Abstract
Free full text

A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates
Abstract
Cell therapy holds promise for tissue regeneration, including in individuals with advanced heart failure. However, treatment of heart disease with bone marrow cells and skeletal muscle progenitors has had only marginal positive benefits in clinical trials, perhaps because adult stem cells have limited plasticity. The identification, among human pluripotent stem cells, of early cardiovascular cell progenitors required for the development of the first cardiac lineage would shed light on human cardiogenesis and might pave the way for cell therapy for cardiac degenerative diseases. Here, we report the isolation of an early population of cardiovascular progenitors, characterized by expression of OCT4, stage-specific embryonic antigen 1 (SSEA-1), and mesoderm posterior 1 (MESP1), derived from human pluripotent stem cells treated with the cardiogenic morphogen BMP2. This progenitor population was multipotential and able to generate cardiomyocytes as well as smooth muscle and endothelial cells. When transplanted into the infarcted myocardium of immunosuppressed nonhuman primates, an SSEA-1+ progenitor population derived from Rhesus embryonic stem cells differentiated into ventricular myocytes and reconstituted 20% of the scar tissue. Notably, primates transplanted with an unpurified population of cardiac-committed cells, which included SSEA-1– cells, developed teratomas in the scar tissue, whereas those transplanted with purified SSEA-1+ cells did not. We therefore believe that the SSEA-1+ progenitors that we have described here have the potential to be used in cardiac regenerative medicine.
Introduction
The embryonic human heart starts beating at around 20 days of gestation and is the target of many genetic and congenital diseases likely originating from the early and complex formation of the myocardium. Over the past decades, cardiac progenitors have been tracked in embryos and appeared as a very early lineage, determined in the primitive streak of chicks (1) as well as mammals (2), when epiblast cells are incorporated within the posterior primitive streak, around day 6 in mice and 16 in humans. Ethical reasons limit the possibility to study such an early stage of development in human embryos. Embryonic stem cells (ESCs) have been long recognized to provide a powerful model to study the early steps of cardiac specification under normal or pathological conditions. Furthermore, ESC-derived cardiomyocytes are a potential cell source for cardiac regeneration. Indeed, cell therapy is a potentially new option for patients with advanced heart failure. However, the clinical trials of bone marrow cell (3) or skeletal myoblast (MAGIC) transplantation (4) have only yielded marginal results. This has been attributed to the limited plasticity of adult stem cells, which precludes their differentiation into functionally integrated cardiomyocytes. Thus, the best substitutes for the missing host cardiomyocytes appear highly plastic cells that can recapitulate cardiomyogenesis. From this standpoint, cardiac stem cells are conceptually attractive but the uncertainties regarding their persistence in adulthood (5) cast serious doubts over their potential therapeutic usefulness. In contrast, pluripotent ESCs emerge as an appealing alternative, but so far their potential use in humans has been hampered by major safety concerns, in that, if undifferentiated and injected in immune depressed mice, they generate teratomas. There is also the possibility that a progenitor cell could stop in the course of differentiation and proliferate in an uncontrolled manner in vivo, as reported with neuronal progenitors grafted in rat brain (6). Thus, definitively committed, early cardiac progenitors could be the best clinical cell source in regenerative cardiology. Several groups have transplanted human ESC–derived (HUESC-derived) cardiomyocytes in postmyocardial infarcted rats (7–9). However, as previously discussed (10), fully differentiated cardiomyocytes have a limited number of divisions, and an important number of cells would be required to replace those lost as a result of disease. Thus, a better option is to use cardiac progenitors still capable of dividing while differentiating.
Using mouse ESCs and HUESCs, several cardiac progenitors have been isolated, searching for a common progenitor at the origin of the whole heart (11). Among them, ISL1+ multipotent progenitors arising in vitro from around E8 of differentiation in mouse give rise to different cell lineages presumed to originate from the secondary heart field, contributing to two-thirds of the myocardium, including formation of the right ventricle, part of atria, and the outflow tract but not the contractile left myocardium (12). However, more recently, ISL1 as a marker restricted to the secondary heart field has been challenged (13). Furthermore, as ISL1 is localized in the nucleus of differentiating ESCs and requires an embryoid bodies environment to be expressed, these cells cannot be easily sorted in great extent as cardiac precursor cells if one foresees a clinical use. More recently, a population of mouse Flk1+ and human CKIT–, KDRlow+, NKX2.5low+, ISL1+ cardiovascular cells, which is capable of generating up to 57% cTNT+ and SMA+ cardiomyocytes, in addition to CD31+ endothelial and SMA+ smooth muscle cells, has been identified from a mouse and later a human ESC line, following sequential treatment for 14 days with a cocktail of growth factors, including activin, BMP4, FGF2, and VEGF (14, 15). Despite these previous findings, the quest for HUESC- or induced pluripotent stem (iPS) cell-derived early progenitors at the origin of both the first and secondary cardiac lineages still remains a relevant issue both for basic developmental biology and clinical application.
Here, we have isolated, sorted, and characterized such a cell population and we report the feasibility of allograft of primate ESC-derived enriched cardiovascular progenitors in a nonhuman primate model of myocardial infarction.
Results
Cardiogenic commitment of pluripotent stem cells and generation and sorting of a large and early population of cardiovascular progenitors.
To drive HUESCs and iPS cells toward a cardiac lineage, we designed experiments to recapitulate the early embryonic developmental pathways from the epiblast stage leading to the precardiac mesoderm lineages. BMP2 together with Wnt3a constitutes a potent combination of factors to induce the mesoderm (16, 17). We thus used the cardiogenic morphogen BMP2 in a defined serum-free medium and in the presence of the FGF receptor inhibitor SU5402 (18). Such a protocol allowed us to combine the action of BMP2 with that of the autocrine Wnt3a ligand, whose expression was strongly induced by BMP2 (9 ± 1–fold increase in mRNA expression). The action of secreted Wnt3a was further demonstrated by adding the Wnt inhibitor, dickkopf homolog 1, in the presence of BMP2. Indeed, this inhibitor significantly decreased the effect of BMP2 on cardiac gene expression (19) and dramatically reduced the number of cells that lost their pluripotency and, in turn, expressed the stage-specific embryonic antigen 1 (SSEA-1) (Supplemental Figure 1; supplemental material available online with this article; 10.1172/JCI40120DS1), an index of differentiation of HUESCs as well as of the human blastocyst (20). As SSEA-1 is one of the earliest markers of HUESC differentiation and loss of pluripotency, and as it is expressed following 4-day treatment of cells with the cardiogenic morphogen BMP2, we reasoned that it might be used to exclude the cells that were not responsive to the morphogen. Indeed, cell sorting using an anti–SSEA-1 antibody allowed for separating cells expressing mRNAs and proteins encoding mesodermal and cardiac markers (21).
We substantially extended this finding and characterized the genetic and epigenetic profiles of BMP2- and WNT3A-induced SSEA-1+–sorted cells as well as their phenotypic fate when engrafted into diseased hearts.
Flow cytometry revealed that 40% (iPS cells) to 53% ± 10% (HUESCs, n = 15) of BMP2-treated cells stained positive for SSEA-1 (Figure (Figure1A1A and Table Table1).1). Sorting out the BMP2-induced SSEA-1+ cell population using an antibody coupled to magnetic beads, revealed that these cells still expressed a high level of OCT4 mRNA (i.e., above the one required to maintain stem cell pluripotency), an index of their cardiac specification (22), but had lost expression of NANOG, CRIPTO, or LEFTY, master stem cell pluripotency genes, not expressed in somatic cells. In contrast, the pluripotent stem cell SSEA-1– population retained expression of these markers (Figure (Figure1B).1B). The BMP2-induced SSEA-1+ cell population could be sorted out from 5 separate HUESC lines, the Rhesus ORMES cell line, and the iPS cell line with different efficiencies (Table (Table1). 1).
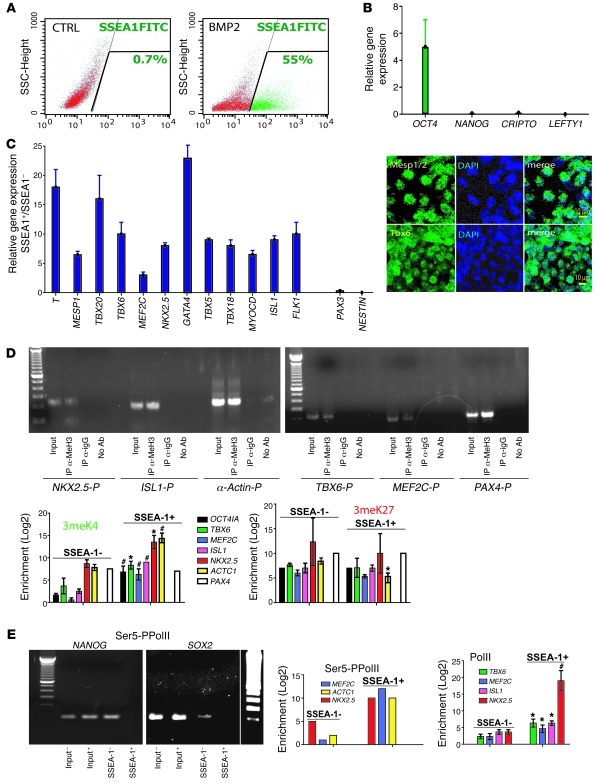
ESCs were treated or not (CTRL) for 4 days with BMP2 (10 ng/ml) and (A) monitored by flow cytometry using a FITC-conjugated anti–SSEA-1 antibody or (B and C) separated using the anti–SSEA-1 Miltenyi kit and the MACS columns. SSEA-1– and SSEA-1+ cDNAs were run in real-time PCR. Data are from 8 experiments, performed on the HUES-24 cell line, and reproduced in different HUESC lines (H9, HUES-9, HUES-24, HUES-26, I3, and I6). The inset in C illustrates SSEA-1+ cells immunostained with anti-Tbx6 and anti-Mesp1/2 antibody 12–24 hours after sorting. (D and E) ChIP assay using (D) anti-H3triMeK4 or anti-triMeK27 antibodies or (E) the anti-PolII or serine-phosphorylated PolII (Ser5-PPolII, left) antibodies. Q-PCR was used to amplify the chromatin-bound DNA, using primers specific for OCT4, TBX6, ISL1, MEF2C, NKX2.5, ACTC1, and PAX4 promoters (sequence within the 700 bp in 3′ of ATG; Table Table2).2). Data (n = 5) show fold enrichment of methylated histones on promoters in the SSEA-1+ versus SSEA-1– population. The gels illustrate the specificity of PCR products (input, SSEA-1+ DNA samples from anti-triMeH3K4 IP, anti-rabbit IgG, or no antibody). OCT4IA, OCT-4 isoform A. (E) The left side shows PCR gel of DNA products after real-time PCR and indicates a complete loss of serine-phosphorylated PolII on both the NANOG and SOX2 promoters. ChiP experiments have been mostly performed using the HUES-24 cell line and were validated in 2 other experiments using the I6 cell line. *P ≤ 0.05; #P ≤ 0.01.
Table 1
Yield of SSEA-1+ cell sorting using anti–SSEA-1–conjugated magnetic beads
We next looked at expression of mesodermal and cardiac lineage marker genes by quantitative PCR (Q-PCR). Freshly sorted SSEA-1+ cells expressed T brachyury-like (T) and FLK1, 2 characteristic genes of the early mesoderm. Furthermore, these cells expressed mesoderm posterior 1 (MESP1), the earliest known marker of cardiopoiesis (23); TBX6, a mesodermal muscle gene upstream of MESP1 (24); TBX5, a ventricular marker; and TBX18, a marker of both epicardium (25) and the left ventricle (26). SSEA-1+ cells also expressed myocardin (MYOCD), GATA4, MEF2C, and NKX2.5, key smooth muscle and cardiac transcription factors of the primary heart field, as well as ISL1 and TBX20, the latter more reminiscent of the secondary heart field. While expressing TBX18, SSEA-1+ whole population expressed, to a small extent, Nestin (NES) or PAX3, 2 markers found in neural crest cells (Figure (Figure1C).1C). TBX3, a marker of the cardiac conduction system (27) among other lineages, was not detected (data not shown). We could not detect any other marker of the hematopoietic or skeletal lineage (CD45, CD34, N-cadherin, MyoD) or any endodermal (SOX17, HEX, FOXA1) (data not shown) or ectodermal (SOX1) marker. Flow cytometry analysis confirmed that a minority (5%–7% in 2 separate experiments) of SSEA-1+ cells were Flk1+ at the time of sorting (data not shown). TBX6 and MESP1/2 were readily detected in most cells at the level of the protein, right after sorting (Figure (Figure1C,1C, right). A similar gene profile could be found in 5 HUES cell lines tested (HUES-24, HUESC-26, HUES-9, H9, I6).
Gene transcription is tuned by the epigenetic status of stem cells (i.e., the histone code). In ESCs, promoters of transcription factors specific for early cell lineages, feature bivalent domains, harboring both epigenetic marks, the activating methylation of lysine 4 of histone 3 (H3K4) and the repressive methylation of lysine 27 (H3K27) (28).Thus, to further characterize the BMP2-induced SSEA-1+ cell population, we designed an epigenetic strategy targeting early transcription factors (Figure (Figure1,1, D and E). Using ChIP assays, combined with real-time PCR, we looked at both activating and repressive epigenetic marks on the OCT4 promoter and on 5 mesodermal and cardiac promoters. We found that the OCT4 promoter was associated with an increase in methylation of H3K4 and was occupied by the polymerase II (PolII), 2 epigenetic marks indicating a transcriptionally active chromatin. TBX6, ISL1, MEF2C, NKX2.5, and cardiac α-actin (ACTC1) promoters shared a common epigenetic status, with a 30- to 2,000-fold increase (NKX2.5) in PolII occupancy. These data were further confirmed in ChIP experiments using an anti-PolII phosphorylated on serine 5, a better mark of activated bivalent promoters (29) (Figure (Figure1E).1E). Ten-fold enrichment in H3K4 methylation on cardiac promoters was observed in SSEA-1+ compared with SSEA-1– cells (Figure (Figure1D),1D), while the H3K27 methylation mark did not significantly change on the same promoters. Interestingly, the promoters of 2 pluripotency genes, SOX2 and NANOG, were not any more occupied by serine 5–phosphorylated RNA PolII in SSEA-1+ cells (Figure (Figure1E).1E). The epigenetic status of the PAX4 promoter, an endodermal gene, was not changed in SSEA-1+ compared with SSEA-1– cells.
Early cardiovascular progenitors feature a unique microRNA signature.
Besides gene transcription, expression of proteins defining a specific cell fate is also regulated at the posttranscriptional level by small non-coding RNAs (miRNAs, miRs), and profiles of miRNAs often reflect a cell-specific signature. We thus used a genomic wide approach (30) to compare the profile of miRNAs in BMP2-induced SSEA-1+ cells and remaining SSEA-1– or undifferentiated HUESCs. This broad approach showed that SSEA-1+ cells gained expression of specific miRNAs, belonging to the cluster miR-17-92. These include miR-20a and miR-20b, known to be turned on when ESCs acquire a cardiac fate (31), and miR-17-92, whose downregulation causes a cardiac defect. Furthermore, miR-106, whose deletion concomitant to the one of miR-17-92 results in severe cardiac defects (32), was also upregulated in SSEA-1+ cells. miR-513, another miRNA not expressed in undifferentiated ESCs, has still unknown functions that were highly upregulated in SSEA-1+ cells; its function is currently under investigation in the laboratory. The gain in expression of these miRNAs was marked in SSEA-1+ cells when compared with the one in SSEA-1– cells and became even more prominent when compared with the profile of undifferentiated stem cells (Figure (Figure2).2). miR-302a, belonging to the cluster 302, whose expression is regulated by OCT4 and potentially SOX2 and NANOG, was also more expressed in SSEA-1+ cells versus SSEA-1– cells or HUESCs. This phenomenon could be related to the dual role of this miRNA in both pluripotency and then in specification of HUESCs toward the mesendoderm (33). Another miRNA expressed in HUESCs was also upregulated in SSEA-1+ cells in agreement with its high expression when HUESCs differentiate within embryoid bodies (30). Expression of miR-615 and miR-135 connected to the pluripotency network in ESCs was unchanged (34). Of note, the miR-302bcd, also involved in the pluripotency transcriptional network (34, 35), was downregulated in SSEA-1+ cells when compared with SSEA-1– cells, while it appeared as upregulated when compared with expression level in HUESCs (Figure (Figure2). 2).
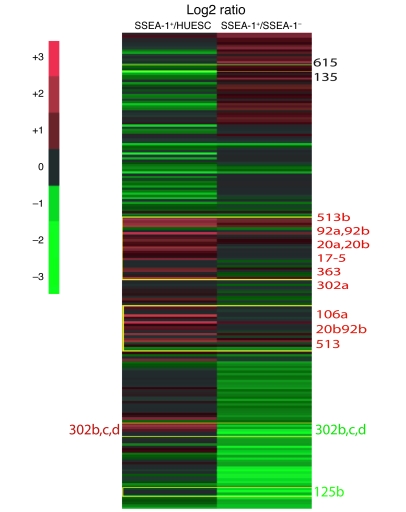
Numbers beside the figure represent miRs. Total RNA was extracted from HUES, SSEA-1+, and SSEA-1– cells and hybridized to a miRNA microarray. Experiments were performed 6 times and log2 ratio values are shown in the heat map.
The cardiovascular population of SSEA-1+ progenitors is clonogenic and multipotential.
To characterize the differentiation potential of SSEA-1+ progenitors toward several cell lineages, we plated isolated SSEA-1+ progenitors as suspensions of single cells on mitotically arrested E14 mouse embryonic fibroblasts (MEFs) in 96-well plates for 5 days, stained them with DAPI, and scanned them using an Arrayscan. Despite being plated by limiting dilution at high or low density (5,000 down to 1 single cell/well), cells generated clones within 5 days. The number of clones per well tightly correlated with the starting number of plated cells (Figure (Figure3A).3A). One single cell (monitored by phase contrast microscopy, using the wild-type HUES-24 cell line, or by GFP expression, using the HUES-9 Oct4GFP cell line) was able to generate a clone within 7 days (Figure (Figure3,3, A–C). In another set of experiments, a single SSEA-1+ cell was observed while carrying out cytokinesis as early as 2 days after plating (Figure (Figure3A,3A, above the graph). The limiting dilution experiments were confirmed by adding a single cell into a well of a 96-well plate using a micropipette and monitoring the formation of a colony (data not shown). Altogether, this demonstrates that SSEA-1+ cell–derived colonies originate from single cells and not from cell clusters, eliminated by thorough trypsinization and filtering (40 μm) before the passage through the sorting magnetic column.
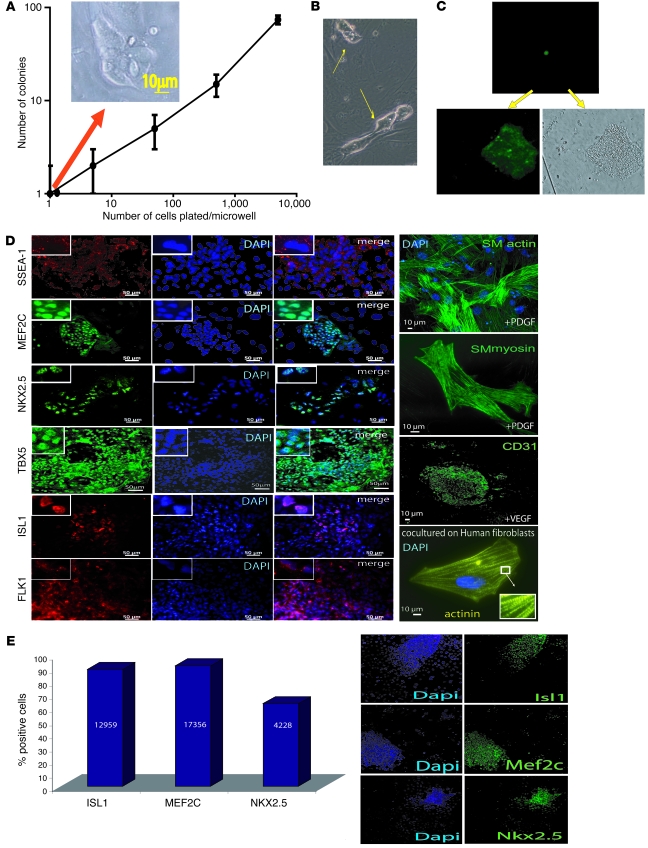
(A–C) Cell clonogenicity. BMP2-induced SSEA-1+–sorted cells were plated at different densities (5,000, 500, 50, 5, and 1 cell/well) in 96-well plates containing MEFs. Five days later, cells were stained with DAPI to be scanned using an Arrayscan. (A) The orange arrow indicates that the colony was generated in a single cell-containing well. Cell colonies were numbered using the Cellomics viewer software. (A and B) A colony generated from a single SSEA-1+ cell (arrows) and visualized 2 (B) or 5 (A) days later (above the graph). (C) A single SSEA-1+ cell derived from the HUES-9 pOct-4-GFP cell line that was plated in a microwell of 96-well plates and visualized by fluorescence right after plating (top image) or 5 days later (bottom GFP and transmitted light images). (D) Immunofluorescence of BMP2-induced SSEA-1+ cells cultured for 5 days on MEFs in the absence of growth factors or challenged by PDGF or VEGF, or cocultured with human cardiac fibroblasts. Top left corner insets show high-magnification images of localization of markers. The inset in the actinin panel shows a high-magnification image of sarcomeres. Experiments were done in triplicate with similar results. (E) BMP2-induced SSEA-1+ cells cultured for 5 days on MEFs immunostained with anti-ISL1, anti-MEF2C, or anti-NKX2.5 antibodies were quantified by HCCI using an Arrayscan. The numbers in the bars of the graph indicate the number of scored and validated cells. Original magnification ×20; ×40 (smooth muscle [SM] myosin in D); ×5 (insets in D).
After 1 week in culture on MEFs in the absence of SU5402, BMP2-induced SSEA-1+–sorted cell–derived colonies still expressed SSEA-1, together with 4 cardiac-restricted transcription factors, ISL1, MEF2C, NKX2.5, and TBX5. A majority of cells from most colonies expressed these markers (Figure (Figure3D),3D), pointing to a rather homogeneous population of cardiac progenitors. ISL1, MEF2C, NKX2.5, and TBX5 were all located into the nucleus. FLK1, poorly expressed in the freshly sorted SSEA-1+ cell population (5%–7%) (data not shown), was found later at the membrane of a proportion, but not in all cells, after 1 week of culture on feeder cells in the absence of SU5402. In contrast, HUES-derived SSEA-1– cells did not express NKX2.5, ISL1, MEF2C, and TBX5 but still expressed OCT4, SOX2, and NANOG as assessed by immunofluorescence (data not shown).
Addition of PDGF (10 ng/ml) or VEGF (50 ng/ml) to BMP2-induced SSEA-1+ cells cultured on MEFs strongly triggered expression of smooth muscle actin and myosin as well as of CD31, respectively, while only 12% of freshly sorted BMP2-induced SSEA-1+ cells (Supplemental Figure 2A) were CD31+ (Supplemental Figure 2B). Plated on collagen 1–coated dishes and treated with VEGF (50 ng/ml for 10 days), 80%–90% of SSEA-1+ cells acquired the morphology of endothelial cells and expressed high levels of CD31, CD34, and FLK1 mRNAs as well as the protein CD31 for 30% of them (Supplemental Figure 2C) but not cardiac troponin T (data not shown), as investigated by immunofluorescence.
When plated on human fibroblasts in the presence of 5% fetal calf serum or in a mix of human cardiac fibroblasts and cardiomyocytes, 10% and 60%–80% of cells, respectively, expressed actinin arranged in sarcomeric units within a week (Figure (Figure3D3D and Figure Figure4A).4A). A similar effect was observed in 70% of cells when cells were treated with the conditioned medium of a coculture of human cardiac fibroblasts and cardiomyocytes.
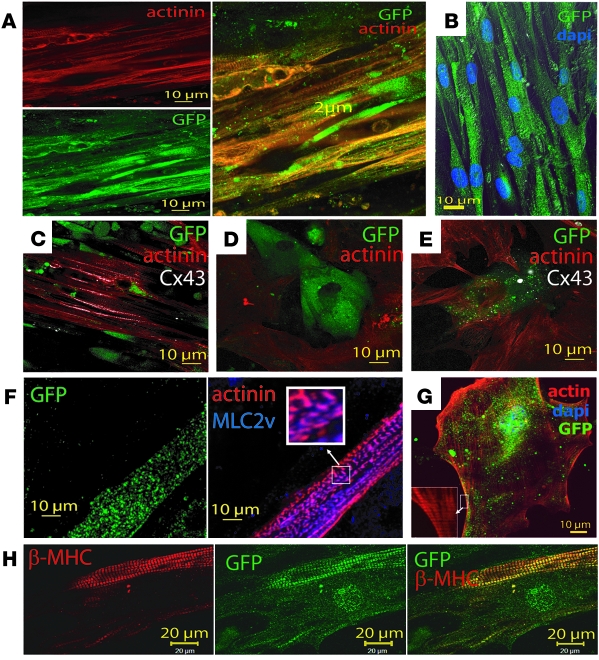
Immunostaining of actin promoter EGFP SSEA-1+ ORMES cells cultured on (A and B) both cardiac fibroblasts and myocytes. (A) Anti-GFP and anti-actinin, with merged image on the right. (B) Anti-GFP immunostaining and DAPI staining of nuclei of SSEA-1+ cells; z-stack of images was acquired in a laser-scanning confocal microscope, with the green and blue channels together with differential interference contrast images. The merged image of a 200-nm focal plane is shown. (C) Merged image of anti-actinin (red), anti-GFP (green), and Cx43 (white) immunostaining. (D) Actin promoter EGFP SSEA-1+ ORMES cells cultured on cardiomyocytes only and (E) fibroblasts only (GFP, green; actinin, red; CX43 or phosphorylated P-CX43, white). (F) Actin promoter EGFP SSEA-1+ ORMES cells cultured on both cardiac fibroblasts and myocytes immunostained with the anti-GFP (green), anti-actinin (red), and anti-MLC2v (blue) antibodies. These images are representative of 3 coculture experiments. Original magnification, ×100 (inset). (G) Actin promoter EGFP SSEA-1+ ORMES cells were cultured for 2 weeks on matrigel in the presence of filtered conditioned medium of a mix of fibroblasts and cardiomyocytes and actin stained with phalloidin. Original magnification, ×63 (inset). (H) Actin promoter EGFP SSEA-1+ ORMES cells cultured on both cardiac fibroblasts and myocytes immunostained with the anti–β-MHC (red) and anti-GFP (green). Merged image is shown on the right. The sarcomeric staining of GFP is due to its binding to myosin (60).
Interestingly, SSEA-1+ cells in culture on MEFs for 4 days expressed some genes specific for the secondary heart lineage. This was further enhanced by FGF8 (5 × 10–8 M) (36) treatment of cells for an additional 4 days. FGF8, indeed, definitively turned on expression of TBX1, retinaldehyde dehydrogenase 2 (RALDH2) (37, 38), HES1 (39), and FOXH1 (40) (Supplemental Figure 3A). The FGF8 effect was also observed when genes were monitored at the level of single colonies (data not shown).
To better quantify the cardiogenic fate of BMP2-induced SSEA-1+ cardiovascular progenitors cultured for 1 week on MEFs, immunostained cells cultured in 96-well plates were scored by high-content cell imaging (HCCI), using an Arrayscan, in 2 experiments. This technological approach allowed us both to quantify and to visualize cells positive for each cardiac marker. The BMP2-induced SSEA-1+ population expanded as cardiovascular progenitors within colonies (Figure (Figure3E,3E, right) expressed NKX2.5 and MEF2C (63% and 91%, respectively) as well as ISL1 (89%). The cells lost expression of OCT4 and SOX2 (data not shown). This quantitative analysis by HCCI was confirmed by flow cytometry (data not shown).
To assess the transcriptomic homogeneity of the clones, individual colonies grown for 7 days on MEFs were randomly picked up through a micropipette and screened for gene expression by RT-Q-PCR. These experiments revealed that each colony expressed, to a similar and high extent, NKX2.5, MEF2C, ACTC1, and MYOCD and, to a lesser degree, FLK1. ISL1 and TBX20 were downregulated in 50% of colonies; NES and PAX3 were expressed in 50% of clones, actually those expressing ISL1 and TBX20. None of the colonies expressed SOX17, an endodermal marker (Supplemental Figure 3B). The multipotentiality of a single clone of SSEA-1+ cells was further demonstrated by cutting single colonies grown on MEFs for 5 days into several cell clusters. Cells were then separately replated on collagen 1–coated dishes and treated with PDGF or VEGF or plated on human fibroblasts. In the later case, cardiomyocytes (Supplemental Figure 4A) were observed 2–3 weeks later, while CD31-positive cells (Supplemental Figure 4B) and smooth muscle actin-positive cells (Supplemental Figure 4C) were present in growth factor treated cells after 1 week (Supplemental Figure 4).
In the perspective of a potential use of SSEA-1+ progenitors in customized regenerative medicine, we used iPS cells. BMP2-induced SSEA-1+ cells could also be derived from iPS cells (Supplemental Figure 5A). A similar pattern of expression of cardiac proteins (i.e., ISL1, MEF2C, and NKX2.5) was found in BMP2-triggered SSEA-1+ cell–derived and sorted cells cultured for 1 week on MEFs, as assessed by immunofluorescence. Furthermore, a minor population of BMP2-induced SSEA-1+ iPS cells (4%) stained positive for CD31 (Supplemental Figure 5B).
Cardiovascular BMP2-induced SSEA-1+ progenitors differentiate into mature cardiomyocytes ex vivo.
To test the cardiogenicity and the functionality of BMP2-induced SSEA-1+ progenitors prior to animal graft experiments, we used an ex vivo experimental set up. In order to track the fate of cells in culture, Rhesus ESCs (ORMES-2 cell line) were genetically engineered to express GFP under the transcriptional control of the human cardiac α-actin promoter. BMP2-induced SSEA-1+ Rhesus cardiac progenitors were then cocultured for 4 weeks with human atrial myocytes, cardiac fibroblasts, or a mix of cardiomyocytes and fibroblasts (Figure (Figure4).4). After 4 weeks, microscopic examination revealed the presence of GFP-positive cells (Figure (Figure4)4) in the 3 cultures, thereby confirming that human cardiac/fibroblastic cells provide the paracrine environment that triggers the cardiomyogenic differentiation of BMP2-induced SSEA-1+ progenitors. The extent of differentiation of BMP2-induced SSEA-1+ cardiac progenitors was prominent and reached 70% ± 10% (in 3 experiments) for cells cocultured on the mix of cardiac fibroblasts and cardiomyocytes or conditioned medium from the coculture (Figure (Figure4G).4G). More interestingly, under this experimental situation, a high proportion of cells (60%–80%), as monitored by concomitant GFP and actinin expression, differentiated in mature cardiomyocytes expressing organized and sarcomeric aligned structures (Figure (Figure4,4, A and B). To investigate a potential cell fusion phenomenon, we acquired z-stacks of images (every 200 nm) of BMP2-induced SSEA-1+ cardiac progenitors cultured on a mix of cardiac fibroblasts and cardiomyocytes and stained with an anti-GFP antibody and DAPI to visualize the nucleus. Examination of images showed that each GFP cell featured a single nucleus in the same focal plane (200 nm) as the one where GFP gave the brightest fluorescence (Figure (Figure4B),4B), excluding a fusion with cells sitting below.
No cells were labelled with an anti-SMA antibody. The remaining non- or poorly actinin-positive cells still acquired a cardiac fate, as they expressed GFP under the transcriptional control of the cardiac α-actin promoter but failed to set a contractile apparatus. When cultured on myocytes alone, cells remains as fetal (sarcomere size, 1.12 ± 0.02; n = 137 sarcomeres, 12 cells) (Figure (Figure4C).4C). On fibroblasts, they started to form more mature sarcomeres (1.80 ± 0.03; n = 70 sarcomeres, 4 cells) (Figure (Figure4D),4D), while on both myocytes and fibroblasts (Figure (Figure4,4, A and B), sarcomeres reached an adult size (2.01 ± 0.03 n = 127, 10 cells). Furthermore, aligned ORMES-derived cardiomyocytes were the only ones to express the ventricular isoform connexin 43 (CX43) (Figure (Figure4C)4C) at their membrane, the ventricular myosin light chain for 80% of them (Figure (Figure4F)4F) and the adult β-MHC isoform (Figure (Figure4H)4H) for 50% of them, an index of acquisition of an adult ventricular phenotype. When cultured alone on matrigel in the only presence of conditioned medium of a mix of fibroblasts and cardiomyocytes, 70% of BMP2-induced SSEA-1+ cardiac progenitors adopted a morphology of embryonic cardiomyocytes and developed within 2 weeks a sarcomeric actin network and became hypertrophied, indicating the presence in the conditioned medium of both differentiating and hypertrophic agents (Figure (Figure4G;4G; n = 6). While quiescent, as no pacemaker cell was present in the dish, when electrically paced, these cells featured a Ca2+ transient (data not shown). CX43 was phosphorylated in the cells in contrast to cytosolic microtubule-associated unphosphorylated CX43, which was expressed in nonmature ORMES-derived cardiomyocytes observed when the latter were cultured on atrial myocytes alone (Figure (Figure4D)4D) or fibroblasts alone (Figure (Figure4E).4E). These data further document that cell transplantation should be performed in the border zone of infarcted myocardium or in patchy scars composed of both fibrosis and reversibly damaged cardiac fibers.
Cardiovascular BMP2-induced SSEA-1+ progenitors engraft in infarcted primate myocardium and differentiate into mature cardiomyocytes.
ORMES cells were treated for 2 days with BMP2. RT-Q-PCR revealed that BMP2 induced expression of cardiac genes (TBX6, TBX20, ISL1, MEF2C, and ACTC1) (Figure (Figure5A),5A), thereby demonstrating a cardiac print of these cells and confirming the strong cardiac inductive action of BMP2 in nonhuman primate cells. To assess the fate of these cells in vivo, we developed a nonhuman primate model of myocardial infarction by subjecting Rhesus monkeys to a percutaneous 90-minute coronary occlusion/reperfusion protocol, followed, 2 weeks later, by open-chest cell transplantation in the infarcted area.
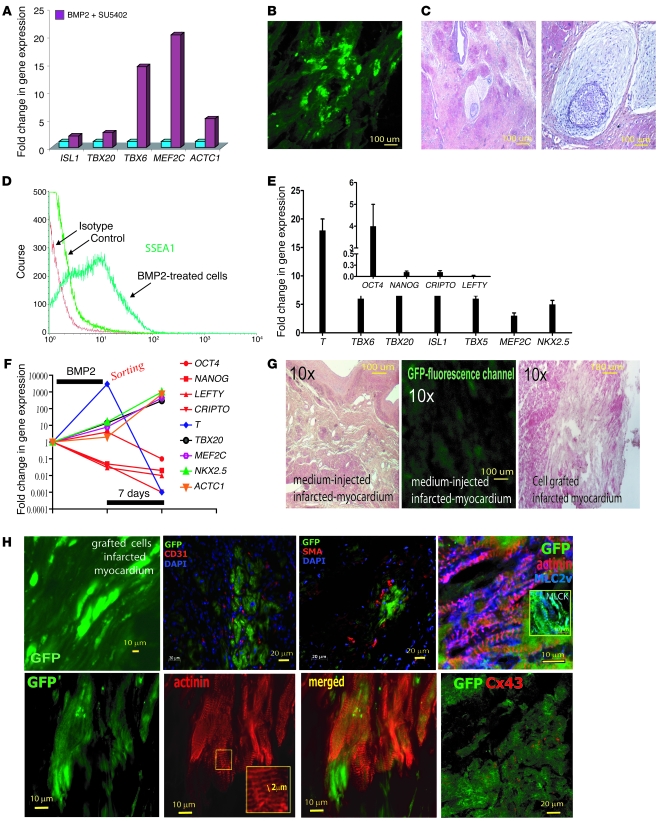
(A) BMP2-induced gene expression in ORMES cells. (B) GFP-expressing cells following engraftment of actin promoter EGFP ORMES cells in the scar area of the myocardium. (C) Teratoma formed after cell engraftment (the left and right images show low and high magnification, respectively). (D) Flow cytometry analysis of FITC-conjugated SSEA-1+ ORMES cells. (E) Q-PCR of mesodermal, cardiac genes (or pluripotency genes, inset) expressed in SSEA-1+ cells sorted after 4 days BMP2 treatment. Data were normalized to GAPDH and to the SSEA-1– population right after sorting. (F) Kinetic of gene expression in SSEA-1+ cells expressed as fold changes versus SSEA-1– cells normalized to undifferentiated ORMES-2, following 4 days BMP2 treatment after sorting and following 7 days culture on MEFs. (G) H&E staining of the myocardial section from a postinfarction primate having received the medium (left image, sham) or the cells together (right image) with an image illustrating the low background of the green GFP fluorescence channel in the non–cell-grafted myocardium (middle panel). (H) Upper panels show, from left to right, GFP-expressing cells 2 months after engraftment of actin promoter EGFP and SSEA-1+ ORMES cells, ×40 original magnification of a myocardial section; costaining of actin promoter EGFP and SSEA-1+ ORMES cells, with anti-CD31 and anti-SMA antibodies; costaining of actin promoter EGFP and SSEA-1+ ORMES cells, with anti-GFP, anti-actinin, and anti-MLC2v antibodies. The inset shows costaining of GFP and MLCK in a myocardial section grafted with SSEA-1+ ORMES.
In a first series of pilot experiments, 107 BMP2 cardiac-committed cells expressing cardiac genes (Figure (Figure5A)5A) were transplanted in the scar area of 2 immunosuppressed primates. The primates were euthanized after 2 months. Confocal microscopy of the explanted hearts showed clusters of GFP-positive cells throughout the injected areas within the scar (Figure (Figure5B).5B). Unfortunately, a careful pathological examination of the myocardium and other organs (e.g., lung, liver) revealed that 1 out of the 2 primates injected with nonsorted cells had developed a microteratoma in the scar area (Figure (Figure5C).5C). Thus, BMP2 nonresponsive SSEA-1– and still pluripotent cells proliferated and differentiated within the tumor and should thus be removed before transplantation.
While untreated ORMES cells featured less than 10% SSEA-1+ cells, 50% ± 5% of BMP2-challenged cells expressed the antigen (Figure (Figure5D).5D). To confirm the phenotype of SSEA-1+ cells, the gene expression profile of both SSEA-1– and BMP2-induced SSEA-1+ cell populations was then compared by real-time RT-PCR. As shown in Figure Figure5E5E and as found in HUESC derivatives, the BMP2-induced SSEA-1+ ORMES cells featured a high expression of T, TBX6, TBX20, ISL1, TBX5, MEF2C, and NKX2.5, while these genes were barely detectable in the SSEA-1– cell population. In contrast, BMP2-induced SSEA-1+ cells dramatically lost expression of most pluripotency genes (i.e., CRIPTO, NANOG, and LEFTY), in comparison with SSEA-1– cells (Figure (Figure4E,4E, inset). Of note, OCT4 was still expressed at a higher level in BMP2-induced SSEA-1+ cells than in the SSEA-1– cell population, as also observed in human BMP2-induced SSEA-1–sorted cells. When cultured on MEFs, the SSEA-1+ ORMES-sorted cells expressed to a great extent after 1 week TBX20, ISL1, MEF2C, NKX2.5, and ACTC1. T, whose expression was high in freshly sorted cells, was downregulated. The pluripotency genes NANOG, LEFTY, CRIPTO (also known as TDGF1), and OCT4 were all downregulated (Figure (Figure5F). 5F).
Infarcted myocardia of 8 immunosuppressed primates were thus transplanted with 2 × 107 SSEA-1+ cardiac progenitors and 3 were transplanted with cell medium. The primates were euthanized after 2 months. Confocal microscopy of the explanted hearts showed clusters of GFP-positive cells throughout the injected areas within the scar (Figure (Figure5H),5H), and this strong cardiac differentiation potential of BMP2-induced SSEA-1+ cardiac progenitors was further confirmed by PCR using GFP-specific primers (data not shown). These cells were surrounded by CD31- and SMA-positive cells, suggesting either a recruitment of these cells toward the grafted cells or a differentiation of SSEA-1+ cells into these lineages as found in vitro. We quantified, by imaging GFP and morphometry, the extent of the graft within the scar in 60 myocardial sections. The scar area was 284,620 ± 82,816 μm2, and the extent of GFP+ cell recolonization was 65,251 ± 46,056 μm2, which represented 20% ± 10% of the scar. The ORMES-derived cardiomyocytes expressed α-actinin, set in sarcomeric structures, featuring an adult size (Figure (Figure5H,5H, inset) and expressing CX43 (Figure (Figure5H).5H). The ventricular phenotype of these cells was demonstrated by expression of the ventricular isoform of myosin light chain 2, associated with the presence of myosin light chain kinase, featuring both diffuse distribution and a sarcomeric organization (41) (Figure (Figure5H,5H, inset). Thus, as shown in a rat model of myocardial infarction (42), the scar environment turned out to be potent enough to trigger differentiation of cardiac-committed primate ESCs. No teratoma was detectable in any of the 8 primates that had received sorted SSEA-1+ cells in any of the investigated organs (myocardium, brain, liver, spleen, pancreas, duodenum, lung, gonads, or bones). This finding strongly suggested that some uncommitted SSEA-1– cells (50% of the injected population), which were not responsive to BMP2 and thus still undifferentiated pluripotent cells, were at the origin of the tumor, thereby confirming that purification of the progenitor cell population is mandatory for the safety of the procedure. The safety of delivering this SSEA-1+ cell population was further supported by another set of experiments performed in 10 humanized immunodeficient Rag–/–, γ chain–/–, C5–/–, MHC–/– mice (43). In this series, none of the intracardiac injections of the SSEA-1+, ISL1+, NKX2.5+, MEF2C+, and TBX5+ cardiac progenitors resulted in teratoma at a 4-month follow-up (180 cardiac and other organs sections examined; data not shown).
Discussion
Herein, we have identified a very early population of OCT4+ cardiac progenitors expressing the membrane antigen SSEA-1. SSEA-1 expression was employed as one of the earliest markers of HUESCs or iPS cell loss in pluripotency, to sort out the cells both responsive and unresponsive to BMP2.
Our findings first revealed that BMP2, in the absence of activin (i.e., in serum-free defined medium), acts in a combinatorial manner with Wnt3a, whose expression was triggered by the morphogen, to drive pluripotent stem cells toward an early mesodermal and cardiogenic fate in vitro, as expected from both their in vivo role in defining the posterior primitive streak and mesoderm (44, 45). Several data suggest that the BMP2-induced SSEA-1+–sorted cell population is likely to be more reminiscent of the early cardiac progenitors allocated to the epiblast. When sorted out, these cells still feature a high level of OCT4 (i.e., above the level required to maintain cell pluripotency), as confirmed by the epigenetic status (high H3K4 methylation) of its promoter. Despite their early stage of specification, SSEA-1+ OCT4+ cells acquired a genetic and epigenetic signature, including a profile of miRNA expression, characteristic of pancardiac progenitors. They lost pluripotency, as indicated by NANOG, CRIPTO, LEFTY, and SOX2 downregulation concomitantly with the expression of SSEA-1. While the remaining presence of OCT4 in the absence of other pluripotency genes might be questionable, its level of expression above the one of undifferentiated stem cells also reflects the first step of cell specification toward a cardiogenic fate (19, 22, 46). Such a gene profile, together with expression of the proteins TBX6 and MESP1/2, indicates that this progenitor somehow reflects the very early cardiac-determined OCT4+ cell population found in the embryonic epiblast (2, 22). In agreement with the last statement, SSEA-1+ cells gained expression of several miRNAs, including 3 (miR-20, miR-17-92, and miR-106) reported to be turned on when ESCs acquire a cardiac fate (31) or are likely important for mesendodermal differentiation (33) and proliferation of cardiac progenitors (32). Interestingly, miRNAs known to be the major targets of the OCT4/SOX2/NANOG transcriptional pluripotency network, miR-302s (34, 47, 48), were dramatically downregulated in SSEA-1+ versus SSEA-1– cells, supporting the idea that SSEA-1+ cells lost their pluripotency to move along a cardiac differentiation pathway. On the other hand, it could be surprising that SSEA-1+ expressed more mRNA-302bcd than HUESCs. However, at the time of sorting (when RNAs were extracted for profiling miRNAs), the progenitors still expressed a level of OCT4 above that of HUESCs. As miRNA-302 as well as miRNA-135 and miRNA-615 are primarily induced by OCT4 (or lost when OCT4 is downregulated in the presence of SOX2 and NANOG) (34), this explains that expression of these miRNAs is maintained or even increased. It should be further stressed that their exact role in pluripotency remains to be established and that like miR-106 and miRNA-363, they might be involved at later stages in cell lineages differentiation (30, 33).
Finally, when sorted they express a series of genes (T, TBX6, MESP, ISL1, TBX20, NKX2.5, MEF2C, MYOCD, and TBX18, etc.) and proteins (TBX6 and MESP1/2) that altogether confer them the status of mesodermal cardiogenic cells. Expression of these genes is expected to be sustained as the epigenetic status of their promoter as well as their occupancy by RNA PolII and phosphorylated serine 5 PolII revealed an active chromatin (H3K4 methylation) and suggest an active transcription, respectively. However, the methylated H3K27 marks still on indicate a differential regulation of both activating (K4) and repressive marks and that the gain in K4 methylation is sufficient to turn on the promoter as shown by gene transcription. They further acquire in culture the expression of the proteins (ISL1, NKX2.5, MEF2C, and TBX5 but not SMA), which altogether indicates that they are cardiac progenitors. NKX2.5, ISL1, MEF2C, and TBX5 are intranuclear. TBX5 cellular localization depends upon the stage of cardiac development (49). Thus, its nuclear localization indicates that despite the early stage of SSEA-1+ cardiac progenitors when sorted, they quickly acquire maturity when in culture.
HCCI, together with cell cytometry analysis, further revealed that the majority of cells expressed these markers, 70%–90% expressing ISL1, MEF2C, and NKX2.5 proteins. Thus, these findings show that the BMP2-induced SSEA-1+ population shares some similarities with the recently reported KDR+low cell population (15). However, only 5%–7% of the BMP2-induced SSEA-1+ population is FLK1+ (or KDR+) at the level of the protein, as assessed by flow cytometry after 4 days of BMP2 treatment, thus suggesting that KDR+ cells represent a minor fraction of the sorted SSEA-1+ progenitors. As in the KDR+low population (15), c-KIT is also absent from SSEA-1+ progenitors as well as from the cardiac field in the embryo (50). However in contrast to KDR+low-sorted cells (15), our progenitors did not express SMA if not challenged by PDGF. In fact, our cardiac cell population was sorted out 2 days earlier than the KDR+low cells, which accounts for the finding that it expressed both OCT4+ and MESP1 proteins in addition to cardiac-specific markers like NKX2.5+ and TBX5+, as assessed by HCCI. SSEA-1+ MESP1/2+ cardiovascular progenitors also stand at a much more early developmental stage than the previously isolated ISL1+ cells (51). In contrast to the latter, they feature a great potential to become ventricular myocytes ex vivo and when engrafted in vivo. The in vivo differentiation of ISL1+ cells (51) remains to be shown. Furthermore, our progenitor population was generated using a short protocol (BMP2-inducing WNT3A), which tends to recapitulate the in vivo induction of the primitive streak and subsequently of cardiogenic mesoderm (17, 44). The most interesting feature of the BMP2-induced SSEA-1+ cells is that they retain the capability to differentiate toward the 3 cardiovascular lineages (i.e., cardiac, endothelial, and smooth muscle). When cultured on fibroblasts or a mix of cocultured human fibroblasts and cardiomyocytes, or in the presence of a conditioned medium of both fibroblasts and cardiomyocytes, they mostly developed a myofibrillogenesis typical of a cardiac cell and expressed CX43, MLC2v, and β-MHC. In contrast, when treated with PDGF or VEGF, they gave rise to smooth muscle and endothelial cells, respectively. Thus, these findings point to a multipotentiality of BMP2-induced SSEA-1+ cardiovascular progenitors. To further characterize the identity of these cells, we carried out single-clone real-time PCR on SSEA-1+ cells in culture on MEFs. Interestingly, while most of the clones expressed pancardiac markers (MESP1, NKX2.5, MEF2C, and ACTC1), 50% expressed ISL1 and more specifically TBX20, a marker of the secondary heart lineage, suggesting that some ISL1+ clones at the level of the protein (Figure (Figure3,3, D and E) started to downregulate expression of this transcription factor, while some other kept it. In line with these findings, SSEA-1+ cells expressed some of the genes specific for the secondary heart lineage (TBX1, RALDH2, HES1, and FOXH1; Supplemental Figure 3A), and this was dramatically enhanced by FGF8. ISL1+ and TBX20+ clones also expressed NES and PAX3, 2 neural crest and/or stem cell markers, and featured the highest levels of TBX18, which marks both the epicardium (25) and left ventricle (26). These data are in agreement with the interplay between the cardiac neural crest and the secondary heart field (52, 53), a process dependent upon BMP receptors and mediated through neural crest derivatives in the epicardium (54). Alternatively, the NKX2.5+, MEF2C+, TBX5+, TBX20+, ISL1+, PAX3+, and NES+ population might represent the most multipotent and early precursor (i.e., cardiovascular stem cell) population among the SSEA-1+ cells. Interestingly, a similar population was recently described as a cardiac neural crest–derived dormant multipotent stem cell in adult heart (55). Thus, these findings suggest that SSEA-1+ early cardiovascular progenitors, each derived from a single cell (Figure (Figure3,3, A–C), can segregate into multiple cardiac lineages (primary, secondary heart lineages and potentially cardiac neural crest and epicardium), when challenged by FGFs and other signals emerging from fibroblasts.
The last and very peculiar property of BMP2-induced SSEA-1+–sorted cells is their potential to engraft in a diseased primate myocardium. When replaced in vivo in a myocardial environment, they differentiate into mature ventricular cardiomyocytes expressing MLC2v and MLCK required for the assembly of the sarcomeres (56) as expected from their early expression of TBX5, while retaining a bi- or tri-potentiality to also potentially generate endothelial and smooth muscle cells, although this remains to be confirmed in a xenogenic graft model. They further express CX43 at their membrane and respond to electrical stimulation, suggesting a potential electrical coupling with the neighboring cells, which remains to be further investigated. Their engraftment turned out to be safe, as their status of definite cardiovascular progenitors prevented them from hyperproliferating, as confirmed by the absence of formation of teratoma in primate treated with sorted SSEA-1+ cells or in immunodeficient Rag–/–, γ chain–/–, C5–/–, Mhc–/– humanized mice, while SSEA-1– still pluripotent cells gave rise to microteratomas in primate myocardium and huge teratomas in mice.
Altogether, our findings demonstrate that pluripotent stem cells give rise to a single pool of early OCT4+ and SSEA-1+/brachyury+/MESP+ cardiogenic mesodermal progenitors. Under specific conditions, the cells further differentiate into cardiac progenitors of the distinct primary and secondary heart lineages or potentially differentiate into cardiac neural crest cells, expressing NKX2.5, MEF2C, and TBX5 with or without ISL1, TBX20, PAX3, and NES. Finally, challenged with signals derived from cardiac myofibroblasts and adult myocytes, they acquire a fate of differentiated cardiomyocytes. Challenged with other growths factors (PDGF, VEGF), they also acquire the fate of smooth muscle cells expressing smooth muscle actin and myosin or of early endothelial cells expressing CD31, CD34, and Flk1, respectively.
Thus, we have identified from pluripotent stem cells and characterized a very early cardiovascular progenitor, segregating later into the first and second heart lineages (57). Furthermore, we have provided the means to easily sort out this cell population to a great extent. How this population is generated in the cell plates before sorting is likely to be a 2-step process. Under BMP2 challenge and in the absence of both FGF (in the presence of SU5402) and activin (in the absence of serum), the OCT4 upregulated SSEA-1+ cells go through an endo/mesendodermal fate, secreting cardiogenic factors including BMP2 and WNT3A (19). The latter directs the cells more specifically toward a cardiogenic fate. The cardiac specification is further enhanced in the presence of FGFs (FGF2, FGF4, FGF8, etc.) secreted by MEF, while VEGF and PDGF redirect the fate of SSEA-1+ cells toward endothelial and smooth muscle lineages (Figure (Figure6). 6).
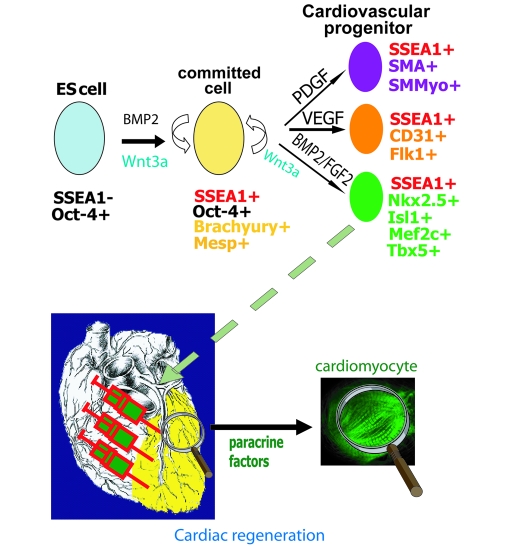
BMP2-induced Oct-4+, SSEA-1+ cells give rise to endo/mesendodermal cells secreting cardiogenic factors, further directing the cell fate toward a cardiac phenotype when plated on MEFs releasing FGF2. Addition of PDGF or VEGF directs the fate of cells toward a smooth muscle and endothelial phenotype, respectively. SMMyo, smooth muscle myosin.
Cardiac development is a dynamic process that is tightly orchestrated by the sequential expression of multiple transcription factors working in a combinatory manner. As such, the different cardiac progenitor cell populations described heretofore are not “competitive” but likely reflect differences in the developmental stage which impacts on the nature and extent of the differentiated derivatives in vitro. Within this complex network, the SSEA-1+ population reported in this paper is appealing because of its ability to give rise to the 3 main cell lineages of the heart, its user-friendly isolation, and its capacity to engraft and to differentiate into mature cardiomyocytes in a clinically relevant large animal model of myocardial infarction. These findings, however, do not preclude that additional studies remain warranted to assess whether still other progenitors might exist that could be endowed with a greater degree of multipotentiality, resulting in a correspondingly greater cardiac repair potential.
Methods
Cell culture.
The HUESC lines (HUES-24 and HUES-9 pOct4/GFP were obtained from Harvard Stem Cell Center, Cambridge, Massachusetts, USA) and the human iPS cell line generated using human dermal fibroblasts infected by lentivirus harboring the cDNAs encoding OCT4, SOX2, LIN 28, KLF4, and NANOG (58) were used throughout this study without any difference in results. HUES-24 and HUES-9 pOCT4/GFP, a transgenic cell line expressing GFP under the transcriptional control of OCT4 promoter (generated using BAC), and iPS cell lines were cultured on MEFs prepared from E14 mouse embryos, using KO-DMEM medium supplemented with β-mercaptoethanol, glutamine, nonessential amino acids, 15% KOSR, and 10 ng/ml FGF2, respectively.
HUESCs and iPS cells were treated for 4 and 6 (iPS) days with 10 ng/ml BMP2 in the presence of 1 μM SU5402, a FGF receptor inhibitor, in RPMI/B27. HUESCs (both HUES-9 and HUES-24) were used within no more than 10 passages (P28–P38). Cells were phenotyped every 15 passages using anti–SSEA-3/4, TRA-1-60, and TRA-1-80 antibodies (Chemicon). Less than 5% of cells were positive for SSEA-1 (Chemicon). Karyotype was found to be normal and stable in the course of the experiments.
Real-time Q-PCR by SYBR Green detection.
RNA was extracted from SSEA-1+ or SSEA-1– cells or single clones using Zymo research kits. One μg of RNA or the total RT reaction (single clone) was reverse-transcribed using the SuperScript II Reverse Transcriptase (Invitrogen) and oligo(16)dT. Q-PCR was performed using a LightCycler LC 1.5 or 480 (Roche Diagnostics). Amplification and specificity of amplicons was carried out as previously described (19). Data were analyzed according to the method described by Pfaffl et al. (59).
Cell sorting.
Trypsinized cells were incubated for 30 minutes with anti–SSEA-1 antibody–coated Miltenyi beads in D-PBS supplemented with 0.5% (wt/vol) BSA and 2 mM EDTA and transferred to a L50 Miltenyi cartridge set on the magnet. Cells were washed 3 times with D-PBS/BSA/EDTA and eluted from the column removed from the magnet using 3 ml of D-PBS/BSA/EDTA (19). FACS, performed 48 hours later when beads detached from cells, revealed a purity of 95% of SSEA-1+ cells.
Flow cytometry analysis.
BMP2-treated cells were trypsinized, washed twice with PBS, and filtered through a 70-μm mesh filter before flow cytometry analysis using FACSCalibur (Becton Dickinson) and CELL QUEST software. Gates were set taking both undifferentiated HUESCs and SSEA-1– cells, stained with both the primary and secondary antibody as negative controls.
Antibodies.
The following antibodies were used for flow cytometry or immunofluorescence: anti-NKX2.5 (mab2444) was from Abcam; anti-TBX6, anti-MESP1/2, and anti-MEF2C were from Aviva System Biology; anti-TBX5 and anti-MYOD were from Abcam; anti-SMA (SMA clone 1A4) and smooth muscle myosin (SMM) were from Dako; anti-ISL1 was from the Developmental Hybridoma Bank of Iowa University, Iowa City, Iowa, USA; the FITC-conjugated anti–SSEA-1, anti-CD31, anti-CD34, and anti-CD45 were from Becton Dickinson. The anti-MLC2v raised against the specific human myosin light chain residues 45–59 was from Biocytex, and the anti-MLCK (clone K36) was from Sigma-Aldrich. Specificity of antibodies was controlled using the respective isotype of the Igs in FACS and using undifferentiated HUESCs for immunostaining.
Cell imaging.
Slides were observed with confocal microscopy (Zeiss LSM 510 META) and low-power laser, allowing us to check for the right spectrum of the fluorochrome that were used (Alexa or GFP), further confirmed by using the META sensitive detector of the LSM 510 scanning microscope. A unique feature of the LSM 510 META scan head is its ability to acquire l stacks in 10-nm increments over a broad spectral range. GFP was detected within a 520–540 nm window. Some myocardial sections have been also treated to decrease autofluorescence (using NaBH4 treatment of sections). GFP was then labelled by an anti-GFP antibody (Covance) and a secondary Alexa Fluor 488– or Texas red–conjugated antibody.
For high-content imaging, after immunostaining, 96-well plates were scanned using the Arrayscan (Cellomics ThemoFisher), using the Cell Health Profiling Bioapplication. The fluorescence threshold was determined from the background obtained in undifferentiated HUESCs and set as a fixed threshold value to scan the wells of SSEA-1+ cells. Thirty fields per well and 18 wells per cardiac marker were scanned as described (19). SSEA-1– HUESCs, stained with the same antibodies as those used for SSEA-1+ cells, were used to set low- and high-intensity thresholds independently for each channel (green, red, blue) and exposure parameters (time of exposure, 300–600 ms). We also set a background correction. Parameters were saved, and all wells were scanned with the same setting. The visualization of images and analysis were then performed using the vHCS software.
Sorted SSEA-1+ cells were plated by limiting dilution into 96-well plates (from 1,000 down to a single cell per well), stained 7 days later with DAPI, and scanned at a magnification of ×10, using the Health Cell Profiling application from Cellomics. The size and fragmentation of objects were set to identify only clusters of cells (cell colonies). The number of colonies was then scored using the software vHCS (Cellomics). To ensure that wells contained a single cell, we also used the HUES-9 pOct-4-GFP cell line and monitored the presence of the cell by epifluorescence.
Culture of human fibroblasts and human atrial myocytes.
Cells were isolated from the right atrial appendage, collected from 3-month-old patients during surgery required to relieve congenital diseases, and cultured as previously described (60). Approval of human tissue collection, care, and use was given by the ethics committee of Hospital Kremlin Bicêtre (AP-HP), Kremlin Bicêtre, France (Committee, protocol 04-26). Parents of patients provided informed consent prior to their participation in the present study.
Generation of α-actin promoter GFP ORMES-2 cell line.
The 106 ORMES-2 cells, cultured as described above, were nucleofected with 3 μg of vector pActin-GFP, purified using a Pharmingen kit and linearized. Cells were then cultured on neomycin-resistant MEFs for 2 days and G418 (100 μg/ml) was added for the next 10 days. Colonies were cut with a needle and picked out of the dish to be further cultured and amplified. Clones were genotyped using GFP primers and further expanded.
Monitoring of gene expression by real-time Q-PCR.
To monitor gene expression in separate single clones, single cell-derived colonies grown for 7 days on MEFs were picked up randomly using a needle and used for RNA extraction (Zymo research kit). RT-Q-PCR was performed using the LightCycler 1.5 as described previously (46) and analyzed according to Pfaffl et al. (59).
ChIP.
The ChIP assay was performed according to the Fast ChIP protocol (61). Briefly, cells were crosslinked for 15 minutes with 1% formaldehyde, and the reaction was stopped for 5 minutes by 125 mM glycine. The cell pellet was then washed with cold PBS, and the cells were lysed with the ChIP buffer (61). One hundred μg chromatin and 3 μg antibody were used in each assay. The antibodies used were the anti-PolII (Santa Cruz Biotechnology Inc.), anti-phospho serine 5 PolII (Covance), the anti-H3triMeK4, and anti-H3triMeK27 (UBI or Abcam), and they were added overnight at 4°C. The Dynabead (protein A–conjugated or anti-IgM–conjugated phosphoserine 5 PolII; Invitrogen) was used to bind the immunocomplex H3-DNA or PolII-DNA. No antibody and nonrelevant rabbit IgG were used as negative controls.
The beads were extensively washed using the ChIP buffer. The protein-DNA interaction was reverse cross-linked by adding chelex to the beads and by boiling the samples for 10 minutes. DNA was then purified on microcartridges. Q-PCR was used to amplify the DNA using primers specific for promoters (sequence within the 700 bp in 3′ of ATG; Table Table2).2). Absolute enrichment was calculated assuming that at most 1% of nucleosomes were immunoprecipitated (62). Genomic region was thus considered enriched if 10 ng IP samples showed a greater enrichment when compared with 0.1 ng of input DNA. Quantification of the immunoprecipitated DNA was normalized against the starting input material.
Table 2
PCR primer sequences

miRNA profiling.
Total RNA was extracted from cells using the mirVANA kit (Ambion). Five μg of RNA for each condition were subjected to dye/swap labelling and were hybridized on printed chips of 2,000 miRs, originating from 44 species; each spot was done in quadruplet. Raw data were analyzed to obtain a log2 intensity value for each spot. Log2 intensity was taken for each spot for both sorted SSEA-1+ cells and control conditions (sorted SSEA-1– cells or HUESCs) and a log2 ratio was obtained. Data were clusterized using Cluster view as previously described (63). The heat map was generated using Tree View software. Data were validated by real-time PCR. Ten ng of total RNA extracted with mirVana kit were used in a RT reaction, using stem-loop microRNA specific primers (Applied Biosystems). Subsequently, the diluted RT product was used in a Q-PCR reaction with TaqMan miR-specific probes (Applied Biosystems). Normalization was done using small RNA RNU48 as a reference.
Myocardial infarction model.
Induction anesthesia of primates was performed with propofol (3 mg/kg) and sufentanyl (0.5 mg/kg), and maintenance anesthesia was ensured with the same drugs, albeit, at a lower dose (1 mg/kg and 0.25 mg/kg, respectively). Intravenous atracurium besilate (1 mg/kg) was added whenever required, and all animals received heparin (50 IU/kg) during the procedure. A myocardial infarction was created in Macaca Mulatta Rhesus monkeys (average weight, 8–10 kg) by a 90-minute balloon inflation in the left circumflex coronary artery, followed by reperfusion and angiographic confirmation of the patency of the revascularized infarct vessel. The occurrence of myocardial necrosis was consistently documented by a postprocedural rise in creatine phosphokinase values. Two weeks later, primates underwent a left thoracotomy and received unsorted or freshly sorted SSEA-1+ ESC-derived progenitors or just cell medium over the scar and at its borders. The monkeys were treated with either FK-506, started 5 days before cell transplantation at the dose of 1 mg/kg/d, or cyclosporine, given intramuscularly for 3 days and then by oral gavage at doses adjusted to target serum drug concentrations in the range of 100 ng/ml. Treatments were continued until sacrifice. Primates were humanely euthanized at 2 and 3 months after transplantation. The area of infarction and cell grafting was identified by sutures that had been placed circumferentially at the time of cell injections. This area was cut into 2 halves, of which 1 was used for assessment for teratoma. The second half was divided into 4 blocks, which were snap frozen in isopentane cooled in liquid nitrogen and subsequently processed as cryosections for identification and characterization of GFP+-grafted cells. In addition, a whole-body autopsy was performed in each animal and used for the search for extracardiac tumors. The experiments with primates were authorized by the Institutional Review Board on Ethics for primate experiments, IMASSA.
Cell transplantation in immunosuppressed mice.
The 105 BMP2-induced SSEA-1+ cells were injected in the myocardium (n = 8), after opening the chest, or subcutaneously in the neck (n = 10) of immunodeficient Rag–/–, γ chain–/–, C5–/–, MHC–/– mice humanized mice. Undifferentiated cells and SSEA-1– cells were also injected subcutaneously in the neck (n = 5 in each group). Four months later, the animals were euthanized and each organ was collected, sliced, and examined for the presence of tumor.
Statistics.
Results are presented as mean ± SEM of 3–8 experiments in each experimental condition. Significance of data was evaluated by a 2-tailed Student’s t test. A P value of less than 0.05 was considered significant.
Acknowledgments
Guillaume Blin is a fellow from the Ministère de la Recherche et de la Technologie. David Nury and Sonia Stefanovic were funded by the grants “Programme Blanc stem cell signature” and “Specistem” of the National Research Agency (to M. Pucéat). This study was funded by the National Research Agency (grants Programmes Blanc stem cell signature and Maladies rares to M. Pucéat), the Genopole Evry, the French Association against Myopathies (AFM grant no. 13968 to C. Rücker-Martin), the LeDucq Foundation (CAPTAA) (Paris, to P. Menasché and M. Pucéat), the Foundation Coeur et Artères, and the Fonds d’Amorçage des Biothérapies (AP-HP, to P. Menasché). Tui Neri is a fellow from the LeDucq Foundation (MITRAL, to M. Pucéat). We thank Laurent Hamon for experimental assistance; S. Garcia (Pasteur Institute) for the teratomas formation in mice; P. Pradeau, D. Lici, and the team of the IMASSA for the care of primates; and Daniel Stockholm (Cell Imaging Facility, Genethon, Evry) for help in cell imaging. We acknowledge Lionel Bonnevie (Hôpital d’Instruction des Armées Bégin) for carrying out the myocardial infarction surgery in primates. We thank Virginie Magnone (Nice, CNRS) for analysis of microarrays.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(4):1125–1139. 10.1172/JCI40120.
See the related Commentary beginning on page 1034.
References
Articles from The Journal of Clinical Investigation are provided here courtesy of American Society for Clinical Investigation
Full text links
Read article at publisher's site: https://doi.org/10.1172/jci40120
Read article for free, from open access legal sources, via Unpaywall:
http://www.jci.org/articles/view/40120/files/pdf
Free to read at www.jci.org
http://www.jci.org/cgi/content/full/120/4/1125
Free to read at www.jci.org
http://www.jci.org/cgi/content/abstract/120/4/1125
Free to read at www.jci.org
http://www.jci.org/cgi/reprint/120/4/1125.pdf
HAL Open Archive
http://www.hal.inserm.fr/inserm-00451770
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1172/jci40120
Article citations
Challenges and perspectives of heart repair with pluripotent stem cell-derived cardiomyocytes.
Nat Cardiovasc Res, 3(5):515-524, 14 May 2024
Cited by: 0 articles | PMID: 39195938
Review
Cell Therapy in the Treatment of Coronary Heart Disease.
Int J Mol Sci, 24(23):16844, 28 Nov 2023
Cited by: 0 articles | PMID: 38069167 | PMCID: PMC10706847
Review Free full text in Europe PMC
Cells and Materials for Cardiac Repair and Regeneration.
J Clin Med, 12(10):3398, 10 May 2023
Cited by: 3 articles | PMID: 37240504 | PMCID: PMC10219302
Review Free full text in Europe PMC
Global research trends in extracellular vesicles based on stem cells from 1991 to 2021: A bibliometric and visualized study.
Front Bioeng Biotechnol, 10:956058, 30 Aug 2022
Cited by: 4 articles | PMID: 36110319 | PMCID: PMC9468424
Genetic Mechanism Study of Auditory Phoenix Spheres and Transcription Factors Prediction for Direct Reprogramming by Bioinformatics.
Int J Mol Sci, 23(18):10287, 07 Sep 2022
Cited by: 2 articles | PMID: 36142199 | PMCID: PMC9499413
Go to all (193) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Monkeying around with cardiac progenitors: hope for the future.
J Clin Invest, 120(4):1034-1036, 24 Mar 2010
Cited by: 6 articles | PMID: 20335651 | PMCID: PMC2846073
Composite cell sheets: a further step toward safe and effective myocardial regeneration by cardiac progenitors derived from embryonic stem cells.
Circulation, 122(11 suppl):S118-23, 01 Sep 2010
Cited by: 71 articles | PMID: 20837902
[A big step forward in the identification of therapeutic human embryonic stem cells-derived progenitors for cardiac cell therapy].
Med Sci (Paris), 26(4):439-441, 01 Apr 2010
Cited by: 2 articles | PMID: 20412753
Pluripotent stem cells for cardiac regeneration: overview of recent advances & emerging trends.
Indian J Med Res, 137(2):270-282, 01 Feb 2013
Cited by: 7 articles | PMID: 23563370 | PMCID: PMC3657850
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NICHD NIH HHS (4)
Grant ID: R01 HD057121
Grant ID: R01 HD057121-02
Grant ID: R01 HD059946
Grant ID: R01 HD059946-03





