Abstract
Free full text

Yeast targets for mRNA methylation
Abstract
N6-Methyladenosine (m6A) is a modified base present in the mRNA of all higher eukaryotes and in Saccharomyces cerevisiae, where there is an increase in m6A levels during sporulation. The methyltransferase, Ime4, is responsible for this modification and has a role in the initiation of meiosis. However, neither the function, nor the extent of distribution of this nucleotide modification is established. We demonstrate that in S. cerevisiae, substantial levels of internal adenosine methylation are present in the GpA context in mRNA from sporulating cells, which is consistent with the preferred methylation consensus of higher eukaryotes. Based upon our quantification data, every second transcript could contain one m6A during meiosis. As methylation is distributed across all mRNA size ranges, it is likely that m6A is not limited to a small population of messages. We developed a new antibody based method for identifying m6A containing messages, and using this method the transcripts of three key, early regulators of meiosis, IME1, IME2 and IME4 itself, were identified as being methylated. The position of m6A in IME2 was narrowed down to a region in the 3′-end. Methylation of these and other targets suggests mechanisms by which IME4 could control developmental choices leading to meiosis.
INTRODUCTION
The process of methylation in eukaryotic DNA is recognized for its fundamental role in regulating gene expression. Methylation in certain RNA species also has an important role during the course of maturation, structural organization and stability. The presence of N6-Methyladenosine has been observed in many RNA species, including tRNA, rRNA and small nuclear RNA (snRNA) (1–6), but the function of this modified nucleotide in mRNA has remained unclear for more than 30 years (7,8).
N6-Methyladenosine (m6A) is detected in the mRNA of some viruses (9,10) and eukaryotes, such as sporulating yeast (11), mammals (12–14), insects (15), monocot plants (16–18) and in the model plant, Arabidopsis thaliana (19). The methyltransferase activity responsible was first purified from HeLa cell extract and was associated with a 70-kD subunit of a 200-kD protein complex (20). After further purification and micro sequencing the protein was identified and named MT-A70.
Four lineages (A–D) of proteins homologous with MT-A70 have been identified in a phylogenetic analysis (21). Members of lineage A, the human MT-A70 (20), the plant homologue MTA (19) and the yeast, Saccharomyces cerevisiae IME4 (11) are the only experimentally proven mRNA m6A methyltransferases.
IME4 has previously been characterized as one of the key regulators of the complex pathways leading to the entry into meiosis in yeast (22). Correct meiosis and sporulation only occurs in diploid yeast, under nitrogen and fermentable carbon source starvation. Under these conditions the expression of IME1 will lead to a complex cascade of gene expression (23). In early sporulating S. cerevisiae mRNA, low levels of m6A were shown and this methylation was IME4-dependent. It is now clear that IME4 exerts its mRNA methyltransferase activity under sporulating conditions (11); however the mechanism by which this regulates early meiotic events remains unknown
An A to m6A conversion in an mRNA molecule would not be expected to alter base pairing specificity and so would not be revealed by cDNA sequencing, thus making the detection and mapping of m6A residues in individual transcripts very difficult. For this reason mapping has been reported for only two highly abundant mRNAs, Rous sarcoma virus and bovine prolactin (24,25). However, analysis of mRNA using ribonuclease fragmentation and labeling methods from animal and maize tissues detected m6A at the central A position in the GAC and AAC sequence context, with a 75% preference for GAC (14,26–28). Similar results were found in A. thaliana and in these experiments the authors quantified m6A levels and showed significant differences between plant tissues (19).
In the R. sarcoma virus genomic RNA, the 1865-nt region studied contains seven m6A in a GACU context. Most of these are within the sequence GGACU, but some are also present in UGACU and AGACU sequences. The bovine prolactin mRNA, contains only one m6A at an AGACU site within the 3′-untranslated region. The extended consensus sequence RRACH (where R = purine and H = A, C or U), suggested by Schibler et al. (26) is consistent with the the mapped methylation sites. However, other structural elements might have a function in selecting the methylation sites, as the suggested consensus for adenosine methylation is far more frequent in an mRNA population than the actual observed frequency of m6A (0.1–0.2% of the nucleotides).
We detected m6A in mRNA from sporulating yeast in a GA but not in a CA or UA sequence context. This is consistent with the consensus sequence found in plants and animals. We quantified the abundance of the methylated dinucleotide Gpm6A in mRNA from sporulating yeast, and we show that methylation is not restricted to a particular mRNA size class. We have also developed a new antibody based method for identifying m6A containing transcripts and found that several early meiotic transcripts are methylated. Further exploitation of this method will allow a global analysis of methylated transcriptomes. In the IME2 message we quantified the m6A and mapped it to the 3′-portion of the mRNA molecule. The identification of methylated mRNA targets of IME4 will likely be an important step in understanding how this base modification regulates gene expression in S. cerevisiae and possibly other eukaryotes.
MATERIALS AND METHODS
Strains and general methods
All analysis was carried out using the rapidly sporulating SK1 (can1), diploid strain (ATCC). Cultures were routinely grown in YPD (1% Bacto-yeast extract, 2% Bacto-peptone, 2% glucose) to a cell density 107 cell/ml, (log phase).
For sporulation experiments, one single colony was inoculated in YPD and grown to 108 cell/ml. This culture was harvested by centrifugation at 1000g for 3 min at room temperature. After washing with sterile water, the pellet was resuspended to a 5 × 106 cell/ml density in PSP2 medium (29). The culture was grown for five generations at 30°C with vigorous shaking, and then harvested by centrifugation. The pellet was washed with sterile water and resuspended in SPM (0.3% potassium acetate, 0.02% raffinose) (29) to a 107 cell/ml density to induce sporulation. Following 3 h vigorous shaking at 30°C, the culture was spun down and the pellet was used for RNA extraction.
RNA extraction method
For total RNA extraction we used a modified, and scaled up, hot phenol extraction method (30) on both vegetative and sporulating cultures. After adding phenol, the cells were disrupted by heating to 65°C, followed by a rapid chill in liquid nitrogen. Glass beads were also added, to facilitate the cell breakage. After centrifugation, the aqueous phase was taken and the phenol extraction step repeated, but omitting glass beads. Poly(A) RNA was purified using oligodT-Cellulose columns (Fluka) following a standard protocol (31). The oligo(dT) chromatography was carried out twice on each sample and the quality of the mRNA was checked on an RNA 6000 LabChip, with Agilent Bioanalyzer (Ambion).
Size fractionation of mRNA
Poly(A) RNA, isolated from sporulating cells, was separated on a 1.3% TBE agarose gel, for half an hour at 100 V. Three fractions of increasing molecular weight were cut out from the gel. The agarose cubes were snap frozen in liquid nitrogen and spun in a micro centrifuge at 7000g for 15 min. The liquid was collected and precipitated with ethanol at −20°C for 1 h. The integrity and the size range of the fractions were confirmed using RNA 6000 LabChip and Agilent Bioanalyser (Ambion). The fractions were subsequently analyzed, using the TLC method and their m6A content was quantified (see below)
Detection of m6A using TLC chromatography
The analysis and the quantification of m6A in different samples were carried out by applying the method from Zhong et al. (19). For each sample, 50 ng of mRNA was digested with 1 µl of Ribonuclease T1 (1000 U/µl; Fermentas) in a final volume of 10 µl 1× polynucleotide kinase (PNK) buffer A for 1 h at 37°C. Another aliquot of the same RNA sample was digested with 1 µl RNaseA (4 μg/µl; Promega) in a final volume of 10 μl 1× PNK buffer A. The 5′-end of the T1 and RNaseA digested mRNA fragments were then labeled using 10 units of T4 PNK (Fermentas) and 1 µl [γ-32P] ATP (6000 Ci/mmol; Perkin-Elmer). The labeled RNA was precipitated, and resuspended in 10 µl of 50 mM sodium acetate buffer (pH 5.5) and digested with P1 nuclease (Sigma-Aldrich) for 1 h at 37°C. Two microliters of each sample was loaded on cellulose TLC plates (20 × 20 cm; Merck) and developed in a solvent system of isobutyric acid: 0.5 M NH4OH (5 : 3, v/v), as first dimension, and isopropanol:HCl:water (70 : 15: 15, v/v/v), as the second dimension. The identification of the nucleotide spots was carried out using m6A containing synthetic RNA. For the quantification of spot intensities, a storage phosphor screen (K-Screen; Kodak) and Bio-Rad Molecular Imager FX in combination with Quantity One 4.6.3. software (Bio-Rad) was applied.
Immunoprecipitation of m6A containing messages
The method used for raising the monoclonal antibody against the N6-methyladenosine-5′-mono-phosphate conjugated to BSA, is described in the supplementary data section (Supplementary Data 1; BioGenes).
The anti-m6A antibody (Ab) was characterized by immunoblot analysis. In vitro transcribed RNA, prepared using RiboMAX RNA Production System-T7 (Promega), (for sequences and constructs see Supplementary Data 2), with and without m6A (N6- methyladenosine-triphosphate was purchased from Trilink) was separated on an agarose gel and transferred to nitrocellulose membrane (Optitran BA-S83, Schleicher and Schuell) using standard methods. The RNA was cross linked by UV light in a Stratalinker (Stratagene) and subjected to western blot analysis, using the anti-m6A antibody (mouse IgG1) and a WesternBreeze anti mouse kit (Invitrogen). Similar RNA samples were mixed (500 ng each, methylated and non-methylated RNA) and incubated with 3.2 μg anti-m6A antibody at 4°C for 1 h in 500 μl PBS. Subsequently 30 μl proteinG magnetic beads (NEB) was added and incubated at 4°C for 1 h. The magnetic beads were separated together with the Ab bound mRNA. Four PBS washes followed and the Ab bound mRNA was extracted by phenol. After ethanol precipitation, the mRNA samples from the Ab bound fraction and from the Ab depleted fraction were subjected to northern blot analysis. The probes were DNA fragments identical to the in vitro transcribed sequences (Supplementary Data 2) and were radio labeled with [α-32P] dCTP using Rediprime Kit (Amersham).
For the immunoprecipitation (IP) experiments on yeast mRNA, 1.2 μg poly(A)RNA was bound to oligo(dT) magnetic beads, using the PolyTtrack System 1000 (Promega) and washed 3× with PBS buffer. Following the washes 3.8 μg Ab and 3 μl RNAguard (Amersham Pharmacia) was added to the oligo(dT) bound mRNA, in a total volume of 500 μl PBS. The mixture was incubated for 1 h at 4°C. The magnetic beads were separated and washed twice with PBS. At this point all mRNA, both Ab bound and unbound, was eluted from the oligo(dT) magnetic beads by adding 50 μl water and incubated for 5 min. This step was repeated and the two eluted volumes were pooled together. The salt concentration was adjusted to 160 mM by the addition of 5 M sodium chloride and the final volume was made 400 μl by adding PBS. This sample was treated with 30 μl proteinG magnetic beads (NEB) for 1 h at 4°C. The magnetic beads were pulled away and the supernatant was kept; this retained supernatant was used as the Ab depleted fraction. The magnetic beads were washed four times with PBS. Finally the Ab bound mRNA was eluted with 10 μl 3 M sodium acetate (pH 5.6), incubated for 2 min. at room temperature. Following the incubation the volume was made up to 200 μl with water and the beads plus mRNA were extracted with an equal volume of acid phenol (pH 5.3) and precipitated with ethanol.
In the initial experiments a mixture of mRNA from sporulating cells (600 ng) and mRNA from log phase cells (600 ng) was subjected to IP. In the second set of experiments, mRNA from sporulating cells only was subjected to IP. Both depleted and pull-down fractions were analyzed by quantitative RT–PCR.
Quantitative RT–PCR
Reverse transcription was carried out using SuperScriptII (Invitrogen) and oligo(dT)25. Quantitative PCR was carried out using an MX3005P qPCR machine and the 2× SensiMx Plus SYBR master mix (Quantace). The 2−ΔΔC′T method (32) was used for data analysis. Samples were run in triplicate, and relative expression levels were determined compared with ACT1 expression. ACT1, the normalizer gene, primers were 5′-CTGCCGGTATTGACCAAACT-3′ and 5′-CGGTGATTTCCTTTTGCATT-3′, the IME2 primers were 5′-AGCCCAAGAGCTTTG TGAAA-3′ and 5′-TTTGTGTTGGTCGCTACTGG-3′, the IME1 primers were 5′-ATGCAAG CGGATATGCATGG-3′ and 5′-TTAAGAATAGGTTTTACTAAAC-3′ and the IME4 primers were 5′-ACGAAATGGATGTCGAGAGG-3′ and 5′-TCCAATACTGCTGCTGATGC-3′.
Mapping m6A sites in IME2
Complementary DNA fragments designed to protect the 5′ or 3′ ends of the IME2 message were amplified by PCR from an IME2 template. For this PCR amplification, the reverse oligonucleotide primer was biotinylated and the forward primer was 5′ phosphorylated prior to use. The primers used for protecting the 5′-end of the IME2 message were as follows, forward: 5′-CAGAAAAGTTAATCGTGTTC3′, reverse: 5′biotinGTAATTGGTAATTTTCAGAGTCAATAAAGACCTCAAAAATTTGTATCAG3′. The following primers were used for the 3′ end protection, forward: 5′-GCAATGTGACTAATACAGAAC3′ and reverse: 5′-biotinCATAAAATGTTCAAGAAAAACAAACATAAAAATATATTGGAGGG-3′. The PCR products were treated with Lambda Exonuclease (NEB) to produce single-stranded biotinylated antisense DNA fragments, which were then hybridized at standard conditions, overnight, at 42°C to poly(A) RNA, isolated from SK1 sporulating cells. The IME2 mRNA molecules bound to the biotinylated probes were separated out with streptavidine magnetic beads (NEB) and washed, following standard methods for northern blots. The quantification of m6A was done on the unprotected part of the mRNA molecule, using the TLC method. The ribonuclease T1 digestion was carried out on the magnetic beads for 1 h, than the magnetic beads were pulled away and the supernatant was subjected to TLC analysis as described previously. The purity of IME2 was confirmed using quantitative RT–PCR. ACT1 was used as the normalizing gene, as previously described.
RESULTS
Quantification of m6A in vegetative and sporulating yeast mRNA
In most animal viruses and higher eukaryotes studied to date, m6A is most commonly found following a G in the mRNA (14,19,24–26). To test whether this is also the case in sporulating yeast, a 2D thin layer chromatography (TLC) method was adapted from Zhong et al. (19). This method was also used to quantify m6A levels. The mRNA was digested with T1 ribonuclease. This enzyme cuts after every guanosine, leaving short oligonucleotides that can be labeled with 32P at their 5′-ends. Thus, only those nucleotides that follow a guanosine will be labeled. However, after T1 digestion the only Gs available for labeling are mononucleotide 3′-phosphates which are poor substrates for polynucleotide kinase. The Gs will therefore be underlabeled relative to the spots corresponding to the other nucleotides as these will always be derived from the 5′-end of oligonucleotide fragments (Figure 1). The labeled polynucleotides were digested to give nucleotide 5′ monophosphates, which were separated by TLC. The efficient labeling and the relative positions of the nucleotides on a TLC plate following separation were established using mixtures of in vitro transcribed RNA either containing, or not containing m6A (Figure 1a and b). This detection method was demonstrated to give quantitative values for m6A relative to A, on mixtures of in vitro transcribed methylated and non-methylated transcripts in combination with phosphor imaging (19).
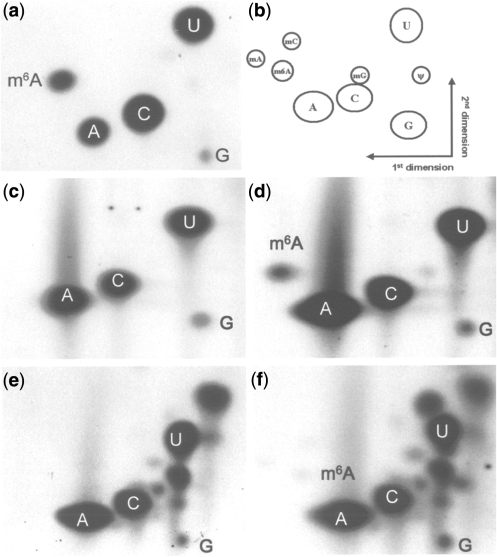
Two-dimensional TLC detection of m6A in mRNA from sporulating SK1 yeast. (a) TLC analysis of in vitro transcribed RNA containing methylated and non-methylated adenosine. (b) Schematic diagram of the relative positions of nucleotide spots. (c) TLC analysis of poly(A) RNA from log phase SK1 strains. (d) TLC analysis of poly(A) RNA from sporulating SK1 strain (3 h after meiosis initiation) m6A:A ratio is 1% (e) TLC analysis of total RNA from log phase SK1 strain. (f) TLC analysis of total RNA from sporulating SK1 strain (3 h after meiosis initiation), m6A:A ratio is 0.07%.
In addition, an RNaseA digestion was also carried out on the mRNA samples. This enzyme cuts after C and U and leaves 5′ OH ends on the following nucleotides which can be labeled as previously described. In these experiments there is an under representation of Cs and Us, because the majority of these nucleotides are mononucleotide 3′-phosphates which are poor substrates for polynucleotide kinase (Supplementary Data 3).
An SK1 diploid yeast culture was grown to log phase and split into two fractions, one of which was used to prepare total RNA. mRNA was prepared from this sample by repeated rounds of oligo(dT) chromatography, and both the poly(A) fraction and the total RNA were subjected to TLC analysis. The rest of the log phase culture was used to inoculate PSP2 medium and was grown for a further five generations to achieve synchrony in sporulation. The cells were harvested and inoculated into sporulation medium (SPM) for 3 h. Total RNA was prepared, the poly(A) fraction isolated and both RNA samples subjected to TLC analysis. The purity of poly(A) RNA samples was always confirmed using RNA 6000 LabChip prior to digestion and labeling (data not shown). An additional indicator of mRNA purity in these experiments is the lack of pseudouridine spots (derived from tRNA or rRNA contamination) on the TLC plates (Figure 1c and d). A spot corresponding to pm6A was clear and substantial in the poly A samples from sporulating yeast (Figure 1d), and was not detectable in the mRNA from mitotic log phase cells (Figure 1c). The presence of m6A is consistent with reported observations of Clancy et al. (11). In addition, the TLC results show that the m6A is present in a Gpm6A and not in a Cpm6A or Upm6A context (Supplementary Data 3). In both the vegetative and the sporulating total RNA samples, the presence of m6A was less obvious and additional modified nucleotides from other RNA species were apparent (Figure 1e and f). Measuring the intensity of the m6A spot relative to the A spot gave an m6A-to-A ratio of 1.0% in the poly(A) sample from sporulating yeast, which is slightly lower than the values for plant and animal systems (19; R. Fray unpublished results).
Quantification of m6A in different mRNA size fractions
Poly(A) RNA from sporulating SK1 diploid cells was separated on a 1.3% TBE agarose gel. Fractions between 4000–2000 nt, 2000–900 nt and 900–200nt were cut out and the mRNA purified from the agarose. The mRNA quality and size was confirmed using RNA 6000 LabChip (Figure 2) and the fractions were subjected to TLC analysis and m6A quantification. All three fractions had high levels of m6A between ~0.7–0.9%, which was similar to the amount in the non-fractionated mRNA samples. No bias was found in the amount of m6A towards any particular molecular weight fractions. These results suggest that the internal adenosine methylation may be a general characteristic of the early meiotic mRNA population.
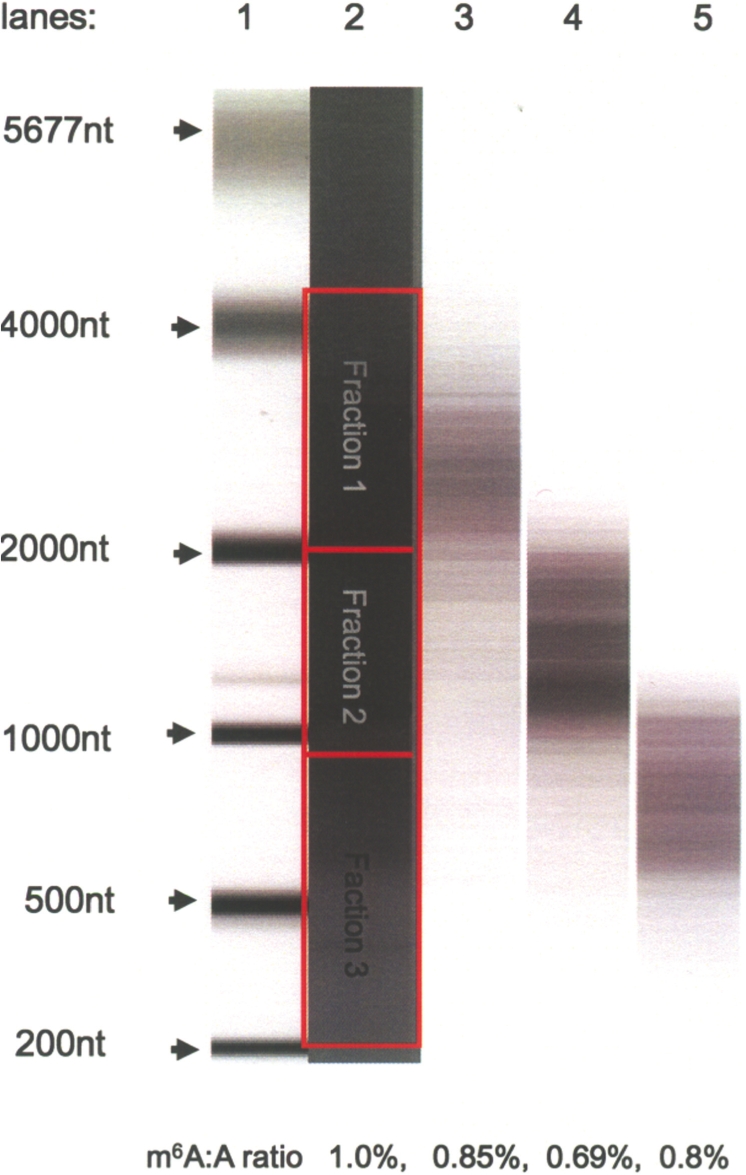
Quantification of m6A in size fractionated poly(A) RNA from sporulating SK1 cells (3 h after meiosis initiation). Poly(A) RNA was separated on a 1.3% agarose TBE gel and three size fractions cut and extracted. Purified RNA samples were analyzed on an RNA6000 LabChip. Lane 1: RNA6000 Ladder. Lane 2: starting poly(A) RNA, 205 ng. Red boxes indicate the positions of the excised gel slices. Lane 3: mRNA population, fraction 1, after purifying from agarose. Lane 4: mRNA, fraction 2, after purification from agarose. Lane 5: mRNA, fraction 3, after purification from agarose. (25–50 ng of RNA was loaded in each lane). The amount of m6A in each fraction was calculated using 2D TLC as described in ‘Materials and Methods’ section.
Isolating methylated messages by immunoprecipitation
A mouse monoclonal anti-m6A antibody, which recognizes m6A in single-stranded nucleic acids and has a strong affinity for m6A containing mRNA, was produced. This was tested against synthetic transcripts containing either A or m6A, and was shown to be able to selectively enrich for the methylated form of RNA (Supplementary Data 4). This Ab was used in immunoprecipitation experiments to identify m6A containing meiotic messages. The relative quantities of early meiotic messages, such as IME2, in the Ab bound fractions were measured by an RT–qPCR method, using ACT1 as a standard. These were compared to the quantities in the Ab depleted fractions. We chose IME2, because Clancy et al. (11) previously hypothesized that this molecule might be one of the IME4 methylation targets due to its key position early in the meiotic pathway. Since ACT1 was used as an internal control and the methylation status of this molecule was unknown, we mixed mRNA from log phase, SK1 cells [does not contain m6A (Figure 1c)] with mRNA from meiotic cells. (The mRNA from meiotic cells was prepared from 3 h sporulating, SK1 cultures.)
The monoclonal Ab was applied to mRNA, previously bound to oligo(dT) magnetic beads. After several washes the mRNA was released from the oligo(dT) beads and the Ab bound mRNA species were pulled out by proteinG magnetic beads.
The relative amount of IME2 mRNA in the Ab bound fraction compared to the depleted fraction was determined. The enrichment of IME2 was nearly sevenfold (Figure 3), indicating that the IME2 message is methylated during early meiosis.
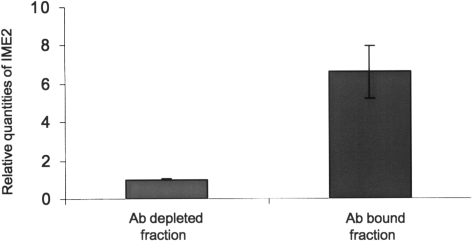
Immunoprecipitation of m6A containing messages enriches for IME2 transcripts. Quantitative RT–PCR of the anti-m6A antibody-bound fraction showed a 7-fold enrichment of IME2 messages relative to the antibody unbound fraction. The starting mRNA was a mixture of poly(A) RNA (non-methylated) from a log phase culture and poly(A) RNA from sporulating cells. Error bars represent SD from three replicates.
The immunoprecipitation method was repeated with an mRNA sample from a 3 h sporulating culture, but without addition of mRNA from vegetative cells. The IME2 message was still enriched in these experiments (Figure 4), but the level of enrichment was threefold less, relative to the enrichment in the immunoprecipitation experiment carried out on mixed vegetative and meiotic mRNA. The threefold decrease in enrichment suggests that in the meiotic mRNA population some ACT1 messages may contain m6A, but to a lesser extent than IME2. We also measured the relative enrichment of IME4 and IME1 mRNA in the Ab bound fraction (Figure 4). The relative amounts of both these messages were also increased in the bound fractions, with IME4 showing the highest level of enrichment. This may indicate that this message is proportionally more methylated than either IME1, or IME2.
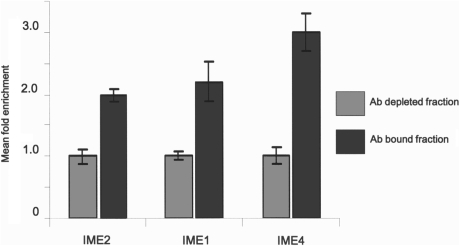
Immunoprecipitation of early meiotic messages. The relative abundance of messages in the anti-m6A antibody bound and unbound fractions were estimated, using quantitative RT–PCR. IME2 and IME1 mRNA species were 2-fold enriched, while IME4 had the highest, 3-fold enrichment in the bound fraction relative to the depleted (Ab unbound) fraction. Error bars represent SD from three replicates.
Mapping m6A containing regions on the IME2 message
The IME2 message was chosen for further analysis, as it is relatively abundant and the distribution of methylation consensus GAC sites are conveniently organized in a group of eight at the 5′-end of the message and a group of 12 at the 3′-end, leaving a 774-nt middle region without GAC (Figure 5A). As for all of the previous experiments, mRNA from a 3 h sporulating culture was used and hybridized to single-stranded DNA probes, containing 5′ biotin tags, which were later bound to streptavidin magnetic beads and used for pulling out the probe bound IME2 transcripts. The DNA probes were designed to protect either the 5′ 526 nt region, or the 3′ 961nt region, thus covering the 5′ 8 GAC or 11 GAC at the 3′-end, respectively (Figure 5B and C). The purity of the IME2 messages was determined using RT-qPCR and in both experiments there was more than a 1000-fold enrichment (Supplementary Data 5). Following T1 ribonuclease digestion and end labeling of the released fragments, the amount of m6A in the non-protected areas of mRNA molecules was analyzed by the TLC quantification method. When the 3′-area was not protected the m6A to A ratio was 0.48% (Figure 5B). However, we found no m6A in the 1291 base region of the IME2 message when the 3′-area was protected (Figure 5C). These results suggest that the methylation must be on one or several of the 11 GACs in the 3′-region, downstream of nucleotide 1291 in the IME2 message.
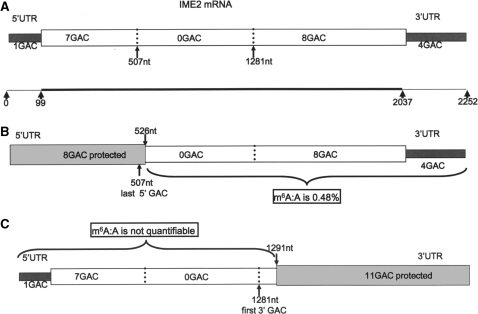
Mapping potential m6A positions in the IME2 message. (A) The GAC consensus sites for adenosine methylation are grouped in two clusters in the IME2 message. The first eight GACs are in the 5′ 507-nt region. There are a further 12 GACs in the 3′-region starting at nt 1281, leaving a 774-nt area in the middle of the message free of GACs. The dark grey boxes indicate the UTRs. (B) When the 5′ GAC containing area is protected (light grey box), only the 3′ 12 GACs are accessible to the T1 nuclease. TLC analysis following T1 digestion revealed the presence of m6A (m6A:A ratio is 0.48%). (C) When the 11 GACs in the 3′-region are protected (light grey box), no m6A was detected by TLC analysis following the T1 nuclease digestion.
DISCUSSION
In both animals and plants the preferred sequence consensus for methylation is GAC. Clancy et al. (11) previously reported that m6A is present within mRNA of sporulating yeast; we show here that such methylation is readily detectable following end labeling of T1 cut mRNA, indicating that, in yeast, m6A is found in a GA and not in the CA or UA context. Thus the site of adenosine methylation in mRNA is conserved among evolutionary distant organisms such as plants animals and yeast, which further highlights the importance of this nucleotide modification.
In S. cerevisiae certain small RNA molecules, such as tRNA and small non-coding RNAs may become polyadenylated (33–37). Often this is the trigger for their degradation. In most cases the protein responsible for the polyadenylation is either the non-canonical poly(A) polymerase Pap2/Trf4 (34–36), or Trf5 (38,39), both are members of the TRAMP complex. Some of these short molecules may contain m6A. Therefore, it remained a possibility that the m6A observed in the poly(A) fraction in this and in earlier experiments was derived from an abundant methylated small non-coding RNA species that had been targeted for degradation, rather than from mRNA. To address this possibility, the poly(A) fraction was size-separated prior to digestion and labeling (Figure 2). Nearly equal values of m6A:A (~0.7–0.9%) were found in the three different poly(A) size fractions, suggesting that the m6A may be distributed across the early meiotic poly(A) population and is not specifically associated with a short non-coding RNA class.
The ratio of labeled m6A to A in our experiments is between 0.9–1% at 3 h after the shift to sporulation. This value is slightly lower, but comparable to other organisms, such as A. thaliana, where the m6A to A ratio varied between 0.4% and 1.5%, depending on developmental stage and tissue type (19) or in mouse and human, where similar results have been found (our unpublished data). Using this experimental approach, only those adenosines in a GA sequence will be labeled, thus the observed m6A:A ratio is really a measure of the Gpm6A:GpA ratio. If all possible 16 dinucleotide pairs occurred with equal frequency, this would be equivalent to m6A being 0.06% of the nucleotides in the mRNA sample as a whole, or 1 m6A nucleotide per 1600 nt. If distributed evenly, this would equate to a little more than half of the mRNAs present being methylated. However, this may be an underestimate of true methylation levels. In plants and animals methylation can also occur in the sequence AAC, if this is also the case in yeast, then the actual frequency of m6A will be higher.
The identification of methylated mRNA species is a pre-requisite for understanding how this base modification regulates gene expression in S. cerevisiae and possibly other eukaryotes. We have had a monoclonal antibody developed, which recognizes the m6A in single-stranded nucleic acids and we established a new method for immunoprecipitating individual mRNA molecules containing m6A. Applying the immunoprecipitation approach, m6A containing individual transcripts, expressed during the first 3 h of meiosis were isolated. In the first instance a mixture of mRNA from log phase, vegetative (does not contain m6A) and sporulating yeast was subjected to IP. In this experiment we set out to specifically test if IME2 message is methylated. This transcript is an attractive target, as it is an abundant, meiosis-specific message that encodes a key early regulator of meiosis. In addition, Ime2 levels have to be tightly regulated for correct meiotic progression (40,41), and Clancy et al. (11) previously speculated that this might be a target for post-transcriptional regulation by Ime4. In this experiment, the enrichment of IME2 in the Ab bound fraction was nearly sevenfold higher relative to the Ab depleted fraction. Measurements were normalized relative to ACT1, which can be assumed to be unmethylated in the RNA fraction derived from the vegetative cells, as methylation was not detectable in the mRNA from mitotic log phase cells (Figure 1c). When mRNA from the meiotic cells alone was used, the enrichment of IME2 was still clear, but less pronounced (2-fold). This is consistent with IME2 being methylated and may also indicate that a small portion of ACT1 transcripts are also modified.
Two further messages, IME1 and IME4, were also tested using IP of meiotic mRNA. Both of these were enriched in the Ab bound fraction, with IME4 messages showing the highest level of enrichment. Until recently only the four messages, bovine prolactin, R. sarcoma virus, SV40 viral mRNA and the mouse dihydrofolate reductase were reported to contain m6A (24,25,42,43). The immunoprecipitation approach allowed us to double the number of the currently known m6A harbouring messages and will allow further global analysis of transcriptomes. Following the immunoprecipitation, IME2 transcript was purified using a biotinilated complementing DNA sequence and streptavidin beads. The m6A to A ratio for the analyzed part of the message was 0.48%. This value is somewhat lower than seen in the mRNA population as a whole, but is not necessarily unexpected. When looking at individual transcripts rather than the total mRNA population, the percentage of end labeled adenosines that are methylated will depend upon the absolute numbers of both Gpm6A and GpA. Thus, within the non-protected fragment of IME2, there are 119 GA sites, of these 12 are in a GAC consensus. The Gpm6A: GpA ratio of 0.48% would mean that out of 208 GpA one would be Gpm6A. Since the unprotected fragment of IME2 contains only 119 GpA sites it takes 1.7 of these fragments to harbor one Gpm6A, which is equivalent of 58% of IME2 messages being methylated. This value is consistent with the observed average for the mRNA population as a whole.
The position of the m6A in a particular message could be indicative of its function. However precise mapping has only been achieved for two highly abundant transcripts (R. sarcoma virus and bovine prolactin) and the methods used are not applicable to the yeast system. In the IME2 mRNA molecule we narrowed down the position of the m6A to a region in the 3′-end containing the last 11 GACs of the molecule [including 3′-UTR (44)]. The 5′ 1291-nt fragment of the IME2 molecule [including 5′UTR (44)] appeared to be free of m6A.
It is thought provoking, that the deletion of the C-terminal 240aa from the Ime2 protein will lead to spore number reduction, as well as the stabilization of the otherwise labile Ime2 protein (40,45). This deletion removes the last 10, potential m6A harboring GAC consensus sites, in the 3′-region.
It has recently been published that Khd1p binds to IME2 mRNA as well as several other mRNA molecules (46). Khd1p differentially affects gene expression, possibly due to combinatorial arrangement with additional factors. However, it is not impossible that the translatability of IME2 message could be affected by altered Khd1p binding, due to the presence of m6A residues. We have manually searched for CNN repeats, the Khd1 binding domains (46), in the IME2 transcript (with UTRs) and two near CNN repeats were found in the 3′-region (Figure 6). However, neither of the repeats overlaps with any of the GAC consensus sites. On the other hand, two potentially methylated GACs are sandwiched between the two CNN repeats, therefore it remains possible that methylation of adenosine(s) in any of these sites could modify the binding of Khd1 or an interacting factor, resulting in altered translatability.

CNN repeat candidates in the IME2 message. The IME2 message contains two groups of CNN repeats, highlighted as red boxes. Additional C-rich regions adjacent to CNN repeats are accentuated by red Cs. Three GACs lie between the CNN repeats (bold and large font). The two GACs indicated in blue are in the region where methylation was detected.
A role for m6A in regulating translation would be consistent with work previously reported by Tuck et al. (47). These authors found that poly(A) RNA, isolated from methotrexate resistant mouse sarcoma cells treated with the methylation inhibitor, cycloleucine, translated less efficiently in an in vitro translation assay, compared to mRNA from the control, untreated cells. However, in their system, cap-associated methylation was also reduced, making the interpretation of their results difficult. The presence of m6A in mRNA might alter splicing during yeast meiosis, as proposed by Clancy et al. (11). However the three transcripts IME1, IME2 and IME4 identified here as methylated messages, are not spliced. Therefore we think, it is unlikely that the primary function of mRNA methylation would be to act as a positive signal for splicing in yeast.
We also would like to speculate that the presence of m6A in meiotic messages could be a flag for message recycling or storage, thus promoting a quick response to sudden changes in conditions.
Our results demonstrate that substantial levels of internal adenosine methylation are present in the GpA context in mRNA of sporulating yeast. This is consistent with the preferred methylation consensus of higher eukaryotes and is conserved between evolutionary distant species, which further highlights the importance of this nucleotide modification in the mRNA. Methylation is homogenously distributed across all mRNA size ranges, indicating that m6A is not limited to a small population of messages. If distributed evenly our measurements imply that slightly more than every second transcript in sporulating yeast is expected to contain m6A. Consistent with this, m6A is found once per 1.7 IME2 message. However, it is likely that transcripts of individual genes will vary in the degree or frequency of m6A. Using a novel immunoprecipitation approach we identify transcripts of three key, early regulators of meiosis, IME1, IME2 and IME4 itself, as being methylated. Methylation of these and other targets suggests mechanisms by which IME4 could control developmental choices leading to meiosis. For example, while a role in promoting splicing seems less likely, a function in translational control or message recycling remains a stronger possibility.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Daphne Jackson Trust Fellowship awarded to Z.B. in conjunction with the Biotechnology and Biological Science Research Council. Biotechnology and Biological Science Research Council grant (BB/C523369/1) to R.G.F. Funding for open access charge: University of Nottingham.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkq266
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/38/16/5327/16766525/gkq266.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/38/16/5327
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/38/16/5327.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/38/16/5327
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Phylogenetic and functional analyses of <i>N</i><sup>6</sup>-methyladenosine RNA methylation factors in the wheat scab fungus <i>Fusarium graminearum</i>.
mSphere, 9(1):e0055223, 12 Dec 2023
Cited by: 3 articles | PMID: 38085094 | PMCID: PMC10826363
Crosstalk between m6A mRNAs and m6A circRNAs and the time-specific biogenesis of m6A circRNAs after OGD/R in primary neurons.
Epigenetics, 18(1):2181575, 01 Dec 2023
Cited by: 1 article | PMID: 36861189 | PMCID: PMC9988353
N6-Methyladenosine mRNA Modification: From Modification Site Selectivity to Neurological Functions.
Acc Chem Res, 56(21):2992-2999, 17 Oct 2023
Cited by: 1 article | PMID: 37847868 | PMCID: PMC10634299
Kar4 is required for the normal pattern of meiotic gene expression.
PLoS Genet, 19(8):e1010898, 28 Aug 2023
Cited by: 5 articles | PMID: 37639444 | PMCID: PMC10491391
Kar4, the yeast homolog of METTL14, is required for mRNA m6A methylation and meiosis.
PLoS Genet, 19(8):e1010896, 21 Aug 2023
Cited by: 5 articles | PMID: 37603553 | PMCID: PMC10470960
Go to all (98) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene.
Nucleic Acids Res, 30(20):4509-4518, 01 Oct 2002
Cited by: 188 articles | PMID: 12384598 | PMCID: PMC137137
The m6A methyltransferase Ime4 epitranscriptionally regulates triacylglycerol metabolism and vacuolar morphology in haploid yeast cells.
J Biol Chem, 292(33):13727-13744, 27 Jun 2017
Cited by: 13 articles | PMID: 28655762 | PMCID: PMC5566527
m6A modification of a 3' UTR site reduces RME1 mRNA levels to promote meiosis.
Nat Commun, 10(1):3414, 30 Jul 2019
Cited by: 35 articles | PMID: 31363087 | PMCID: PMC6667471
The Ime2 protein kinase family in fungi: more duties than just meiosis.
Mol Microbiol, 80(1):1-13, 01 Mar 2011
Cited by: 23 articles | PMID: 21306447
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (2)
Grant ID: BB/C523369/1
An investigation into the role of post-transcriptional methylation of plant mRNA during development
Dr Fray, University of Nottingham
Grant ID: BB/C513369/1






