Abstract
Free full text

Imbalances in p97 co-factor interactions in human proteinopathy
Abstract
The ubiquitin-selective chaperone p97 is involved in major proteolytic pathways of eukaryotic cells and has been implicated in several human proteinopathies. Moreover, mutations in p97 cause the disorder inclusion body myopathy with Paget disease of bone and frontotemporal dementia (IBMPFD). The molecular basis underlying impaired degradation and pathological aggregation of ubiquitinated proteins in IBMPFD is unknown. Here, we identify perturbed co-factor binding as a common defect of IBMPFD-causing mutant p97. We show that IBMPFD mutations induce conformational changes in the p97 N domain, the main binding site for regulatory co-factors. Consistently, mutant p97 proteins exhibit strongly altered co-factor interactions. Specifically, binding of the ubiquitin ligase E4B is reduced, whereas binding of ataxin 3 is enhanced, thus resembling the accumulation of mutant ataxin 3 on p97 in spinocerebellar ataxia type 3. Our results suggest that imbalanced co-factor binding to p97 is a key pathological feature of IBMPFD and potentially of other proteinopathies involving p97.
Introduction
p97—also known as cell division cycle 48 (Cdc48) and valosin-containing protein (VCP)—is a chaperone-related, ring-shaped AAA ATPase that has a central role in many pathways of the ubiquitin–proteasome system; for instance, endoplasmic reticulum-associated protein degradation (ERAD), transcription factor processing and control of cell-cycle progression and developmentally regulated degradation of the myosin chaperone UNC-45 (Schuberth & Buchberger, 2008; Meyer & Popp, 2008). These diverse cellular functions of p97 are controlled by many co-factors that recruit specific ubiquitinated substrates to p97 and regulate their fate, often by altering their ubiquitination state (Rumpf & Jentsch, 2006; Schuberth & Buchberger, 2008). p97 consists of an amino-terminal N domain and two ATPase domains, D1 and D2. Most co-factors bind to the N domain, either by virtue of a ubiquitin regulatory X (-related) domain or through one of several linear binding motifs (Schuberth & Buchberger, 2008; Yeung et al, 2008).
The central role of p97 in protein homeostasis is underscored by its presence in pathological protein aggregates of several neurodegenerative diseases, including Parkinson, Alzheimer, amyotrophic lateral sclerosis and polyglutamine diseases (Hirabayashi et al, 2001; Mizuno et al, 2003; Ishigaki et al, 2004). The latter have also been functionally linked to p97 (Matsumoto et al, 2004; Boeddrich et al, 2006; Duennwald & Lindquist, 2008). The protein deposits that characterize these so-called proteinopathies are frequently positive for ubiquitin or ubiquitinated proteins, suggesting that the aggregated protein(s) cannot be degraded efficiently by the ubiquitin–proteasome system.
Besides links between these proteinopathies and p97, missense mutations in the VCP gene encoding p97 give rise directly to the late-onset familial disorder inclusion body myopathy with Paget disease of bone and frontotemporal dementia (IBMPFD; Online Mendelian Inheritance in Man 167320). IBMPFD is a multisystemic disease that manifests in muscle weakness, abnormal rates of bone growth and behavioural alterations (Kimonis et al, 2008; Weihl et al, 2009). At the cellular level, IBMPFD is characterized by an impaired degradation of p97 target proteins and an accumulation of ubiquitinated protein aggregates (Weihl et al, 2006; Hubbers et al, 2007; Janiesch et al, 2007; Ju et al, 2008). Specifically, IBMPFD-associated mutant p97 proteins are defective in the degradation of ERAD substrates (Weihl et al, 2006) and of UNC-45, a target of the ubiquitin ligase E4B (UFD-2; Janiesch et al, 2007). Furthermore, they were shown recently to be impaired in a yet uncharacterized, crucial function of p97 in autophagosome maturation (Ju et al, 2009; Vesa et al, 2009; Tresse et al, 2010). The molecular basis underlying these cellular defects has, however, remained elusive. IBMPFD-associated mutant p97 proteins were reported to retain a normal hexameric structure and ATPase activity (Weihl et al, 2006), although specific mutants might hydrolyse ATP at an elevated rate (Halawani et al, 2009).
In this study, we show that IBMPFD mutations induce conformational alterations in the N domain of p97, which result from impaired communication between the D1 and N domains. Importantly, these alterations result in significantly perturbed co-factor interactions of p97, suggesting that imbalanced co-factor binding is an important determinant of IBMPFD pathology.
Results
IBMPFD mutations cause structural alterations in p97
To elucidate the molecular basis of IBMPFD, we initially analysed potential structural defects of four representative, purified p97 mutant proteins causing IBMPFD. We did not observe alterations in the overall structure or ATPase activity (supplementary Figs S1–S3 online; Weihl et al, 2006; Hubbers et al, 2007). Notably, however, limited proteolysis experiments revealed significant differences between wild-type and IBMPFD-causing p97 proteins. In the presence of a small amount of trypsin, wild-type p97 was cleaved rapidly, leading to the transient appearance of a 75 kDa fragment that was degraded further to yield a metastable fragment of about 52 kDa (Fig 1A, upper panel). The addition of nucleotides slightly slowed degradation kinetics, without affecting the fragment pattern (Fig 1A, upper panel; Wang et al, 2003; Halawani et al, 2009). In the absence of nucleotides, IBMPFD-causing p97 mutant proteins exhibited degradation patterns and kinetics very similar to those of wild-type p97 (Fig 1A). Surprisingly, however, on addition of ADP, the metastable 52 kDa fragment was hardly detectable for IBMPFD-causing p97 mutant proteins, despite an apparently normal decrease of the 75 kDa fragment. In the presence of ATP, the 52 kDa fragment and an additional, slightly larger fragment were generated from IBMPFD-causing p97 mutant proteins. Importantly, this ATP-induced fragment doublet was observed in all IBMPFD-causing p97 mutant proteins analysed, but not in the p97K251A and p97K524A mutant proteins unrelated to IBMPFD (Fig 1B). Edman analysis identified the amino-terminus of p97 in the upper tryptic fragment that is observed exclusively with IBMPFD-causing p97 mutant proteins, whereas the 52 kDa fragment started carboxy-terminal of residue Lys 18 (Fig 1C; supplementary Fig S4 online). Masses of about 54 kDa for the upper fragments and about 52 kDa for the lower fragments were determined by electrospray ionization mass spectrometry (ESI-MS), with the mass differences matching the mass of residues 1–18 (supplementary Table S1 online). From these data, residue Arg 487 in the first helix of the D2 domain was calculated to be the tryptic target site giving rise to both fragments (Fig 1C; supplementary Table S1 online). In conclusion, the 54 kDa fragment observed in IBMPFD-causing p97 mutant proteins in the presence of ATP results from a structural rearrangement that protects the N-terminal region around residue Lys 18 from tryptic cleavage. Conversely, the lack of the 52 kDa fragment observed in the presence of ADP in IBMPFD-causing 97 mutant proteins could be due either to protection of residue Arg 487, or, more probably, to a strongly increased susceptibility of the N and/or D1 domains to tryptic degradation. Thus, our results reveal significant, nucleotide-induced structural alterations in IBMPFD-causing p97 mutant proteins.
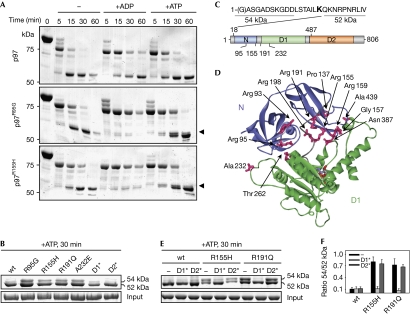
Nucleotide-induced N domain rearrangements in IBMPFD-causing p97 mutant proteins. (A) Time course of partial tryptic degradation of recombinant p97 in the absence and presence of the indicated nucleotides. Reaction products were separated by SDS–PAGE and stained with Coomassie brilliant blue. Arrowheads mark an additional fragment of IBMPFD-causing mutant p97. (B) Details of the tryptic pattern of various p97 mutants in the presence of ATP. The constitutive 52 kDa and IBMPFD-specific 54 kDa fragments are labelled. D1*, p97K251A; D2*, p97K524A. (C) Edman analysis of the 52 and 54 kDa tryptic p97 fragments. Top: N terminal sequence of p97. Residues identified by Edman degradation of the indicated fragment are underlined; residue Lys 18, the cleavage site giving rise to the 52 kDa fragment, is in bold. Bottom: domain organization of p97. Positions of the tryptic cleavage sites Lys 18 and Arg 487 and of IBMPFD-causing mutations analysed in this study are indicated. (D) IBMPFD-causing mutations localize to the N–D1 domain interface. The three-dimensional structure of the ND1 fragment of p97 (Protein Data Bank entry 1E32) is coloured according to panel (C). Mutated residues (magenta sticks) at the domain interface are labelled. The structure lacks residues 1–20. (E) Details of the tryptic pattern of p97 variants combining IBMPFD-causing mutations (R155H, R191Q) with mutations in the D1 (D1*) or D2 (D2*) ATPase domains. (F) Quantification of the results in (E). The mean (n=3) with s.d. values for the ratio between the 54 and 52 kDa fragments are shown. IBMPFD, inclusion body myopathy with Paget disease of bone and frontotemporal dementia; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis; wt, wild type.
The three-dimensional structure of the N-terminal region of p97 is unknown, precluding a more detailed structural interpretation of the differential accessibility of residue Lys 18 in wild-type and mutant p97. Remarkably, however, most VCP mutations in IBMPFD patients affect p97 residues that localize to the interface between the N and D1 domains (Fig 1C,D). Moreover, the 54 kDa tryptic fragment was also observed for the p97A232E mutant protein possessing a mutation in the D1 domain (Fig 1B–D), suggesting that the nucleotide-induced conformational alterations at the N-terminus might result from perturbed interactions with the adjacent D1 ATPase ring. Indeed, recent structural studies of wild-type p97 revealed a direct interaction of nucleotides bound to D1 with the linker connecting the N and D1 domains, supporting the hypothesis that N domain conformation is dictated by D1 nucleotide state (Zhang et al, 2000; DeLaBarre & Brunger, 2005). To explore the involvement of the D1 domain in structural alterations of mutant p97, we examined the appearance of the 54 kDa tryptic fragment in two p97 variants that combined IBMPFD mutations with Walker A motif mutations preventing nucleotide binding to the D1 or D2 ATPase domains, respectively. Intriguingly, the 54 kDa fragment was lost on mutation of the Walker A motif in the D1, but not in the D2, domain (Fig 1E,F). These results indicate that nucleotide binding to the D1 domain is a prerequisite for structural alterations at the N-terminus of IBMPFD-causing p97 mutant proteins, most probably because of altered transmission of conformational changes from the D1 ring to the N domain of p97.
Ufd1–Npl4 impairs binding of E4B to p97 mutants
The finding that IBMPFD-causing p97 mutant proteins exhibit structural rearrangements of the N domain prompted us to analyse their interaction with purified, recombinant N domain-binding co-factors in vitro. Consistent with a previous report (Hubbers et al, 2007), we failed to detect differences between wild-type and mutant p97 proteins in their ability to bind to two major N domain co-factors, p47 and the heterodimer Ufd1–Npl4 (data not shown). We also investigated the binding of IBMPFD-causing p97 mutant proteins to the ubiquitin chain-elongating co-factor E4B, the human homologue of Caenorhabditis elegans UFD-2. UFD-2 and E4B have been implicated in the degradation of the myosin chaperone UNC-45 in worm larvae and IBMPFD patient myoblasts, respectively (Janiesch et al, 2007). Interestingly, the Slow Wallerian degeneration protein (WldS), a murine chimeric fusion protein between an E4B fragment and Nmnat1, was reported recently to bind to the N domain of p97 through a VCP-binding motif (VBM) within its E4B portion (Morreale et al, 2009). We found that full-length human E4B binds directly to the N domain of p97 (supplementary Fig S5 online) through its VBM (data not shown; Boeddrich et al, 2006). However, as with p47 and Ufd1–Npl4, we could not detect significant differences in the direct binding of wild-type and mutant p97 to recombinant E4B (data not shown).
We reasoned that the ability of p97 to function in various cellular pathways should not only require proficient binding to isolated co-factors but also an intact balance of co-factor interactions in their simultaneous presence, that is, under competitive conditions. As Ufd1–Npl4 and p47 are known to bind to p97 in a mutually exclusive manner (Meyer et al, 2000), we analysed binding of IBMPFD-causing p97 mutant proteins to Ufd1–Npl4 in the presence of increasing amounts of p47. Again, wild-type and mutant p97 did not differ significantly with respect to the mutual exclusive binding of Ufd1–Npl4 and p47 (Fig 2A). We then tested whether binding of E4B to p97 is mutually exclusive with p47 or Ufd1–Npl4. We found that increasing amounts of p47 competed for the binding of wild-type and mutant p97 to E4B with similar efficiency (Fig 2B, upper panel; Fig 2D). Under the same conditions, Ufd1–Npl4 was unable to compete with E4B for the binding to wild-type p97. Intriguingly, however, increasing amounts of Ufd1–Npl4 resulted in significantly reduced binding of E4B to IBMPFD-causing p97 mutant proteins, indicating efficient competition (Fig 2B, middle panel; Fig 2D). This difference between wild-type and mutant p97 in competitive binding of Ufd1–Npl4 and E4B was specific, because addition of a large excess of bovine serum albumin did not affect E4B binding (Fig 2B, lower panel; Fig 2D). These data provide the first evidence that IBMPFD-causing p97 mutant proteins are impaired in E4B binding in the presence of Ufd1–Npl4.
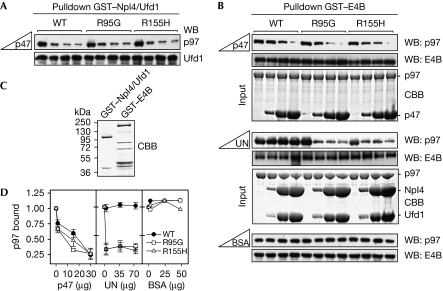
IBMPFD-causing p97 mutant proteins exhibit reduced binding to E4B in the presence of Ufd1–Npl4. (A) Mutually exclusive binding of Ufd1–Npl4 and p47 to wild-type and mutant p97. Binding of p97 to glutathione–Sepharose beads decorated with GST–Npl4/Ufd1 heterodimer in the presence of increasing amounts of p47 was analysed by western blot analysis using the indicated antibodies. (B) Binding of wild-type and mutant p97 to glutathione–Sepharose beads decorated with GST–E4B in the presence of increasing amounts of p47, Ufd1–Npl4 or BSA (upper, middle and lower panels, respectively) was analysed by western blot analysis using the indicated antibodies. For the competition with p47 and Ufd1–Npl4, CBB-stained gels of the protein(s) added to the GST–E4B-decorated beads are shown (input). (C) CBB-stained gel showing equal amounts of the input used in (A) and (B)—GST–Npl4/Ufd1 and GST–E4B, respectively. (D) Quantification of bound p97 in (B). n=2 for competition with p47; n=3 for competition with Ufd1–Npl4; n=1 for BSA control. BSA, bovine serum albumin; CBB, Coomassie brilliant blue; GST, glutathione-S-transferase; IBMPFD, inclusion body myopathy with Paget disease of bone and frontotemporal dementia; WB, western blot.
Imbalanced p97 co-factor interactions in vivo
We next investigated co-factor interactions of IBMPFD-causing p97 mutant proteins in living cells, in which various N-domain-binding co-factors compete for p97 (Alexandru et al, 2008; Schuberth & Buchberger, 2008). To this end, p97 was immunoprecipitated from human embryonic kidney (HEK) 293T cells ectopically expressing wild-type or mutant p97, and immunoprecipitates were analysed for associated endogenous co-factors. Intriguingly, this approach revealed marked differences in the level of co-factors bound to wild-type p97 compared with IBMPFD-associated p97 (Fig 3A,E). When compared with wild-type p97, significantly elevated amounts of p47 and Ufd1–Npl4 were co-precipitated with IBMPFD-causing mutant p97. By contrast, and in line with the results of in vitro experiments, the amount of bound E4B was reduced significantly. Notably, binding of these co-factors to the p97E305A mutant protein deficient in ATP hydrolysis by the D1 ring, but unrelated to IBMPFD, was similar to that of wild-type p97, suggesting that the differences observed for IBMPFD-associated p97 are not merely a consequence of altered ATP binding or hydrolysis. Furthermore, binding of peptidyl:N-glycanase, a co-factor binding to the C-terminus of p97 (Yeung et al, 2008), was not affected by any of the mutations tested (Fig 3A,E). Together, these data demonstrate for the first time, to our knowledge, altered interactions of IBMPFD-causing p97 mutant proteins with N domain-binding co-factors in vivo, including an impaired interaction with the ubiquitin ligase E4B.

Perturbed co-factor interactions of IBMPFD-causing p97 mutant proteins in vivo. (A) Lysates (Input) of HEK293T cells expressing the indicated Flag-tagged p97 variants were subjected to anti-Flag immunoprecipitation. Immunoprecipitated p97 and co-precipitated endogenous co-factors were detected by western blot analysis using the indicated antibodies. Tubulin was the input control. (B) The same experiment as in (A) was performed to compare the binding of the VBM-containing co-factors E4B, Atx3 and Hrd1 to the indicated p97 variants. (C) Lysates (Input) of healthy (H1 and H2) and IBMPFD patient (R155H, R155C and R155S) myoblasts were subjected to immunoprecipitation using p97 or control (IgG) antibodies. Immunoprecipitated p97 and co-precipitated co-factors were detected by western blot analysis using the indicated antibodies. (D) The same lysates as in (C) were subjected to immunoprecipitation using Atx3 or control (IgG) antibodies. Immunoprecipitated Atx3 and co-precipitated p97 were detected by western blot analysis with the indicated antibodies. An unspecific band in the input recognized by the 1H9, but not the MJD, Atx3 antibody is marked by an asterisk. (E) Quantification of co-immunoprecipitations. Top, for panels (A) and (B): for wt, R95G and R155H, n=3 for E4B, p47, Np14 and Atx3, and n=2 for PNGase and Hrd1; for E305A, n=2 for E4B and PNGase, and n=1 for p47, Np14, Atx3 and Hrd1. Middle, for panel (C): n=2. Bottom, for panel (D): n=1. Atx3, ataxin 3; HEK, human embryonic kidney; IBMPFD, inclusion body myopathy with Paget disease of bone and frontotemporal dementia; IP, immunoprecipitation; VBM, valosin-containing protein-binding motif; WB, western blot; wt, wild type.
As E4B binds to p97 through a VBM (Boeddrich et al, 2006), we asked whether two other VBM-containing co-factors involved in ERAD, the E3 ubiquitin ligase Hrd1 and the deubiquitinating enzyme ataxin 3, would likewise exhibit impaired binding to IBMPFD-causing mutant p97 proteins. Surprisingly, however, significantly higher amounts of endogenous ataxin 3 bound to IBMPFD-causing and to D1 ATPase-deficient p97 mutant proteins, compared with wild-type p97 (Fig 3B,E). By contrast, equal amounts of Hrd1 were co-immunoprecipitated with wild-type, IBMPFD-causing and D1 ATPase-deficient p97 (Fig 3B,E). Thus, IBMPFD-associated mutations have significant but opposite effects on the binding of the two co-factors, E4B and ataxin 3 implicated in proteinopathies, but not on Hrd1 binding.
Finally, we analysed p97–co-factor interactions under pathophysiologically relevant conditions—that is, in cultured primary myoblasts from three IBMPFD patients expressing p97 proteins altered in the mutational hotspot Arg 155, in comparison with myoblasts from two healthy individuals. On immunoprecipitation of endogenous p97 from these cells, significantly more p47, Npl4 and ataxin 3 bound to p97 from IBMPFD myoblasts, whereas there was no difference in the interaction with Hrd1 (Fig 3C,E). We failed to detect E4B in immunoprecipitates from either healthy or patients' myoblasts (data not shown). The stronger binding of ataxin 3 to p97 was confirmed further by elevated amounts of p97 in immunoprecipitates of endogenous ataxin 3 from IBMPFD myoblasts (Fig 3D,E). These results demonstrate that IBMPFD-causing mutant p97 proteins exhibit imbalanced co-factor associations in the pathophysiological context of patients' myoblasts.
Discussion
Nucleotide binding to the D1 ring of p97 has important regulatory functions with respect to N domain conformation and co-factor binding (Zhang et al, 2000; DeLaBarre & Brunger, 2005; Beuron et al, 2006). This study, to the best of our knowledge, provides the first evidence that IBMPFD-causing mutations affect the normal control of N domain conformation by the D1 nucleotide state, as indicated by the protection of the N-terminal region from proteolytic attack. This region has so far eluded structural characterization by crystallography and nuclear magnetic resonance spectroscopy, suggesting that it is conformationally flexible. Interestingly, however, its amino-acid sequence is highly conserved and residues 13–17 are predicted, although with low confidence, to form a short α-helix (data not shown). We thus speculate that the N-terminal region of p97 is not completely unstructured, but rather involved in defined, D1-controlled conformational changes of the N domain, which in turn regulate co-factor interactions.
In vivo, even minor defects in the control of co-factor interactions are potentially toxic, because p97 might become trapped in unproductive co-factor complexes in various cellular degradation pathways. IBMPFD manifests as a late-onset disease, and it has been speculated that IBMPFD-causing mutations in VCP might have relatively subtle, but ultimately fatal, effects on p97 function (Watts et al, 2004). Our finding that IBMPFD-causing mutations disturb, but do not completely abolish, p97 co-factor interactions provides evidence in support of such a pathogenesis mechanism. It is noteworthy that we observed perturbed co-factor binding in primary myoblasts from IBMPFD patients that lack pathological protein aggregates (Hubbers et al, 2007), suggesting that this is an early event in IBMPFD pathogenesis rather than a consequence of the disease.
Speculation
Unexpectedly, our study revealed an increased interaction of IBMPFD-causing mutant p97 with the deubiquitinating enzyme ataxin 3. Enhanced binding of polyglutamine-expanded ataxin 3 to wild-type p97 has been observed in the neurodegenerative disease spinocerebellar ataxia type 3 (Hirabayashi et al, 2001; Matsumoto et al, 2004), and p97 and Ufd1–Npl4 have recently been shown to be trapped by polyglutamine-expanded proteins (Duennwald & Lindquist, 2008). The analogous accumulation of wild-type ataxin 3 and Ufd1–Npl4 on mutant p97 in IBMPFD illustrates that mutations in either interaction partner can have similar consequences at the molecular level. Thus, our results might be a first hint to a potential involvement of ataxin 3 in the neurodegenerative manifestation of IBMPFD. Future studies on IBMPFD might shed light on pathogenesis mechanisms in yet other proteinopathies that similarly affect p97 function through mutations in regulatory co-factors.
Methods
Proteins. Information about cloning procedures and protein purification can be found in supplementary information online.
Partial proteolysis. Tryptic degradation was performed at 25°C using 25 μg p97 and 0.625 μg trypsin (Roche, Mannheim, Germany) in 50 μl buffer (50 mM Tris–HCl (pH 7.8), 10 mM MgCl2, 1 mM dithiothreitol; 3 mM nucleotide as indicated). Reactions were stopped at different time points and samples were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis. ESI-MS analysis and Edman sequencing of tryptic fragments were performed as described in supplementary information online.
In vitro binding assays. Glutathione–Sepharose pulldown experiments were performed as described previously (Kern et al, 2009) using 0.04 nmol glutathione-S-transferase fusion proteins and 20 μl bead slurry. p97 (0.04 nmol) in the absence or presence of co-factors (at 1-, 12.5- and 25-fold molar excess) was added for 1 h in a total volume of 120 μl buffer (50 mM Tris–HCl (pH 7.8), 150 mM NaCl, 1 mM dithiothreitol, 0.1% Triton X-100).
Immunoprecipitation. HEK293T cells were transiently transfected with pCMV-Tag2B-p97 by using the calcium phosphate method. Human primary normal and IBMPFD patient myoblasts (Muscle Tissue Culture Collection/EuroBioBank; Friedrich-Baur-Institute, Munich, Germany; Hubbers et al, 2007; Vesa et al, 2009) were maintained in skeletal muscle growth medium (PromoCell, Heidelberg, Germany) containing 15% fetal calf serum and split when less than 70% confluent. Cell lysis and immunoprecipitations were performed as described previously (Kern et al, 2009), using Flag–M2 beads (Sigma-Aldrich, Munich, Germany), or p97 (Novus Biologicals, Cambridge, UK), ataxin 3 (MJD; a gift from H. Paulson) and control (Bethyl Laboratories, Montgomery, TX, USA) antibodies bound to Protein G–Sepharose beads.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Acknowledgments
We thank S. Jentsch for continued support; E. Weyher and J. Kellermann for ESI-MS and Edman analysis, respectively; O. Stemmann, E. Wiertz, T. Suzuki and H. Paulson for antibodies; S. Krause and the Muscle Tissue Culture Collection for IBMPFD patient and control myoblasts; and S. Müller and S. Böhm for comments on the paper. This work was supported financially by the Deutsche Forschungsgemeinschaft (to A.B.) and the Basque Government (postdoctoral fellowship to V.F-S).
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134: 804–816 [Europe PMC free article] [Abstract] [Google Scholar]
- Beuron F et al. (2006) Conformational changes in the AAA ATPase p97–p47 adaptor complex. EMBO J 25: 1967–1976 [Europe PMC free article] [Abstract] [Google Scholar]
- Boeddrich A et al. (2006) An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J 25: 1547–1558 [Europe PMC free article] [Abstract] [Google Scholar]
- DeLaBarre B, Brunger AT (2005) Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol 347: 437–452 [Abstract] [Google Scholar]
- Duennwald ML, Lindquist S (2008) Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev 22: 3308–3319 [Europe PMC free article] [Abstract] [Google Scholar]
- Halawani D, Leblanc A, Rouiller I, Michnick SW, Servant MJ, Latterich M (2009) Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2-domain and the D1 ring modulate p97/VCP ATPase activity and D2 AAA+ ring conformation. Mol Cell Biol 29: 4484–4494 [Europe PMC free article] [Abstract] [Google Scholar]
- Hirabayashi M et al. (2001) VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ 8: 977–984 [Abstract] [Google Scholar]
- Hubbers CU et al. (2007) Pathological consequences of VCP mutations on human striated muscle. Brain 130: 381–393 [Abstract] [Google Scholar]
- Ishigaki S, Hishikawa N, Niwa J, Iemura S, Natsume T, Hori S, Kakizuka A, Tanaka K, Sobue G (2004) Physical and functional interaction between Dorfin and Valosin-containing protein that are colocalized in ubiquitylated inclusions in neurodegenerative disorders. J Biol Chem 279: 51376–51385 [Abstract] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T (2007) The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol 9: 379–390 [Abstract] [Google Scholar]
- Ju JS, Miller SE, Hanson PI, Weihl CC (2008) Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J Biol Chem 283: 30289–30299 [Europe PMC free article] [Abstract] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC (2009) Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 187: 875–888 [Europe PMC free article] [Abstract] [Google Scholar]
- Kern M, Fernandez-Saiz V, Schafer Z, Buchberger A (2009) UBXD1 binds p97 through two independent binding sites. Biochem Biophys Res Commun 380: 303–307 [Abstract] [Google Scholar]
- Kimonis VE, Fulchiero E, Vesa J, Watts G (2008) VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta 1782: 744–748 [Abstract] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI (2004) Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J 23: 659–669 [Europe PMC free article] [Abstract] [Google Scholar]
- Meyer HH, Popp O (2008) Role(s) of Cdc48/p97 in mitosis. Biochem Soc Trans 36: 126–130 [Abstract] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (2000) A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19: 2181–2192 [Europe PMC free article] [Abstract] [Google Scholar]
- Mizuno Y, Hori S, Kakizuka A, Okamoto K (2003) Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci Lett 343: 77–80 [Abstract] [Google Scholar]
- Morreale G, Conforti L, Coadwell J, Wilbrey AL, Coleman MP (2009) Evolutionary divergence of valosin-containing protein/cell division cycle protein 48 binding interactions among endoplasmic reticulum-associated degradation proteins. FEBS J 276: 1208–1220 [Abstract] [Google Scholar]
- Rumpf S, Jentsch S (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21: 261–269 [Abstract] [Google Scholar]
- Schuberth C, Buchberger A (2008) UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci 65: 2360–2371 [Abstract] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP (2010) VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6: 217–227 [Europe PMC free article] [Abstract] [Google Scholar]
- Vesa J, Su H, Watts GD, Krause S, Walter MC, Martin B, Smith C, Wallace DC, Kimonis VE (2009) Valosin containing protein associated inclusion body myopathy: abnormal vacuolization, autophagy and cell fusion in myoblasts. Neuromuscul Disord 19: 766–772 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Q, Song C, Yang X, Li CC (2003) D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97–VCP. J Biol Chem 278: 32784–32793 [Abstract] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36: 377–381 [Abstract] [Google Scholar]
- Weihl CC, Dalal S, Pestronk A, Hanson PI (2006) Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum Mol Genet 15: 189–199 [Abstract] [Google Scholar]
- Weihl CC, Pestronk A, Kimonis VE (2009) Valosin-containing protein disease: inclusion body myopathy with Paget's disease of the bone and fronto-temporal dementia. Neuromuscul Disord 19: 308–315 [Europe PMC free article] [Abstract] [Google Scholar]
- Yeung HO, Kloppsteck P, Niwa H, Isaacson RL, Matthews S, Zhang X, Freemont PS (2008) Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans 36: 62–67 [Abstract] [Google Scholar]
- Zhang X et al. (2000) Structure of the AAA ATPase p97. Mol Cell 6: 1473–1484 [Abstract] [Google Scholar]
Articles from EMBO Reports are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/embor.2010.49
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2892324?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains.
Nat Struct Mol Biol, 08 Jul 2024
Cited by: 0 articles | PMID: 38977901
Valosin-Containing Protein (VCP): A Review of Its Diverse Molecular Functions and Clinical Phenotypes.
Int J Mol Sci, 25(11):5633, 22 May 2024
Cited by: 1 article | PMID: 38891822 | PMCID: PMC11172259
Review Free full text in Europe PMC
Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes.
Int J Mol Sci, 25(6):3371, 16 Mar 2024
Cited by: 0 articles | PMID: 38542345 | PMCID: PMC10970294
p37 regulates VCP/p97 shuttling and functions in the nucleus and cytosol.
Sci Adv, 10(18):eadl6082, 03 May 2024
Cited by: 1 article | PMID: 38701207 | PMCID: PMC11068011
Advances in the study of protein folding and endoplasmic reticulum-associated degradation in mammal cells.
J Zhejiang Univ Sci B, 25(3):212-232, 01 Mar 2024
Cited by: 1 article | PMID: 38453636 | PMCID: PMC10918413
Review Free full text in Europe PMC
Go to all (78) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases
- (1 citation) OMIM - 167320
Protein structures in PDBe
-
(1 citation)
PDBe - 1E32View structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A unique IBMPFD-related P97/VCP mutation with differential binding pattern and subcellular localization.
Int J Biochem Cell Biol, 45(4):773-782, 16 Jan 2013
Cited by: 24 articles | PMID: 23333620
Structural and functional deviations in disease-associated p97 mutants.
J Struct Biol, 179(2):83-92, 08 May 2012
Cited by: 14 articles | PMID: 22579784 | PMCID: PMC4788498
Review Free full text in Europe PMC
Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy.
Hum Mol Genet, 19(r1):R38-45, 21 Apr 2010
Cited by: 87 articles | PMID: 20410287 | PMCID: PMC2875057
Review Free full text in Europe PMC
Valosin-Containing Protein (VCP)/p97 Oligomerization.
Subcell Biochem, 104:485-501, 01 Jan 2024
Cited by: 0 articles | PMID: 38963497
Review




