Abstract
Free full text

Presenilins and the γ-secretase: still a complex problem
Abstract
The presenilins form part of a complex of membrane proteins that are involved in the proteolytic cleavage of cell-surface molecules. This article reviews the history of the discovery of the presenilins, their role in the pathogenesis of Alzheimer's disease and in the metabolism of the amyloid-β precursor protein. Unanswered questions about their biochemical mechanism of action and their effects on Ca2+ homeostasis are examined.
Alzheimer's disease (AD) is the most common cause of dementia in the elderly. Typically 5-10% of the population over the age of 65 have dementia, and of these cases, a large percentage have AD [1]. AD is characterised by the presence of proteinaceous deposits in the brain [2]. The extracellular amyloid deposits, which are found in the neuropil (amyloid plaques) and in association with small-medium size cerebral blood vessels (cerebral amyloid angiopathy), are composed of a 4 kDa polypeptide known as the amyloid-β protein (Aβ) which is derived by proteolytic cleavage from a much larger amyloid-β precursor protein (APP) [3]. Aβ displays a spontaneous ability to aggregate into oligomers and larger fibrillar structures, and it is generally thought that the accumulation of oligomeric Aβ is chiefly responsible for the neurodegeneration that occurs in AD [4].
For the generation of Aβ, APP is first cleaved on the N-terminal side of the Aβ sequence by the β-site APP cleaving enzyme-1 (BACE1), a transmembrane aspartyl protease [3]. The resulting 99-amino acid residue C-terminal fragment (C99) is then cleaved by the γ-secretase to yield Aβ and a C-terminal APP intracellular domain (AICD) fragment (Fig. (Fig.1).1). The function of the AICD fragment is unclear, although it is thought to have a role in intracellular signalling. For example, AICD may be involved in the regulation of gene transcription, synaptic plasticity and cytoskeletal dynamics [5].
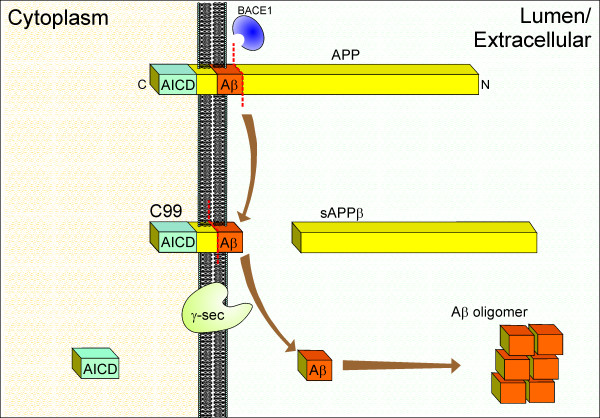
Amyloidogenic processing of the β-amyloid precursor protein (APP) by BACE1 and γ-secretase. Initially, BACE1 cleaves APP on the N-terminal end of the Aβ sequence to yield a large secreted N-terminal fragment (sAPPβ) and a smaller membrane-associated C-terminal stub (C99), which is then cleaved by the γ-secretase complex to yield Aβ and an APP intracellular domain (AICD). Secreted Aβ aggregates in the extracellular environment to form neurotoxic oligomers.
The major form of Aβ possesses 40 amino-acid residues (Aβ1-40). However, other minor species are also produced which vary in the C-terminal sequence. Production of a longer 42-residue species (Aβ1-42) is thought to be intimately associated with AD pathogenesis [6]. Aβ1-42 aggregates more readily than Aβ1-40, and increased production of Aβ1-42 may seed aggregation of Aβ1-40 or other Aβ species [4].
Genetic clues to the pathogenesis of AD
Approximately 5% of all AD cases are autosomal dominant [7]. Soon after the complete APP sequence was cloned in 1987 [8], it became clear that at least one familial AD (FAD) locus was located on chromosome 21 [9] and attention turned to the APP gene that had previously been localised to a region within chromosome 21. The first FAD mutation was identified within the APP gene [10], and soon after, a number of other APP mutations were also identified [11-13]. All of the FAD mutations in the APP gene cluster around the region encoding the Aβ sequence, suggesting that they have some effect on the aggregation or proteolytic processing of APP.
APP mutations on chromosome 21 account for only a small fraction of the total number of FAD cases. It was clear that multiple FAD loci existed on other chromosomes. The first evidence for an FAD locus on chromosome 14 [14] came well before the identification of the locus on chromosome 21. Then, in 1995, FAD mutations in two presenilin (PS) genes located on chromosome 14 (presenilin-1, PS1) and chromosome 1 (presenilin-2, PS2) were reported [15-17]. The PS genes encode proteins that are homologous to the C. elegans sel-12 gene, which is known to be involved in Notch signalling [18]. This observation provided that first clue that PS1 may be involved in cell-surface receptor signalling. To date, >100 FAD mutations have been found in PS1 and 11 mutations in PS2 [19]. The PS genes encode proteins with 8 or 9 transmembrane domains. The proteins are synthesized as ~50 kDa proteins that are subsequently cleaved by a presenilinase into a ~30 kDa N-terminal fragment and a ~20 kDa C-terminal fragment, which remain associated with each other [20].
PS mutations are linked to γ-secretase activity [21]. Specifically, it has been observed that FAD mutations in PS1 increase the proportion of C99 that is cleaved by γ-secretase at position 42 of the Aβ sequence. The altered cleavage pattern causes increased production of the more pathogenic Aβ1-42. While the first impression may be that these are "gain-of-function" mutations, such a conclusion is difficult to reconcile with the large number of FAD mutations that have been identified, particularly in PS1. Instead, mutations in PS are more likely to be "loss-of-function" mutations [22] in which a decrease in the rate of γ-secretase cleavage of APP leads to an increase in the proportion of Aβ1-42. PS1 knockout has been shown to cause an 80% decrease in Aβ production [23], while combined PS1 and PS2 knockout abolishes γ-secretase activity and hence Aβ production [24]. In addition, γ-secretase activity co-purifies with a high molecular weight complex that contains PS1 and several other proteins (nicastrin, aph-1 and pen-2). It is now known that the γ-secretase consists of a complex of proteins of which PS, nicastrin, aph-1 and pen-2 are the principal components. Expression of all 4 proteins in cells is necessary for γ-secretase activity [25].
Inhibitor studies demonstrate that the γ-secretase is a member of the aspartyl protease family [26]. All members of this family require two aspartyl residues for enzyme activity [27]. Some aspartyl proteases (e.g. BACE1) have two aspartyl residues within a single subunit, but other proteases have only one aspartyl residue, and therefore dimerization is needed to activate the enzyme. The amino-acid sequence of both PS1 and PS2 contains two conserved aspartyl residues within two domains predicted to be membrane spanning. These two aspartyl residues are thought to form part of the catalytic domain [28]. In support of this idea, mutation of these two residues has been shown to cause loss of γ-secretase activity [29]. In addition, affinity labelling experiments demonstrate that γ-secretase inhibitors bind directly to PS [30,31].
While the exact number of γ-secretase substrates is unknown, a large number of transmembrane proteins are reportedly cleaved by the enzyme [32,33]. Some of the γ-secretase substrates (other than APP) include APLP2, Notch, Delta, and tumour necrosis factor-α converting enzyme (TACE). Of these proteins, Notch and Delta may be the most important as some abnormalities or toxicities associated with γ-secretase inhibition or knockdown could be due to failure of the Notch/Delta signalling pathway [34]. Cleavage of Notch by γ-secretase produces a Notch intracellular domain fragment (NICD), the counterpart of the AICD produced from APP. Like AICD, the NICD has an important signalling functions. For example, NICD can translocate to the nucleus where it activates the transcription factor CBF1/JBP-Jkappa, regulating downstream gene expression [35]. Because γ-secretase inhibition could lead to unwanted side effects or toxicities, its potential as a therapeutic target for AD is uncertain. Unless a method can be found to inhibit γ-secretase processing without inhibiting other proteolytic cleavage events, it may be difficult to develop a successful AD therapeutic based on γ-secretase inhibition,.
However, there may be ways around this problem. For example, if a successful AD therapy can be achieved by only partially lowering Aβ production, rather than by abolishing Aβ production, then it may possible to use doses of a γ-secretase inhibitor that are low enough to produce sufficient inhibition of the γ-secretase for therapeutic purposes, but which avoid some of the unwanted side effects. Such a strategy could conceivably be employed in combination with other anti-Aβ agents (e.g. β-secretase inhibitors), if they are available.
Unanswered questions about PS
While there is now clear consensus that PS forms part of the γ-secretase complex, there are still many unanswered questions. One question is how the PS family of proteins evolved. Although there are superficial morphological similarities between PSs and some other proteases, the PSs and their homologues do not share any significant amino-acid sequence homology with known proteases or hydrolases. Presumable, any similarities PS shares with other aspartyl proteases (e.g. mechanism, substrate specificity, inhibitor profile etc.) must have arisen through convergent evolution. The evolutionary history of the PSs is further complicated by the finding that a PS homologue is present in plants where it performs functions apparently unrelated to γ-secretase activity [36]. Is it possible that PS has a function unrelated to proteolytic cleavage? There is certainly evidence for this idea (see below).
A second question is how intramembranous proteolysis of APP occurs. Both the scissile bond in APP and the active site aspartates in PS are buried deep in the lipid membrane [28], and it is difficult to understand how hydrolytic cleavage can occur in such a non-aqueous environment. Attempts to determine a 3-dimensional structure for the γ-secretase complex using cryo-electron microscopy have not yet resolved this issue [37,38].
Four major families of proteins which catalyse regulated intramembranous proteolysis (RIP) have been reported, the presenilin/γ-secretase family and the signal peptide proteases, which are aspartyl proteases, the site 2 proteases (metalloproteases) and the rhomboid proteases (serine proteases) [39]. The crystal structure of a rhomboid protease [40] provides some clues as to how the γ-secretase could cleave APP. It is clear that the active site in the rhomboid proteases is not buried deep within the lipid bilayer, but is instead formed by a V-shaped opening that faces laterally on one site of the lipid membrane and is exposed to the aqueous environment. The structure of rhomboid proteases suggests that substrates may extend into the catalytic domain where they are cleaved. Thus, although the region that is cleaved may be within the transmembrane domain, the scissile bond must be outside a lipid environment for cleavage to occur.
A similar mechanism could occur within the γ-secretase/PS complex (Fig. (Fig.2).2). Indeed, the idea that the transmembrane domain of APP must be partially uncoiled in order to be cleaved is quite attractive, because it could explain why α-secretase or β-secretase cleavage occurs first, before γ-secretase cleavage. It is tempting to speculate that α- or β-secretase cleavage could destabilise the structure of APP within the membrane, causing the protein to slip in such a manner that it can enter the active site of the γ-secretase complex (Fig. (Fig.22).
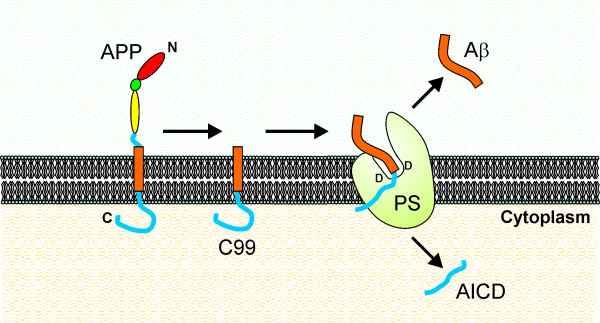
Hypothetical model showing how γ-secretase/PS may cleave C99 to yield Aβ. In this model, PS contains 2 catalytic aspartyl residues (D) that form part of the active site. The two residues are in an aqueous environment formed by a pocket on one side of the membrane. After cleavage of APP by BACE1, the product, C99, is destabilised and slips into the active site where it is cleaved to form Aβ and the APP intracellular domain (AICD).
The possibility that C99 needs to partially slip out of the membrane may also explain why γ-secretase cleaves APP at multiple sites. While the commonly held view is that γ-secretase cleavage involves the two main cleavage sites (positions 40 and 42 of the Aβ sequence), the actual cleavage pattern is much more complex. Several different cleavage sites close to the C-terminal end of the Aβ sequence have been identified. For example, several C-terminally truncated Aβ species can be produced, indicating that other cleavage sites exist [41]. It might be expected that if the first γ-secretase cleavage occurs at position 40 or 42, that the AICD fragment would then commence its N-terminus at position 41 or 43. However, this is not the case. Most studies indicate that AICD begins at or close to position 49, which is referred to as the ε cleavage site [42]. In addition to this site, a ζ-cleavage site has been identified at position 46 [43]. It is highly likely that the γ-secretase cleaves at this ζ-site as well, as ζ-cleavage is inhibited by γ-secretase inhibitors [43]. Cleavage of C99 could occur sequentially with the total amount of slippage of the C99 peptide dictating the final cleavage position. Of course, such a possibility must remain speculative until the idea is tested experimentally.
Another question is whether PSs have functions that are entirely unrelated to proteolytic activity. Numerous studies suggest that PS functions to regulate diverse activities such as Wnt signalling, neurogenesis, cell adhesion, synapse formation and apoptosis [44]. It is not the aim of this review to cover this extremely diverse group of studies, as it has been well reviewed previously [44]. The central question is whether these diverse functions can be explained by a γ-secretase activity. In this context, it is worth noting that some functions of PS do appear to be unrelated to γ-secretase. For example, although PS1 interacts with β-catenin, γ-secretase activity is not needed for this interaction as it is not affected by γ-secretase inhibitors [45].
The plot thickens - the role of calcium
Despite the evidence that PS forms the catalytic subunit of the γ-secretase complex, another putative role of PS has emerged. Recent studies show that mutations in PS disrupt Ca2+ homeostasis; however, the cause of this disruption remains obscure. Dysregulation of intracellular Ca2+ levels is thought to be an important mechanism in AD pathogenesis [46]. Therefore, PS-mediated effects on Ca2+ may be important for understanding the pathogenesis of AD. However, the question is whether PS mutations cause AD by influencing APP metabolism, Ca2+ levels, or both. It has been proposed that PS is an ion channel in the endoplasmic reticulum (ER) and that the FAD mutations increase ion channel permeability [47]. However, other mechanisms seem likely. Studies have shown that PS-regulates the release of Ca2+ via ryanodine or inositol 1,4,5-trisphosphate (IP3) channels [48,49], although, once again, the mechanism is unclear. PS mutations alter PIP2 metabolism and regulate cation flux through transient receptor potential M7 channels [49], and more recently, Cheung et al. [50] have shown that PS regulates Ca2+ channel gating via a mechanism involving the IP3 receptor. PS mutations may also decrease the activity of the sarco ER Ca2+ ATPase (SERCA) pump [51].
However, it is the link between γ-secretase cleavage and intracellular Ca2+ stores which is the most intriguing aspect. Cheung et al. [50] found that PS mutant-induced enhancement of Aβ secretion can be abolished by IP3 receptor knockout, indicating that γ-secretase activity is controlled downstream of PS by IP3. How this finding relates to the hypothesis that PS is the catalytic subunit of the γ-secretase needs to be explored further.
Conclusions
While much progress has been made in elucidating the structure and metabolism of PS, its binding partners and the relationship between FAD mutations and Aβ production, many questions remain unanswered. Experiments involving gene knockout, overexpression and mutagenesis, and affinity labelling argue strongly that PS is the catalytic subunit of the γ-secretase. However, the role of other components of the γ-secretase complex (nicastrin, aph1, pen2) remains uncertain. The mechanism by which γ-secretase cleaves within the transmembrane domain also remains unclear. Does PS cleave in the hydrophobic environment of the lipid membrane, or does the substrate of cleavage (C99 or C83) slip out of the lipid bilayer prior to cleavage? The latter mechanism seems like a real possibility as it may explain why α-secretase or β-secretase cleavage of APP is required before γ-secretase cleavage. It is certainly possible that cleavage at the N-terminus of the Aβ sequence may destabilise the C-terminal peptide, allowing for some slippage within the membrane.
Finally, the role of PS in the release of intracellular calcium stores needs to be understood. Of particular significance here are the findings that FAD mutations influence inositol phosphate signalling and that inositol phosphate signalling can, in turn, regulate Aβ production. While it is possible that FAD mutations have multiple effects which converge on Aβ metabolism, until the precise mechanism by which PS mutations influence Aβ is understood, there will continue to be more questions than answers.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DHS, DWK and LF wrote this article jointly. All authors read and approved the final manuscript.
Acknowledgements
DHS and LF are funded by project grants from the National Health and Medical Research Council of Australia.
References
- Storey E, Kinsella GJ, Slavin MJ. The neuropsychological diagnosis of Alzheimer's disease. J Alzheimers Dis. 2001;3:261–285. [Abstract] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. 10.1073/pnas.82.12.4245. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nunan J, Small DH. Regulation of APP cleavage by alpha-, beta- and gamma-secretases. FEBS Lett. 2000;483:6–10. 10.1016/S0014-5793(00)02076-7. [Abstract] [CrossRef] [Google Scholar]
- Jarrett JT, Lansbury PT Jr. Seeding "one-dimensional crystallization" of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73:1055–1058. 10.1016/0092-8674(93)90635-4. [Abstract] [CrossRef] [Google Scholar]
- Muller T, Meyer HE, Egensperger R, Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics - relevance for Alzheimer's disease. Prog Neurobiol. 2008;85:393–406. 10.1016/j.pneurobio.2008.05.002. [Abstract] [CrossRef] [Google Scholar]
- Walsh DM, Selkoe DJ. A beta oligomers - a decade of discovery. J Neurochem. 2007;101:1172–1184. 10.1111/j.1471-4159.2006.04426.x. [Abstract] [CrossRef] [Google Scholar]
- Bertram L. Alzheimer's disease genetics current status and future perspectives. Int Rev Neurobiol. 2009;84:167–184. full_text. [Abstract] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. 10.1038/325733a0. [Abstract] [CrossRef] [Google Scholar]
- Patterson D, Gardiner K, Kao FT, Tanzi R, Watkins P, Gusella JF. Mapping of the gene encoding the beta-amyloid precursor protein and its relationship to the Down syndrome region of chromosome 21. Proc Natl Acad Sci USA. 1988;85:8266–8270. 10.1073/pnas.85.21.8266. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. 10.1038/349704a0. [Abstract] [CrossRef] [Google Scholar]
- Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease. Science. 1991;254:97–99. 10.1126/science.1925564. [Abstract] [CrossRef] [Google Scholar]
- Chartier-Harlin MC, Crawford F, Houlden H, Warren A, Hughes D, Fidani L, Goate A, Rossor M, Roques P, Hardy J. Early-onset Alzheimer's disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature. 1991;353:844–846. 10.1038/353844a0. [Abstract] [CrossRef] [Google Scholar]
- Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–347. 10.1038/ng0892-345. [Abstract] [CrossRef] [Google Scholar]
- Weitkamp LR, Nee L, Keats B, Polinsky RJ, Guttormsen S. Alzheimer disease: evidence for susceptibility loci on chromosomes 6 and 14. Am J Hum Genet. 1983;35:443–453. [Europe PMC free article] [Abstract] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. 10.1038/375754a0. [Abstract] [CrossRef] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. 10.1126/science.7638622. [Abstract] [CrossRef] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–778. 10.1038/376775a0. [Abstract] [CrossRef] [Google Scholar]
- Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature. 1995;377:351–354. 10.1038/377351a0. [Abstract] [CrossRef] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. 10.1038/nrn2494. [Abstract] [CrossRef] [Google Scholar]
- Ward RV, Davis JB, Gray CW, Barton AJ, Bresciani LG, Caivano M, Murphy VF, Duff K, Hutton M, Hardy J. Presenilin-1 is processed into two major cleavage products in neuronal cell lines. Neurodegeneration. 1996;5:293–298. 10.1006/neur.1996.0040. [Abstract] [CrossRef] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–870. 10.1038/nm0896-864. [Abstract] [CrossRef] [Google Scholar]
- Shen J, Kelleher RJ. The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA. 2007;104:403–409. 10.1073/pnas.0608332104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. 10.1038/34910. [Abstract] [CrossRef] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Total inactivation of gamma-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol. 2000;2:461–462. 10.1038/35017105. [Abstract] [CrossRef] [Google Scholar]
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. 10.1038/ncb960. [Abstract] [CrossRef] [Google Scholar]
- Evin G, Cappai R, Li QX, Culvenor JG, Small DH, Beyreuther K, Masters CL. Candidate gamma-secretases in the generation of the carboxyl terminus of the Alzheimer's disease beta A4 amyloid: possible involvement of cathepsin D. Biochemistry. 1995;34:14185–14192. 10.1021/bi00043a024. [Abstract] [CrossRef] [Google Scholar]
- Beher D, Graham SL. Protease inhibitors as potential disease-modifying therapeutics for Alzheimer's disease. Expert Opin Investig Drugs. 2005;14:1385–1409. 10.1517/13543784.14.11.1385. [Abstract] [CrossRef] [Google Scholar]
- Wolfe MS. Therapeutic strategies for Alzheimer's disease. Nat Rev Drug Discov. 2002;1:859–866. 10.1038/nrd938. [Abstract] [CrossRef] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. 10.1038/19077. [Abstract] [CrossRef] [Google Scholar]
- Esler WP, Kimberly WT, Ostaszewski BL, Diehl TS, Moore CL, Tsai JY, Rahmati T, Xia W, Selkoe DJ, Wolfe MS. Transition-state analogue inhibitors of gamma-secretase bind directly to presenilin-1. Nat Cell Biol. 2000;2:428–434. 10.1038/35017062. [Abstract] [CrossRef] [Google Scholar]
- Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. 10.1038/35015085. [Abstract] [CrossRef] [Google Scholar]
- Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of gamma-secretase substrates and mapping of substrate requirements. PLoS Biol. 2008;6:e257. 10.1371/journal.pbio.0060257. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Magold AI, Cacquevel M, Fraering PC. Gene expression profiling in cells with enhanced gamma-secretase activity. PLoS One. 2009;4:e6952. 10.1371/journal.pone.0006952. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Imbimbo BP. Therapeutic potential of gamma-secretase inhibitors and modulators. Curr Top Med Chem. 2008;8:54–61. 10.2174/156802608783334015. [Abstract] [CrossRef] [Google Scholar]
- Lai EC. Notch signalling: control of cell communication and cell fate. Development. 2004;131:965–973. 10.1242/dev.01074. [Abstract] [CrossRef] [Google Scholar]
- Khandelwal A, Chandu D, Roe CM, Kopan R, Quatrano RS. Moonlighting activity of presenilin in plants is independent of γ-secretase and evolutionarily conserved. Proc Natl Acad Sci USA. 2007;104:13337–13342. 10.1073/pnas.0702038104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Osenkowski P, Li H, Ye W, Li D, Aeschbach L, Fraering PC, Wolfe MS, Selkoe DJ. Cryoelectron microscopy structure of purified gamma-secretase at 12 A resolution. J Mol Biol. 2009;385:642–652. 10.1016/j.jmb.2008.10.078. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci USA. 2006;103:6889–6894. 10.1073/pnas.0602321103. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. 10.1146/annurev.genet.38.072902.093416. [Abstract] [CrossRef] [Google Scholar]
- Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. 10.1038/nature05255. [Abstract] [CrossRef] [Google Scholar]
- Wang R, Sweeney D, Gandy SE, Sisodia SS. The profile of soluble amyloid beta protein in cultured cell media. Detection and quantification of amyloid beta protein and variants by immunoprecipitation-mass spectrometry. J Biol Chem. 1996;271:31894–31902. 10.1074/jbc.271.50.31894. [Abstract] [CrossRef] [Google Scholar]
- Weidemann Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Evin G. A novel epsilon-cleavage within the transmembrane domain of the amyloid precursor protein demonstrates homology with Notch processing. Biochemistry. 2002;41:2825–2835. 10.1021/bi015794o. [Abstract] [CrossRef] [Google Scholar]
- Xu X. γ-Secretase catalyzes sequential cleavages of the Aβ PP transmembrane domain. J Alzheimer Dis. 2009;16:211–224. [Europe PMC free article] [Abstract] [Google Scholar]
- Thinakaran G, Parent AT. Identification of the role of presenilins beyond Alzheimer's disease. Pharmacol Res. 2004;50:411–418. 10.1016/j.phrs.2003.12.026. [Abstract] [CrossRef] [Google Scholar]
- Meredith JE Jr, Wang Q, Mitchell TJ, Olson RE, Zaczek R, Stern AM, Seiffert D. Gamma-secretase activity is not involved in presenilin-mediated regulation of beta-catenin. Biochem Biophys Res Commun. 2002;299:744–750. 10.1016/S0006-291X(02)02747-X. [Abstract] [CrossRef] [Google Scholar]
- Small DH, Gasperini R, Vincent AJ, Hung AC, Foa L. The role of Abeta-induced calcium dysregulation in the pathogenesis of Alzheimer's disease. J Alzheimers Dis. 2009;16:225–233. [Abstract] [Google Scholar]
- Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer's disease-linked mutations. Cell. 2006;126:981–993. 10.1016/j.cell.2006.06.059. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chan SL, Mayne M, Holden CP, Geiger JD, Mattson MP. Presenilin-1 mutations increase levels of ryanodine receptors and calcium release in PC12 cells and cortical neurons. J Biol Chem. 2000;275:18195–18200. 10.1074/jbc.M000040200. [Abstract] [CrossRef] [Google Scholar]
- Landman N, Jeong SY, Shin SY, Voronov SV, Serban G, Kang MS, Park MK, Di Paolo G, Chung S, Kim TW. Presenilin mutations linked to familial Alzheimer's disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci USA. 2006;103:19524–19529. 10.1073/pnas.0604954103. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cheung KH, Shineman D, Muller M, Cardenas C, Mei L, Yang J, Tomita T, Iwatsubo T, Lee VM, Foskett JK. Mechanism of Ca2+ disruption in Alzheimer's disease by presenilin regulation of InsP3 receptor channel gating. Neuron. 2008;58:871–883. 10.1016/j.neuron.2008.04.015. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Green KN, Demuro A, Akbari Y, Hitt BD, Smith IF, Parker I, LaFerla FM. SERCA pump activity is physiologically regulated by presenilin and regulates amyloid beta production. J Cell Biol. 2008;181:1107–1116. 10.1083/jcb.200706171. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from Molecular Brain are provided here courtesy of BMC
Full text links
Read article at publisher's site: https://doi.org/10.1186/1756-6606-3-7
Read article for free, from open access legal sources, via Unpaywall:
https://molecularbrain.biomedcentral.com/counter/pdf/10.1186/1756-6606-3-7
Citations & impact
Impact metrics
Citations of article over time
Article citations
Novel Applications of NSAIDs: Insight and Future Perspectives in Cardiovascular, Neurodegenerative, Diabetes and Cancer Disease Therapy.
Int J Mol Sci, 22(12):6637, 21 Jun 2021
Cited by: 11 articles | PMID: 34205719 | PMCID: PMC8235426
Review Free full text in Europe PMC
Concordance of Several Subcellular Interactions Initiates Alzheimer's Dementia: Their Reversal Requires Combination Treatment.
Am J Alzheimers Dis Other Demen, 32(3):166-181, 17 Mar 2017
Cited by: 11 articles | PMID: 28423937 | PMCID: PMC10852791
Review Free full text in Europe PMC
Analyzing and Modeling the Kinetics of Amyloid Beta Pores Associated with Alzheimer's Disease Pathology.
PLoS One, 10(9):e0137357, 08 Sep 2015
Cited by: 21 articles | PMID: 26348728 | PMCID: PMC4562663
Role of gamma-secretase in human umbilical-cord derived mesenchymal stem cell mediated suppression of NK cell cytotoxicity.
Cell Commun Signal, 12:63, 30 Sep 2014
Cited by: 30 articles | PMID: 25266361 | PMCID: PMC4195898
Discovery of candidate genes for muscle traits based on GWAS supported by eQTL-analysis.
Int J Biol Sci, 10(3):327-337, 10 Mar 2014
Cited by: 31 articles | PMID: 24643240 | PMCID: PMC3957088
Go to all (12) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Presenilins: how much more than γ-secretase?!
Biochem Soc Trans, 38(6):1474-1478, 01 Dec 2010
Cited by: 9 articles | PMID: 21118110
Presenilins as endoplasmic reticulum calcium leak channels and Alzheimer's disease pathogenesis.
Sci China Life Sci, 54(8):744-751, 24 Jul 2011
Cited by: 8 articles | PMID: 21786197
Review
Beyond γ-secretase activity: The multifunctional nature of presenilins in cell signalling pathways.
Cell Signal, 28(1):1-11, 21 Oct 2015
Cited by: 59 articles | PMID: 26498858
Review
Nicastrin is dispensable for gamma-secretase protease activity in the presence of specific presenilin mutations.
J Biol Chem, 284(19):13013-13022, 02 Mar 2009
Cited by: 28 articles | PMID: 19254953 | PMCID: PMC2676034

 1,2
1,2 


