statistic can be obtained from Equation 1 by choosing
and using the estimator
Outline of ROADTRIPS Approach for Unknown Structure
The idea behind ROADTRIPS is to extend tests of the form given in Equation 1 to the situation when there could be unknown population structure and/or cryptic relatedness in the sample. To do this, we use genome-screen data to form an appropriate estimator
where we can allow V to take into account any known pedigree information, in addition to phenotype information, while simultaneously accounting for pedigree errors and additional unknown structure through
Population Genetic Modeling Assumptions
The modeling assumptions we make are weak and are satisfied by commonly used models of population structure and commonly used models for related individuals. We consider S SNPs in a genome screen, and we generalize the notation of the previous subsections to a set of S SNPs by letting Ys be the genotype vector corresponding to SNP s, namely, Ys = (Y1s, …, Yns)T, s = 1, …, S, where Ysi = ½ × (the number of alleles of type 1 at SNP s in individual i). Our modeling assumption on the null mean, generalized from the preceding subsections, can be stated as
We make the following assumption regarding the null covariance matrix:
where Ψ is an arbitrary, positive semidefinite matrix, and σs2 > 0 for all 1 ≤ s ≤ S. Here, the key point is that the correlation structure, captured by Ψ, is assumed to be the same across SNPs, whereas the scalar multiplier σs2 is allowed to vary across SNPs. (Of course, this presumes that the same individuals are genotyped at all SNPs. When some individuals have missing genotypes at SNP s, the entries of Ys and the rows and columns of Ψ that correspond to individuals with missing genotypes would be deleted.)
Note that Ψ and σs2 are defined only up to a constant multiple, in the sense that cΨ and c−1σs2 would give the same value of Σs. By convention, σs2 is usually chosen to be the variance of an outbred individual in the absence of population structure. We now give two examples of population genetic models that satisfy our assumptions.
Example 1: Related Individuals without Additional Population Structure
An example of a simple model that satisfies the assumptions of the previous subsection is the model for related individuals in an unstructured population. In this model, individuals in the sample can be related by pedigrees, where the pedigree founders are assumed to be independently drawn from a population that is in Hardy-Weinberg equilibrium (HWE). Mendelian inheritance is assumed in the pedigrees. In this case, it has previously been shown15 that Equations 9 and 10 hold, where ps is interpreted as the allele frequency of SNP s in the population from which the founders are drawn,
If the pedigrees are fully known, then the structure matrix Ψ is known, but if some genealogical information is missing, then Ψ might be partially or completely unknown.
Example 2: Balding-Nichols Model with Admixture
In the Balding-Nichols model6,28,29 with admixture, we let ps denote the “ancestral” allele frequency at SNP s and let qks denote the allele frequency of SNP s in subpopulation k, 1 ≤ k ≤ K. We assume that the qks are random variables that are independent across both k and s, with qks ~Beta(ps(1 − fk)/fk, (1 − ps)(1 − fk)/fk), where fk ≥ 0 can be viewed as Wright's standardized measure of variation30 for subpopulation k. For SNP s, let qs = (q1s, …, qKs)T denote the vector of subpopulation-specific allele frequencies. Individual i is assumed to have admixture vector ai = (ai1, …, aiK)T, where aik ≥ 0 for all i and k, and
Estimation of the Matrix Ψ
The matrix Ψ is a function of the genealogy of the sampled individuals, where genealogy is broadly interpreted as including both population structure and the pedigree relationships of close relatives. The matrix Ψ will be unknown when there is hidden population structure and/or cryptic relatedness in the sample. We allow a completely general form for Ψ, assuming only that it is positive semidefinite (psd). When genome-screen data are available on the sampled individuals, this information can be used to estimate Ψ. For any pair of individuals i and j, let 𝒮ij be the set of markers for which both i and j have nonmissing genotype data. Then if the allele frequencies ps were known, and if σs2 = 0.5ps(1 − ps) as in Examples 1 and 2, an unbiased estimator of Ψij would be
where |𝒮ij| is the number of elements of 𝒮ij. If one assumed, for example, that genotypes at different SNPs were independent with
The estimator
If every sampled individual were genotyped at the same markers, with no missing genotypes, then
Estimation of σ2 when Structure Is Unknown
In this subsection, we drop the subscript s and use notations Y, p, and σ2 for the SNP being tested; e.g., we assume E0(Y) = p1 and Var0(Y) = Σ = σ2Ψ. As we did for the case of known structure, we consider two general approaches to estimation of σ2 when structure is unknown. The first approach is to take estimators of the form
As we did in the case of known structure, we also consider an estimator of σ2 that does not assume σ2 = 0.5p(1 − p) at the SNP being tested. When
where
Association Tests when Structure Is Partially or Completely Unknown
We apply Equation 8 to extend association tests developed for the situation of known structure (as described in subsection Examples of Association Tests with Known Structure) to association tests that are appropriate for situations of partially or completely unknown structure. We call the tests based on Equation 8 the ROADTRIPS versions of the corresponding tests given by Equation 1, and we now give several examples. Table 1 gives the weight vector for each statistic defined below.
Table 1
Weight Vectors V for the ROADTRIPS Statistics
| Statistic | V |
|---|---|
| Rχ | |
| RM | AN + Φ−1ΦN,MAM − (AN + Φ−1ΦN,MAM)T1(1TΦ−11)−1Φ−11 |
| RMNI | |
| RWNI |
Rχ1 and Rχ2, the ROADTRIPS Versions of the Corrected χ2 and Corrected Armitage Tests
To extend the corrected χ2 and corrected Armitage tests to the situation of unknown structure, we apply Equation 8 with V given in Equation 5 and
RM, the ROADTRIPS Version of MQLS when Structure Is Partially Known
As the MQLS is generally the most powerful of the statistics for complex trait mapping when structure is known,16 we expect the ROADTRIPS version of MQLS to be powerful when structure is unknown. We consider separately the cases of partially known structure and completely unknown structure. An example of partially known structure occurs when reliable pedigree information on sampled individuals is available, but one wants to allow for the possibility of additional cryptic relatedness or unknown population structure not captured by the pedigree information. In the context of partially known structure, we compute the matrix Φ of Equation 4 as a function of the known pedigree information. At the same time, we also calculate the estimator
RMNI, the ROADTRIPS Version of MQLS when Structure Is Completely Unknown
For the case of completely unknown structure, we define the RMNI test, which is a ROADTRIPS version of the MQLS, where “NI” stands for “no information.” We form RMNI from Equation 8, where we take
RWNI, the ROADTRIPS Version of WQLS when Structure Is Completely Unknown
We form RWNI from Equation 8, where we take
GAW 15 Rheumatoid Arthritis Data
We apply ROADTRIPS to perform association analysis of RA data provided for GAW 15 by a UK group led by Jane Worthington and Sally John (these data are described in detail elsewhere).22 Data are available on 157 nuclear families, where 156 of these have at least two affected individuals. Individuals were diagnosed as affected according to the American College of Rheumatology (ACR) criteria. There are 550 individuals with available genotype data. After exclusion of 2 duplicate individuals and 4 outlier individuals who have estimated inbreeding coefficients more than 3 standard deviations (SDs) above the average (where the estimated inbreeding coefficient of individual i is taken to be
GAW 14 COGA Data
We apply ROADTRIPS to identify SNPs associated with alcohol dependence in data provided by the COGA for GAW 14.23 These data were previously analyzed with association methods that assume known structure.16 There are a total of 1614 individuals from 143 pedigrees, with each pedigree containing at least three affected individuals. We include in our analysis only those individuals who are coded as “white, non-Hispanic.” We designate as cases those individuals who are affected with ALDX1 or who have symptoms of ALDX1, where ALDX1 is defined to be DSM-III-R alcohol dependence with the Feighner Alc Definite phenotype. By these criteria, there are 830 cases with available SNP data. We designate as “unaffected controls” those individuals who are labeled as “pure unaffected,” and we designate as “controls of unknown phenotype” those individuals who are labeled as “never drank alcohol.” Among individuals with available SNP data, these criteria result in 187 unaffected controls and 13 unknown controls. The data set includes 10,810 autosomal SNPs. We exclude 403 SNPs that are not polymorphic and analyze the remaining 10,407 SNPs.
Results
Simulation Studies
We perform simulation studies, in which population structure and related individuals are simultaneously present in the case-control sample, in order to compare the performance of ROADTRIPS to that of previously proposed association methods that correct in some way for either population structure or related individuals or both. The methods to which we compare ROADTRIPS are GC, FBAT, the method of Rakovski and Stram,13 EIGENSTRAT, MQLS, and the corrected Armitage χ2 test. We also include the standard (uncorrected) Armitage test in the type 1 error study. We simulate four different settings of population structure, including admixture, and two different settings of relationship configuration, where the latter refers to pedigree relationships among sampled individuals.
Relationship Configurations
Both relationship configurations 1 and 2 include 100 unrelated affected individuals, 400 unrelated unaffected individuals, and individuals sampled from 120 outbred, three-generation pedigrees, where each pedigree has a total of 16 individuals, the phenotypes of all individuals in the pedigrees are observed, and genotypes are observed for only a subset of individuals in each pedigree. We sample six types of these pedigrees; the types are described in Table 2. Relationship configuration 1 has 40 pedigrees of type 1, 40 of type 2, and 40 of type 3, as well as 100 unrelated affected and 400 unrelated unaffected individuals. Relationship configuration 2 contains all six types of pedigrees in Table 2, with 20 of each type, and it also contains 100 unrelated affected and 400 unrelated unaffected individuals.
Table 2
Pedigree Configuration Types Used in Simulations
| Type | Naf | Nun | Genotyped Individuals |
|---|---|---|---|
| 1 | 4 | 12 | Unaffected sib pair and their unaffected first cousin |
| 2 | 5 | 11 | 1 affected parent, 2 affected offspring |
| 3 | 6 | 10 | 1 aff. parent, 2 aff. offspr., unaff. sib pair who are 1st cousins to the latter |
| 4 | 4 | 12 | 1 affected parent with 2 affected and 1 unaffected offspring |
| 5 | 5 | 11 | 1 affected and 2 unaffected sibs, unaffected aunt and her affected spouse |
| 6 | 6 | 10 | 1 aff. and 1 unaff. parent with 2 unaff. offspr., 2 other affecteds |
Naf and Nun are the total numbers of affected and unaffected individuals in the pedigree, respectively, among whom only the indicated individuals are genotyped.
Population Structure Settings
Each simulation setting specifies a particular relationship configuration combined with a particular setting of population structure. Each setting of population structure is a special case of the Balding-Nichols model with admixture described in Example 2, in which we take fk = 0.01 for every subpopulation. Population structure 1 has individuals sampled from two subpopulations, with 60% of the pedigrees and affected unrelated individuals sampled from subpopulation 1 and the remaining 40% of the pedigrees and affected unrelated individuals sampled from subpopulation 2. Among the unrelated unaffecteds, 40% are sampled from subpopulation 1 and 60% from subpopulation 2. Population structure 2 is similar to population structure 1, except that the proportions 60% and 40% are replaced by 80% and 20%, respectively. Population structure 3 is similar to population structure 1, except that there are three subpopulations, with all of the unrelated unaffecteds sampled from subpopulation 3. Population structure 4 has individuals sampled from an admixed population, formed from two subpopulations. Individuals in the admixed population are assumed to have i.i.d. admixture vectors of the form (a, 1 − a), where a is a Uniform(0,1) random variable.
Within a given setting of population structure, to simulate the unrelated individuals needed in relationship configurations 1 and 2, we first simulate genotypes according to the chosen setting of population structure, simulate phenotypes conditional on genotypes, and then randomly ascertain 100 affected and 400 unaffected individuals. To sample particular pedigree types within a given setting of population structure, we first simulate genotypes for pedigree founders according to the chosen setting of population structure, drop alleles down the pedigree, simulate phenotypes conditional on genotypes, and then do rejection sampling to obtain 100 replicates of each of the pedigree configuration types. Then, in the simulations, pedigrees of each type are sampled with replacement from the 100 previously obtained replicates of that pedigree type.
Random and Causal SNPs
We use a trait model that has two unlinked causal SNPs (which we call “SNP 1” and “SNP 2”) with epistasis between them.16 The ancestral frequencies of the type 1 alleles at SNPs 1 and 2 are taken to be 0.1 and 0.5, respectively. Individuals with at least one copy of allele 1 at SNP 1 and at least one copy of allele 1 at SNP 2 have a penetrance of 0.3. All other individuals have a penetrance of 0.05. In the power studies, association is tested with SNP 2. In contrast, “random” SNPs are assumed to be unlinked and unassociated with the trait, and their ancestral allele frequencies are obtained as i.i.d. draws from a uniform (0.1, 0.9) distribution.
Assessment of Type 1 Error
For each of the eight combinations of population structure and relationship configuration, we generate genotype data for 100,000 random SNPs that are neither linked nor associated with the trait, and we test each of them for association at the 0.0001 level, using various test statistics. In Table 3, for each test statistic, we report the empirical type 1 error, which we calculate as the proportion of simulations in which the test statistic exceeds the χ12 quantile corresponding to nominal type 1 error level 0.0001. The statistics compared are the four ROADTRIPS statistics, Rχ2, RM2, RMNI2, and RWNI2, where the subscript “2” denotes use of the estimator
Table 3
Empirical Type 1 Error, at Level 0.0001, in the Presence of Both Related Individuals and Population Structure
| Empirical Type 1 Error of Testsa | ||||||||
|---|---|---|---|---|---|---|---|---|
| Settingb | Rχ or RMNI | RM | RWNI | GC | EIG | MQLS | Corr Arm | Arm |
| (1,1) | 0.00009 | 0.00004 | 0.00011 | 0.00007 | 0.00118 | 0.00027 | 0.00018 | 0.00202 |
| (1,2) | 0.00011 | 0.00012 | 0.00011 | 0.00010 | 0.00059 | 0.00043 | 0.00016 | 0.00073 |
| (2,1) | 0.00008 | 0.00012 | 0.00010 | 0.00004 | 0.00116 | 0.01690 | 0.00485 | 0.01970 |
| (2,2) | 0.00013 | 0.00015 | 0.00012 | 0.00005 | 0.00054 | 0.02281 | 0.00723 | 0.01375 |
| (3,1) | 0.00010 | 0.00010 | 0.00010 | 0.00007 | 0.00155 | 0.06752 | 0.02409 | 0.06094 |
| (3,2) | 0.00015 | 0.00012 | 0.00008 | 0.00008 | 0.00058 | 0.08464 | 0.03189 | 0.05028 |
| (4,1) | 0.00007 | 0.00011 | 0.00012 | 0.00007 | 0.00086 | 0.00056 | 0.00032 | 0.00277 |
| (4,2) | 0.00008 | 0.00007 | 0.00012 | 0.00012 | 0.00036 | 0.00028 | 0.00019 | 0.00069 |
Empirical type 1 error rates are calculated based on 100,000 simulated random SNPs. Rates that are significantly different from the nominal 0.0001 level are in bold. Rχ and RMNI are equivalent statistics for all the settings shown. For the ROADTRIPS statistics, the
For GC and all of the ROADTRIPS statistics, empirical type 1 error is not significantly different from the nominal level. In contrast, type 1 error is inflated for EIGENSTRAT, MQLS, the corrected Armitage χ2, and the Armitage trend test. This is to be expected, because these tests either (1) correct for related individuals but not for population structure (MQLS, corrected Armitage χ2), (2) correct for population structure but not for related individuals (EIGENSTRAT), or (3) correct for neither (Armitage trend test). In particular, the top principal components in EIGENSTRAT are not able to capture the complicated covariance structure due to the related individuals in the samples. The results for EIGENSTRAT in Table 3 are obtained with the default setting of ten principal components and with outlier removal. The results without outlier removal and with different numbers of principal components are similar (results not shown). We also performed all the ROADTRIPS tests with variance estimator
Type 1 Error with Missing Genotypes: Rχ2 and GC
When all SNPs have the same pattern of missing genotypes, we expect Rχ2 to perform similarly to GC, because in this case, both Rχ2 and GC are equivalent to correcting all the Armitage χ2 statistics across the genome by a common factor, though this factor differs between the two methods. However, when different SNPs have different rates of missing genotypes, we expect the Rχ2 statistic to have better control of type 1 error than GC, because the Rχ2 statistic allows different SNPs to have different correction factors appropriate to the level of genotype information available, whereas GC applies the same correction factor to all SNPs.
To assess the magnitude of this effect, we perform a simulation study under four different settings of population structure and relationship configuration, given in columns 1 and 2 of Table 4. For each combination of settings, genotype data are generated for 100,000 random SNPs that are neither linked nor associated with the phenotype. The proportions of individuals with missing genotype data at different SNPs are taken to be i.i.d. random variables drawn from a Beta(3, 12) distribution, which has a mean of 0.2 and a SD of 0.1. Given the proportion of missing genotypes at a marker, the individuals whose genotypes will be set to missing for that marker are chosen uniformly at random from the sample.
Table 4
Empirical Type 1 Error at Level 0.0001 when Genotypes are Missing at Random in the Presence of Both Related Individuals and Population Structure
| Empirical Type 1 Error | |||
|---|---|---|---|
| Population Structure | Relationship Configuration | Rχ2 | Genomic Control |
| 2 | 1 | 0.00008 | 0.00031 |
| 2 | 2 | 0.00007 | 0.00036 |
| 3 | 1 | 0.00006 | 0.00057 |
| 3 | 2 | 0.00013 | 0.00058 |
Empirical type 1 error rates are calculated based on 100,000 simulated random SNPs. Rates that are significantly different from the nominal 0.0001 level are in bold.
The empirical type 1 error rates for Rχ2 and GC are given in Table 4. GC has inflated type 1 error for all of the simulation settings, because of undercorrection of test statistics from SNPs that have relatively low levels of missing genotypes, whereas the empirical type 1 error for Rχ2 is not significantly different from the nominal 0.0001 level. The results illustrate that ROADTRIPS is not only robust to cryptic population and pedigree structure, but also to varying rates of randomly missing genotype data. This results from the fact that the entire empirical covariance matrix is estimated in ROADTRIPS, so when some individuals have missing genotype data at the SNP being tested, the corresponding rows and columns can be deleted from the empirical covariance matrix, allowing one to obtain a variance estimator that accounts for missing genotype data at the marker being tested.
Power Comparison
We assess power to detect association in the presence of population and pedigree structure only for those tests that maintain correct nominal type 1 error, namely the four ROADTRIPS tests, FBAT, and GC (although, as can be seen in Table 4, the type 1 error of GC might not be correct when the rate of missing genotype data varies across markers). The simulations are performed with relationship configuration 2 under each of the four different settings of population structure. For each setting, 5000 simulated replicates are performed, and SNP 2 of the trait model is tested for association with the trait by each method. Table 5 reports power for each statistic for settings 1, 2, and 4 of population structure. Here, power is calculated as the proportion of simulations for which the statistic exceeds the χ12 quantile corresponding to nominal type 1 error level 0.0001. Power for population structure 3 is close to 0 for all of the statistics (data not shown). The ROADTRIPS statistics are all calculated with estimator
Table 5
Power to Detect Association in the Presence of Both Related Individuals and Population Structure
| Population Structure | Power (Standard Error) | ||||
|---|---|---|---|---|---|
| Rχ or RMNI | RM | RWNI | GC | FBAT | |
| 1 | 0.79 (0.006) | 0.94 (0.003) | 0.59 (0.007) | 0.78 (0.006) | 0.0012 (0.0005) |
| 2 | 0.42 (0.007) | 0.48 (0.007) | 0.36 (0.007) | 0.43 (0.007) | 0.0016 (0.0006) |
| 4 | 0.70 (0.007) | 0.80 (0.006) | 0.48 (0.007) | 0.70 (0.007) | 0.0002 (0.0002) |
Power is assessed at significance level 0.0001 on the basis of 5000 simulated replicates. The highest power for each simulation setting is in bold. Relationship configuration 2 is used in each case. Rχ and RMNI are equivalent statistics for all the settings shown. For the ROADTRIPS statistics, the
We also assess power to detect association when there is pedigree structure but no population structure. The simulation is carried out as in the preceding paragraph except that relationship configuration 2 with no population structure is simulated. In addition to the tests compared in Table 5, we also calculate power for the MQLS and corrected Armitage χ2 tests, both of which have correct type 1 error in this setting, provided that the pedigree structure is known. Estimated power for this setting is given in Table 6. The MQLS and RM tests are the most powerful among the tests considered. The RM test is able to match the power of MQLS even though RM uses the estimator
Table 6
Power to Detect Association with Related Individuals in the Absence of Population Structure
| Power (Standard Error) | ||||||
|---|---|---|---|---|---|---|
| Rχ or RMNI | RM | RWNI | GC | FBAT | MQLS | Corr Arm |
| 0.90 (0.004) | 0.98 (0.002) | 0.81 (0.006) | 0.91 (0.004) | 0.0002 (0.0002) | 0.98 (0.002) | 0.90 (0.004) |
Power is assessed at significance level 0.0001 on the basis of 5000 simulated replicates of relationship configuration 2 with no population structure. The highest power is in bold. Rχ and RMNI are equivalent statistics in this simulation setting. For the ROADTRIPS statistics, the
GAW 15 Rheumatoid Arthritis Data
We apply RM, Rχ, GC, and FBAT to the GAW 15 data to test for association of SNPs with RA. Using a previously reported22 prevalence of 0.8% for RA in people of European descent, we set both the offset value in FBAT and the prevalence value in RM to 0.008. The entries of the empirical covariance matrix
Table 7
Rheumatoid Arthritis Data Results: SNPs with p Value <0.00005 for at Least One of the Four Tests
| Chr | Marker | Pos. (cM) | NCA | NCO | p | p Value | |||
|---|---|---|---|---|---|---|---|---|---|
| Rχ | RM | GC | FBAT | ||||||
| 11 | snp264363 | 105.57721 | 208 | 133 | 0.86 | 4.7e-3 | 5.2e-7a | 1.7e-2 | 1.2e-2 |
| 9 | snp152076 | 77.047858 | 267 | 162 | 0.92 | 3.0e-2 | 5.4e-7a | 1.1e-1 | 9.9e-3 |
| 11 | snp547632 | 63.954804 | 230 | 135 | 0.80 | 1.2e-6a | 6.2e-2 | 1.8e-4 | 3.0e-1 |
| 3 | snp151721 | 114.80384 | 279 | 174 | 0.84 | 2.8e-4 | 2.2e-6 | 6.8e-4 | 3.0e-3 |
| 18 | snp511091 | 107.60685 | 219 | 130 | 0.57 | 4.2e-2 | 1.1e-5 | 6.6e-2 | 6.6e-2 |
| 14 | snp51741 | 68.2942 | 298 | 188 | 0.95 | 1.4e-3 | 1.2e-5 | 1.3e-3 | 1.5e-2 |
| 15 | snp66639 | 51.864772 | 296 | 187 | 0.97 | 1.5e-5 | 1.0e-2 | 1.4e-4 | NA |
| 3 | snp71651 | 40.817259 | 188 | 118 | 0.74 | 3.5e-2 | 1.8e-5 | 6.3e-2 | 3.6e-2 |
| 4 | snp570108 | 66.453602 | 224 | 142 | 0.71 | 5.1e-2 | 3.9e-5 | 1.2e-1 | 2.4e-1 |
| 8 | snp261673 | 67.427701 | 192 | 131 | 0.61 | 2.9e-1 | 4.6e-5 | 4.0e-1 | 3.7e-1 |
The chromosome (Chr), the name of the marker (Marker), the position of the marker on the chromosome (Pos.), the number of genotypes available in cases (NCA) and controls (NCO), and the major allele frequency in the case-control sample as a whole (p) are displayed. An insufficient number of informative families for the FBAT analysis are indicated by NA.
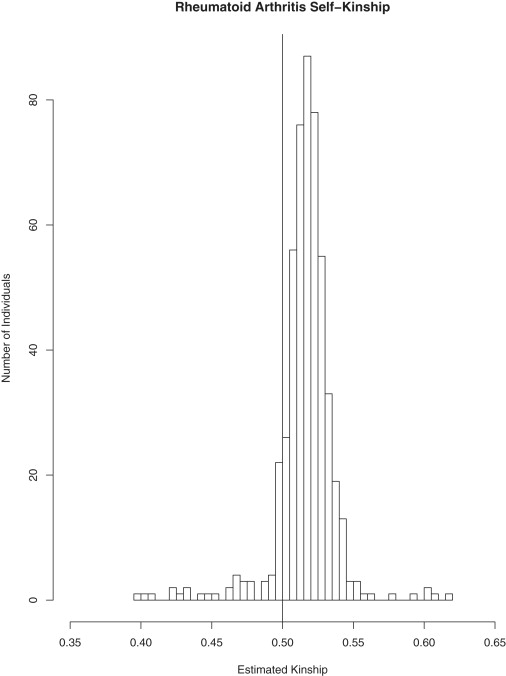
A histogram of the estimated self-kinship values (where the estimated self-kinship value of individual i is taken to be
GAW 14 COGA Data
We apply RM, Rχ, and GC to test for association with an alcoholism-related phenotype in the GAW 14 COGA data. A previous analysis16 of these data used the MQLS (with k set to 0.05) and corrected χ2 tests; these tests correct for known pedigree information, but do not make any correction for unknown structure. In our reanalysis, we compare the results of the tests with and without the correction for unknown structure. To make the results comparable, we set k = 0.05 in the RM test. Table 8 lists SNPs for which at least one of the RM, Rχ, and GC tests has a nominal p value <1.0 × 10−5. For 11 of these 12 SNPs, the RM test has the smallest p value among the three tests used. After Bonferroni correction to adjust for three different tests of association at each of 10,407 SNPs, the RM test is significant at the 5% level for 4 SNPs: tsc1750530 on chromosome 16 (p = 2.3 × 10−8 uncorrected, 0.0007 corrected), tsc0046696 on chromosome 18 (p = 1.4 × 10−7 uncorrected, 0.004 corrected), tsc1177811 on chromosome 1 (p = 1.5 × 10−7 uncorrected, 0.005 corrected), and tsc1637642 on chromosome 5 (p = 6.8 × 10−7 uncorrected, 0.02 corrected). The Rχ test is significant at the 5% level for an additional SNP, tsc0571038 on chromosome 11 (p = 8.0 × 10−7 uncorrected, 0.025 corrected). Of the 5 SNPs that were previously16 identified as genome-wide significant by MQLS or
Table 8
COGA Data Results: SNPs with p Value <0.00001 for at Least One of the Three Tests
| Chr | Marker | Pos. (cM) | NCA | NCO | p | p Value | ||
|---|---|---|---|---|---|---|---|---|
| Rχ | RM | GC | ||||||
| 16 | tsc1750530 | 59.8297 | 644 | 145 | .85 | 3.6e-4 | 2.3e-8a | 1.6e-3 |
| 18 | tsc0046696 | 104.665 | 459 | 118 | .60 | 4.0e-1 | 1.4e-7a | 4.8e-1 |
| 1 | tsc1177811 | 105.535 | 587 | 149 | .68 | 2.9e-2 | 1.5e-7a | 3.4e-2 |
| 5 | tsc1637642 | 95.4901 | 419 | 159 | .84 | 3.6e-1 | 6.8e-7a | 4.3e-2 |
| 11 | tsc0571038 | 95.3968 | 581 | 122 | .56 | 8.0e-7a | 4.5e-3 | 5.4e-5 |
| 18 | tsc0057290 | 33.9594 | 497 | 126 | .71 | 6.1e-2 | 1.8e-6 | 5.3e-2 |
| 6 | tsc0808295 | 47.1522 | 681 | 162 | .76 | 9.4e-1 | 1.8e-6 | 9.4e-1 |
| 11 | tsc0569292 | 6.78451 | 455 | 127 | .74 | 6.0e-1 | 3.6e-6 | 5.7e-1 |
| 19 | tsc1189131 | 68.94 | 478 | 121 | .55 | 7.4e-1 | 4.2e-6 | 7.3e-1 |
| 3 | tsc0175005 | 158.199 | 594 | 152 | .82 | 1.6e-2 | 4.8e-6 | 2.2e-2 |
| 3 | tsc1519933 | 167.431 | 515 | 134 | .64 | 9.5e-2 | 5.3e-6 | 7.6e-2 |
| 13 | tsc0056748 | 73.9934 | 530 | 133 | .84 | 6.5e-1 | 6.5e-6 | 6.7e-1 |
The chromosome (Chr), the name of the marker (Marker), the position of the marker on the chromosome (Pos.), the number of genotypes available in cases (NCA) and controls (NCO), and the major allele frequency in the case-control sample as a whole (p) are displayed.
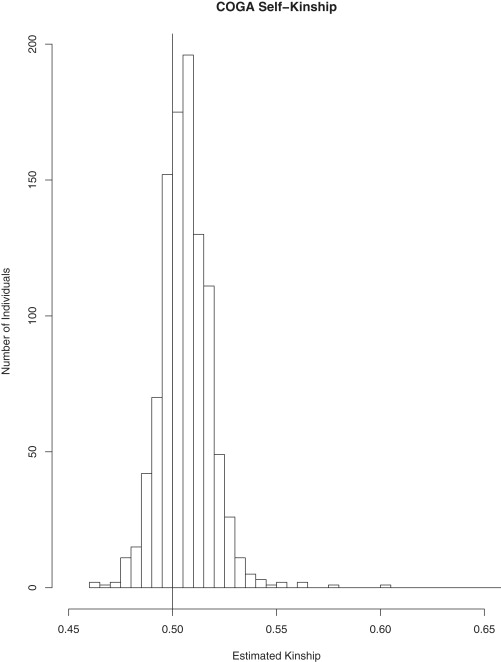
Figure 2 gives a histogram of the estimated self-kinship coefficient values for the genotyped individuals who were included in the analysis. The center of the histogram is shifted from 0.5 (the self-kinship coefficient in the absence of population structure and inbreeding). Seventy-one percent of the values are greater than 0.5, and the mean self-kinship value is 0.506. Just as there were in the UK RA data, in the COGA data there are a few pairs of individuals who appear to have misspecified relationships or be cryptically related. As previously mentioned, ROADTRIPS adjusts for this in the variance correction.
Assessment of Computation Time
Using a single processor on a shared machine with eight quad-core AMD Opteron 8384 25 GHz processors with 64 GB RAM, analysis of 10,156 SNPs from the RA data and 10,810 SNPs from the COGA data with four tests (Rχ, RM, GC, and a ROADTRIPS version of WQLS) took approximately 5 and 21 min, respectively. The large difference in computing time is due to the COGA data having extended pedigrees and a sample size that is almost twice that of the RA data. The slowest step is the Cholesky decomposition33 of Φ (for the calculation of RM and a ROADTRIPS version of WQLS), which we compute at every SNP because the pattern of missing genotype data varies. The computing time scales linearly with the number of SNPs. The speed could presumably be improved, as we have not made serious attempts to optimize the code.
Discussion
Technological advances in high-density genome scans have made it feasible to perform case-control association studies on a genome-wide basis with hundreds of thousands or millions of markers. The observations in these studies can have several sources of dependence, including population structure and relatedness among the sampled individuals, some of which might be known and some unknown. Failure to properly account for this structure can lead to spurious association or reduced power. We develop ROADTRIPS, a case-control association testing method that simultaneously corrects for both pedigree and population structure, including admixture, where some or all of the structure can be unknown. The method also automatically adjusts for pedigree errors and sample switches. ROADTRIPS is computationally feasible for the analysis of genome-screen data with millions of markers, and it is applicable to association studies with completely general combinations of family and case-control designs. The method does not require the genealogy of the sampled individuals to be known, but when pedigree data are available, ROADTRIPS can incorporate this information to improve power. Our simulation studies indicate that including known pedigree information in ROADTRIPS (by use of the RM test) provides an overall and, in some cases, substantial improvement in power over other available methods. In an analysis of GAW 15 RA data from small UK pedigrees, ROADTRIPS detected three SNPs that have significant association with a RA phenotype. In a reanalysis of the GAW 14 COGA data, ROADTRIPS detected five SNPs that have significant association with alcoholism, one of which had not been identified as significant in the previous analysis, and another SNP identified as significant in the previous analysis is no longer identified when cryptic structure is accounted for.
We have shown that when different SNPs have different rates of missing genotype data, ROADTRIPS is still valid, whereas GC is not properly calibrated for this setting. The ROADTRIPS method takes into account both the structure in the data and the particular missing genotype pattern at each SNP to construct a valid test. In contrast, the uniform inflation factor applied to all SNPs by GC can result in both an increase in type 1 error, due to undercorrection of SNPs with lower rates of missing genotype data, as well as a loss of power, due to overcorrection of SNPs with higher rates of missing genotype data. One approach to dealing with this problem in samples of unrelated individuals is to impute missing genotype data. However, there are special difficulties that arise with the use of imputation methods in samples with related individuals and hidden structure. First, Mendelian errors and incompatible genotypes can be introduced with this approach, unless the imputation is performed jointly among related individuals. Second, in samples with hidden population structure, e.g., samples from admixed populations, it is often unclear what the reference population should be, because an individual's ancestry at a particular SNP will generally be unknown. Finally, imputed genotypes are dependent among relatives, where the dependence among imputed genotypes differs from the ordinary dependence among genotypes and is affected by the type and amount of information available for each individual for each SNP. Thus, unlike the pedigree and population structure we consider, the dependence structure among imputed genotypes for different individuals differs across SNPs. This complex dependence among imputed genotypes for related individuals would need to be taken into account in the analysis in order to construct a valid test. However, to our knowledge, the current generation of imputation methods gives information only on the marginal accuracy (e.g., marginal posterior probabilities and not joint posterior probabilities) of imputed genotypes across individuals, so these methods would not allow valid assessment of uncertainty in the general setting of case-control association testing with related individuals.
The ROADTRIPS method uses an estimator of Ψ that is closely related to the estimated covariance matrix used in EIGENSTRAT.6 Recently, Choi et al.17 have used a different estimated kinship matrix in the context of association testing when pedigree information is missing. The kinship estimation approach of Choi et al. is suitable for close relatives in the absence of population structure but is not particularly well suited to accounting for population structure. To make this statement more precise, we consider the case of unrelated individuals with population structure based on the Balding-Nichols model with or without admixture. In this context, if one assumed that genotypes at different SNPs were independent with
Web Resources
The URLs for data presented herein are as follows:
ROADTRIPS source code, http://www.stat.uchicago.edu/~mcpeek/software/index.html
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Acknowledgments
This study was supported in part by the University of California President's Postdoctoral Fellowship and the Lamond Family Foundation Postdoctoral Fellowship (to T.T.) and by National Institutes of Health grant R01 HG001645 (to M.S.M.). Data on alcohol dependence were provided by the Collaborative Study on the Genetics of Alcoholism (U10AA008401) through the Genetics Analysis Workshop (R01GM031575). Data on RA were provided by a UK group led by Jane Worthington and Sally John through the Genetics Analysis Workshop (R01GM031575).
References
Articles from American Journal of Human Genetics are provided here courtesy of American Society of Human Genetics
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ajhg.2010.01.001
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0002929710000029/pdf
Citations & impact
Impact metrics
Article citations
Using GWAS and Machine Learning to Identify and Predict Genetic Variants Associated with Foodborne Bacteria Phenotypic Traits.
Methods Mol Biol, 2852:223-253, 01 Jan 2025
Cited by: 0 articles | PMID: 39235748
Review
Estimation of inbreeding and kinship coefficients via latent identity-by-descent states.
Bioinformatics, 40(2):btae082, 01 Feb 2024
Cited by: 0 articles | PMID: 38364309 | PMCID: PMC10902678
RETROSPECTIVE VARYING COEFFICIENT ASSOCIATION ANALYSIS OF LONGITUDINAL BINARY TRAITS: APPLICATION TO THE IDENTIFICATION OF GENETIC LOCI ASSOCIATED WITH HYPERTENSION.
Ann Appl Stat, 18(1):487-505, 31 Jan 2024
Cited by: 2 articles | PMID: 38577266 | PMCID: PMC10994004
A resource of induced pluripotent stem cell (iPSC) lines including clinical, genomic, and cellular data from genetically isolated families with mood and psychotic disorders.
Transl Psychiatry, 13(1):397, 16 Dec 2023
Cited by: 4 articles | PMID: 38104115 | PMCID: PMC10725500
Genetic association models are robust to common population kinship estimation biases.
Genetics, 224(1):iyad030, 01 May 2023
Cited by: 1 article | PMID: 36843304 | PMCID: PMC10474929
Go to all (112) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (2)
- (1 citation) OMIM - 103780
- (1 citation) OMIM - 180300
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Fast Genome-Wide QTL Association Mapping on Pedigree and Population Data.
Genet Epidemiol, 41(3):174-186, 12 Dec 2016
Cited by: 7 articles | PMID: 27943406 | PMCID: PMC5340631
A comparison of association methods correcting for population stratification in case-control studies.
Ann Hum Genet, 75(3):418-427, 31 Jan 2011
Cited by: 57 articles | PMID: 21281271 | PMCID: PMC3215268
Application of a new method for GWAS in a related case/control sample with known pedigree structure: identification of new loci for nephrolithiasis.
PLoS Genet, 7(1):e1001281, 20 Jan 2011
Cited by: 6 articles | PMID: 21283782 | PMCID: PMC3024262
Relatedness in the post-genomic era: is it still useful?
Nat Rev Genet, 16(1):33-44, 18 Nov 2014
Cited by: 134 articles | PMID: 25404112
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: K01 CA148958
NHGRI NIH HHS (1)
Grant ID: R01 HG001645
NIAAA NIH HHS (1)
Grant ID: U10 AA008401
NIGMS NIH HHS (2)
Grant ID: R01 GM031575-28
Grant ID: R01 GM031575