Abstract
Free full text

CCRXP: exploring clusters of conserved residues in protein structures
Associated Data
Abstract
Conserved residues forming tightly packed clusters have been shown to be energy hot spots in both protein–protein and protein–DNA complexes. A number of analyses on these clusters of conserved residues (CCRs) have been reported, all pointing to a crucial role that these clusters play in protein function, especially protein–protein and protein–DNA interactions. However, currently there is no publicly available tool to automatically detect such clusters. Here, we present a web server that takes a coordinate file in PDB format as input and automatically executes all the steps to identify CCRs in protein structures. In addition, it calculates the structural properties of each residue and of the CCRs. We also present statistics to show that CCRs, determined by these procedures, are significantly enriched in ‘hot spots’ in protein–protein and protein–RNA complexes, which supplements our more detailed similar results on protein–DNA complexes. We expect that CCRXP web server will be useful in studies of protein structures and their interactions and selecting mutagenesis targets. The web server can be accessed at http://ccrxp.netasa.org.
INTRODUCTION
Conserved residues forming tightly packed clusters might correspond to energy hot spots in both protein–protein and protein–DNA complexes. The role of conserved residues in determining interface residues has been well documented (1–3). A number of studies have pointed to the crucial nature of clusters of conserved residues (CCRs) (4–8). CCRs have been shown to be distributed in protein–DNA and protein–protein interfaces. Clusters of hot spots have been shown to contribute in a major way to the stability of protein cores and interfaces, and are useful in understanding both protein–protein and protein–DNA interactions. They further assist in predicting interfaces. Obtaining clusters is time consuming and requires several computational steps. An objective protocol and tool to determine CCRs should therefore be useful. CCRXP automates the detection and analysis of such clusters in protein structures. In this article, we describe the working principles of CCRXP and also present additional statistics, which shows that energy hot spots are more abundant in clusters detected by this server compared with other residues.
IMPLEMENTATION
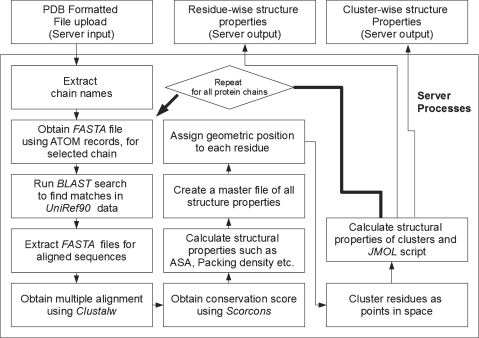
CCRXP consists of two input modules, whose implementation is detailed in Figure 1. Default module CCRXP_lite can be accessed directly from the server’s top page by entering a valid protein data bank (PDB) ID. The alternative module CCRXP_ADV allows users to select a number of filtering and clustering options (Supplementary Data). Both versions allow users to upload their coordinate files.
CCRXP uses a number of publicly available tools as well as those developed by us. The main input of the server is PDB formatted file. Only the latest PDB format (version 3.0 onwards) compliant files (9) will provide complete results.
Some of the most important tools used are as follows:
BLAST: the standard blastall program (10) is used to search similar sequences in the UniRef90 database, derived from UniProt (11). The top N alignments are saved for further processing (N is selected by users, with 20 as default). By far, this is the slowest component of the server processes and determines the speed of CCRXP. An alternative could be to use alignments provided by users. However, PDB files often have modified or missing residues in the terminal or intermediate positions, causing a mismatch between sequence and structure data sets. Therefore, a comprehensive system starting from all information derived from a single source (PDB ATOM records) is found to be less error-prone and retained.
Clustalw is used to generate multiple alignments of sequences found by blastall (12).
Scorecons: a publicly available program scorecons, provided by Valdar (13) is used to compute the conservation score from multiple alignments.
DSSP: this program is used to calculate solvent accessibility and secondary structure (14).
Packing density, the geometric positions of residues used for clustering and some other structural properties of the clusters are obtained by dedicated C programs written earlier [e.g. (8)]. The clustering algorithm essentially uses a single linkage criterion in which a tree is cut into branches at a fixed distances cutoff (default is 5 Å). The clustering program starts with all residue positions as seeds and successively adds other residues to the evolving clusters using single linkage criterion. Specifically, residues are scanned sequentially (in multiple iteration) and attached to a growing cluster if the distance between an atom of the residue to any atom of any residue in the cluster is less than the distance cutoff. Many seeds will generate identical clusters and only one of them is finally selected. Users are allowed to choose the maximum linkage distance in the advanced version of CCRXP. In addition, buried residues may be filtered out from clusters by choosing a solvent accessibility cutoff. Results can also be filtered by cluster size.
Server-side load distribution is performed by open source workload management system PBS (www.openpbs.org).
Final cluster outputs are also rendered in the form of a Jmol script (http://www.jmol.org), which allows users of Java-enabled browsers to manually examine them on the fly.
CLUSTERS IN PROTEIN–DNA, PROTEIN–PROTEIN AND PROTEIN–RNA COMPLEXES
We have shown earlier that the residues belonging to a conserved cluster contribute more to the stability of protein–DNA complexes (8). To evaluate the applicability of this principle, we present statistics of free energy changes in mutations taking place in positions characterized by conservation scores, number of conserved neighbors and the cluster size to which a given mutant residue belongs (parent cluster size). To do so, we first classified mutant data for protein–protein and protein–RNA complexes into hot spots and non-hot spots, using a common definition, i.e. positions at which a mutation to Ala (in protein–protein complex alanine-scanning data in protein–protein complexes) or any other residue (protein–RNA complex data derived from ProNIT) caused a loss of stability (ΔΔG) by >2.0 kcal/mol (15). Then we calculated the expected values for three types of residues in hot spots namely (i) the number of conserved residues (a cutoff for conservation score was fixed at 0.6 for all statistics in this work); (ii) the number of conserved neighbors (within 5 Å from the target residue in complex structure); and (iii) the number of residues in the parent cluster (size of the cluster to which a query mutant position belongs, as computed by CCRXP using default settings). Expected values were calculated by computing the per residue values for the entire data and multiplying by the total number of hot spot mutations. Expected values of these parameters were compared with the observed number of residues in each category. Inspections of the statistical results establishes that the residues belonging to these clusters are more likely to be energy hot spots (most stabilizing residues) compared to all other residues, including conserved residues. A summary of statistics is presented below.
Clusters in protein–RNA complexes
A total of 157 single-residue mutations in RNA-binding protein, which had sufficient homologs to calculate conservation scores were obtained (complete data in Supplementary Data). All single mutation data with known values of free energy change and entry in PDB from ProNIT were used for this study (9,15). Table 1 (upper panel) shows the main results of the statistics using a chi-squared test of significance. As observed from the table, conserved residues are more abundant in hot spots compared to non-hot spots (70% or 0.70 per residue compared with 55% or 0.55 per residue), leading to a χ2-value of 0.9 (corresponding P-value is 3.3e-1). When we look at the number of conserved residue neighbors in hot spots, these values are 4.1 versus 2.5 in hot spots and non-hot spots, respectively. This increased the χ2-value to 20.0 and improved P-value to 7.8e-6. However, when we look at the size of the parent clusters, we find that hot spot residues lie in clusters whose average size is 15.6 compared with 11.0 for non-hot spots, thereby increasing the χ2-value to 52.1 and substantially improving P-value to 5.1e-13. Thus, we conclude that looking at the CCRs, we are more likely to pick residues with higher contribution to stability and that is where CCRXP will be useful.
Table 1.
Chi-square statistics of conserved residues, number of conserved neighbors and size of the parent cluster in hot spots residues (ΔΔG ≥ 2.0 kcal/mol)
| Frequency in hot spots (per mutant position) | Frequency in non-hot spots (per mutant position) | Expected counts (in all hot spots) | Observed counts (in all hot spots) | χ2-value | P-value | |
|---|---|---|---|---|---|---|
| Protein–RNA complexesa | ||||||
    Conserved residues Conserved residues | 0.70 | 0.55 | 37.1 | 43 | 0.9 | 3.3e-01 |
    Conserved neighbors Conserved neighbors | 4.1 | 2.5 | 186.9 | 252 | 20.0 | 7.8e-06 |
    Residues in parent clusters Residues in parent clusters | 15.6 | 11.0 | 767.0 | 967 | 52.1 | 2.0e-11 |
| Protein-protein complexesb | ||||||
    Conserved residues Conserved residues | 0.47 | 0.35 | 22.8 | 27 | 0.8 | 3.8e-01 |
    Conserved neighbors Conserved neighbors | 2.4 | 1.7 | 108.3 | 125 | 2.6 | 1.1e-01 |
    Residues in parent clusters Residues in parent clusters | 7.4 | 2.7 | 262.9 | 428 | 96.4 | 2.4e-24 |
aHot spot mutations = 60; conserved residue mutations = 96; total mutations = 157.
bHot spot mutations = 58; conserved residue mutations = 59; total mutations = 150.
Clusters in protein–protein complexes
For the protein–protein complexes, we analyzed 150 mutations in protein–protein interfaces, taken from our recent study (16). Complete statistics is provided in Supplementary Data. Statistics, identical to the previous section on protein–RNA complexes is presented in Table 1 (lower panel). In the case of protein–protein complexes, conserved residue populations in hot spots were only weakly higher than non-hot spots (0.47 per residue or 47% residues are conserved in hot spots, compared with 0.35 per residue or 35% in non-hot spot regions, with χ2 = 0.8; P = 0.38), whereas the number of conserved neighbors had a slightly better separation (2.2 per residue compared with 1.7 in non-hot spot residues; χ2 = 2.6; P = 0.11), and most significantly the number of residues in parent clusters of hot spots were much more abundant than in non-hot spot residues (χ2 = 96.4; P = 2.4e-24). The average parent cluster size of hot spot residues was found to be 7.4 compared with 2.7 for non-hot spot residues.
The above results show that the significance of conserved clusters is not limited to DNA-binding proteins, but extends to protein–RNA and protein–protein complexes. We should note that CCRXP is complementary to a previous database, HotSprint, documenting computational hot spots in the protein–protein interfaces combining conservation, packing density and solvent accessibility of residues in the protein interfaces. In HotSprint, only individual hot spots are provided whereas CCR XP is a server that finds CCRs in protein–protein, protein–RNA and protein–DNA complexes that are tightly packed in 3D protein structures. We further show that these CCRs comprise hot spots.
INTERPRETATION OF THE CCRXP OUTPUTS
A number of structural features for residues in conserved clusters are returned by CCRXP. As shown above and in our previous works, we conclude that the most important residues are the ones that occur in large clusters, have higher conservation scores and are also surrounded by more conserved neighbors. Solvent accessibility values returned by the server also identify residues on the surface as well as more solvent-accessible members of a cluster. The number of positively and negatively charged residues is also provided to roughly estimate the electrostatic nature of a cluster. Further information on the electrostatic nature is provided by the dipole moment values, calculated by selecting only the cluster members and assigning charges to selected residue positions as in our earlier work (17). Although explicit prediction scores are not provided, it has been shown that positively charged clusters are often found in the DNA interface and such clusters can be detected by this server. Similarly, hydrophobic clusters, often present in protein–protein interfaces, can also be identified.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Federal funds from the National Cancer Institute; National Institutes of Health (contract number HHSN261200800001E); Intramural Research Program of the NIH, National Cancer Institute and Center for Cancer Research; Industrial Technology Research Grant Program in 2007 from New Energy and Industrial Technology Development Organization (NEDO) of Japan (to K.M.). Funding for open access charge: Institute's internal funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. OK acknowledges TUBITAK (Research Grant: 109T343).
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkq360
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2896124?pdf=render
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/38/suppl_2/W398
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/38/suppl_2/W398.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/38/suppl_2/W398
Citations & impact
Impact metrics
Citations of article over time
Article citations
Host-pathogen protein-nucleic acid interactions: A comprehensive review.
Comput Struct Biotechnol J, 20:4415-4436, 04 Aug 2022
Cited by: 5 articles | PMID: 36051878 | PMCID: PMC9420432
Review Free full text in Europe PMC
Analyses on clustering of the conserved residues at protein-RNA interfaces and its application in binding site identification.
BMC Bioinformatics, 21(1):57, 17 Feb 2020
Cited by: 4 articles | PMID: 32066366 | PMCID: PMC7027071
Discrete structural features among interface residue-level classes.
BMC Bioinformatics, 16 Suppl 18:S8, 09 Dec 2015
Cited by: 3 articles | PMID: 26679043 | PMCID: PMC4682381
Interolog interfaces in protein-protein docking.
Proteins, 83(11):1940-1946, 29 Sep 2015
Cited by: 2 articles | PMID: 25740680 | PMCID: PMC5054918
Communication routes in ARID domains between distal residues in helix 5 and the DNA-binding loops.
PLoS Comput Biol, 10(9):e1003744, 04 Sep 2014
Cited by: 18 articles | PMID: 25187961 | PMCID: PMC4154638
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
DR_bind: a web server for predicting DNA-binding residues from the protein structure based on electrostatics, evolution and geometry.
Nucleic Acids Res, 40(web server issue):W249-56, 31 May 2012
Cited by: 20 articles | PMID: 22661576 | PMCID: PMC3394278
VoroMQA web server for assessing three-dimensional structures of proteins and protein complexes.
Nucleic Acids Res, 47(w1):W437-W442, 01 Jul 2019
Cited by: 15 articles | PMID: 31073605 | PMCID: PMC6602437
PredHS: a web server for predicting protein-protein interaction hot spots by using structural neighborhood properties.
Nucleic Acids Res, 42(web server issue):W290-5, 22 May 2014
Cited by: 28 articles | PMID: 24852252 | PMCID: PMC4086081
HotPoint: hot spot prediction server for protein interfaces.
Nucleic Acids Res, 38(web server issue):W402-6, 05 May 2010
Cited by: 87 articles | PMID: 20444871 | PMCID: PMC2896123
Funding
Funders who supported this work.
Intramural NIH HHS
PHS HHS (1)
Grant ID: HHSN261200800001E