Abstract
Free full text

LRRK2 Is Involved in the IFN-γ Response and Host Response to Pathogens
Abstract
LRRK2 was previously identified as a defective gene in Parkinson’s disease, and it is also located in a risk region for Crohn’s disease. In this study, we aim to determine whether LRRK2 could be involved in immune responses. We show that LRRK2 expression is enriched in human immune cells. LRRK2 is an IFN-γ target gene, and its expression increased in intestinal tissues upon Crohn’s disease inflammation. In inflamed intestinal tissues, LRRK2 is detected in the lamina propria macrophages, B-lymphocytes, and CD103-positive dendritic cells. Furthermore, LRRK2 expression enhances NF-κB–dependent transcription, suggesting its role in immune response signaling. Endogenous LRRK2 rapidly translocates near bacterial membranes, and knockdown of LRRK2 interferes with reactive oxygen species production during phagocytosis and bacterial killing. These observations indicate that LRRK2 is an IFN-γ target gene, and it might be involved in signaling pathways relevant to Crohn’s disease pathogenesis.
Genome-wide association studies (GWASs) have identified new risk factors for complex diseases, including Crohn’s disease (CD) (1). CD is a chronic inflammatory bowel disease (IBD) thought to result from dysregulated immune response to commensal intestinal microbiota (2). Meta-analysis of three independent studies of CD highlighted 31 susceptibility loci that were replicated in an independent study (3). The large number of distinct susceptibility loci reflects the complexity of the intestinal mucosal system and suggests that a number of distinct cell circuits are likely perturbed in IBD. These signal transduction pathways need to be finely tuned to sense and respond to microbes with minimal damage to the host mucosa. The single nucleotide polymorphism (SNP) rs11175593, located in a noncoding region on chromosome 12, is one of the loci identified as a risk factor for CD with a p value of 3.08 × 10−10 and an odds ratio of 1.54 (3). LRRK2 is located downstream from SNP rs11175593. LRRK2 has been previously identified as the defective gene at the PARK8 susceptibility locus responsible for the autosomal dominant form of Parkinson’s disease (PD) (4, 5). LRRK2 mutations are associated with ~10% of familial PD cases and 2% of sporadic cases (6), and their effects have been mainly studied in neurons despite reports indicating LRRK2 expression in the microglia, brain-resident macrophages (7).
LRRK2 is a 285-kDa protein containing a Ras of complex GTPase domain, a C-terminal of Ras of complex domain, and an MAPK kinase kinase domain. It possesses three potential protein interaction domains: an ankyrin domain, leucine-rich repeats, and WD40 repeats (4, 5). Based on the presence of these catalytic domains and scaffold domains, it has been speculated that LRRK2 may be involved in the assembly of multiprotein signaling complexes. LRRK2 can exist as a dimeric complex dependent on its WD40 domain (8). In vitro studies indicate that the GTPase and kinase domains of LRRK2 are functional (9–11), although its physiologic substrates have not been identified. Studies to elucidate the pathophysiology of PD have shown that LRRK2 PD-associated mutant proteins induce a broad range of cellular outcomes such as cell death, inclusion bodies, and autophagy (12–15). However, wild type LRRK2 has not been clearly implicated in these processes. Although recent data have indicated that LRRK2 plays a role in synaptic vesicle endocytosis, the physiologic role of LRRK2 in a cellular context remains poorly understood (16).
In this study, we aimed to assess whether LRRK2 might have some functions in the inflammatory responses and the response to pathogens. We found that LRRK2 is expressed in immune cells and in the mucosa of patients with CD. LRRK2 is an IFN-γ target gene able to activate signaling pathways involved in immune responses in response to pathogens. This study clearly indicates that LRRK2 is involved in immune response.
Materials and Methods
Abs
Anti-human LRRK2 Ab (NB300-267) and its corresponding blocking peptide were purchased from Novus Biological (Littleton, CO). Anti-mouse LRRK2 Ab (AL106) was obtained from Alexis Biochemical (Plymouth Meeting, PA). Anti–β-actin and anti-myc tag abs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CD56 (MEM-188). Anti-mannose receptor (15-2) Abs were obtained from BioLegend (San Diego, CA). Anti-NSO, anti–NF-H and anti-NSE abs were obtained from Neuromics (Edina, MN). Anti-myeloperoxidase Ab (2C7) was obtained from Gene Tex (Irvine, CA). Anti-CD3 and anti–dendritic cell-specific intercellular adhesion molecule-3–grabbing nonintegrin (DC-SIGN) Abs were obtained from BD Pharmingen (San Diego, CA). Anti-mast cell tryptase Ab was obtained from Affinity BioReagents (Golden, CO). Anti-CD20 (L26) Ab was obtained from ScyTek Laboratories (Logan, UT). Anti-CD103 Ab was obtained from Srotec (Oxford, United Kingdom).
Plasmids and siRNA
Human wild type LRRK2 cDNA cloned in pcDNA3.1 myc/his Avector from Invitrogen (Carlsbad, CA) was provided by Dr. Andrew West (Department of Neurology, University of Alabama, Tuscaloosa, AL). LRRK2 kinase-dead mutant K1906M and PD-associated mutant G2019S were generated by two-step PCR using primer pairs that contained the appropriate mutations and produced a final product corresponding to a fragment of LRRK2 between the two unique restriction enzyme sites ClaI and PacI. This wild type fragment was then excised from the plasmid wild type LRRK2 template and replaced by the fragment containing the mutations. Plasmids were sequenced to confirm the presence of the expected mutations and the absence of additional mutations.
Dominant negative IκB kinase (IKK)-β construct IKK-β-K44AE442G was a gift from Dr. Brian Seed (Massachusetts General Hospital, Boston, MA), and was cloned into pCMV-3Xmyc vector. A 3×HA-tagged K-Ras-V12 construct was cloned into the pKH3 vector by PCR. Stealth control siRNA and siRNA directed against mouse LRRK2 were obtained from Invitrogen (90 and 92). All the siRNA were transfected at 100nM using Hiperfect (Qiagen, Valencia, CA).
RT-PCR
RNA extraction was performed using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions. One microgram of total RNA was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA).
Real-time quantitative PCR was performed in a Bio-Rad iCycler thermal cycler equipped with an iQ5 optical module using the iQSYBRGreen super mix (Bio-Rad). One hundred nanograms of reverse-transcribed cDNA was used for each PCR with 250 nM forward and reverse primers. The thermal cycling conditions were 4 min at 95°C, followed by 40 cycles at 94°C for 15 s, and 59°C for 1 min. The threshold cycle (CT) values were obtained for the reactions, reflecting the quantity of the template in the sample. LRRK2 ΔCT was calculated by subtracting the calibrator gene GAPDH CT value from the LRRK2 CT value and thus represented the relative quantity of the target mRNA normalized to GAPDH mRNA. If not specified, the lowest mRNA content of LRRK2 condition was defined as one arbitrary unit.
The PCR assays were performed using the following primer sets:human GAPDH (generated amplicon of 440 bp), forward primer 5′-TCATCTCTGCCCCCTCTGCT-3′, reverse primer 5′-CGACGCCTGCTTCACCACCT-3′; and human LRRK2 (generated amplicon of 288 bp), forward primer 5′-TGGGTTGGTCACTTCTGTGC-3′, reverse primer 5′-CATTGGCTGGAAATGAGTGC-3′
Patient intestinal biopsies
Human colonic biopsies were obtained from patients with CD (inactive disease state) and patients with ulcerative colitis (UC; inactive disease state) undergoing colonoscopy. Informed consent was obtained from all patients prior to the procedure, and the protocol was approved by the Human Research Committee of the Massachusetts General Hospital. Tissues were either processed for RNA extraction and RT-PCR as described above or embedded and frozen in OCT compound (Fisher, Pittsburgh, PA).
Immunofluorescence staining
For tissue staining, frozen sections (7 μm) were prepared from OCT-embedded CD patient intestinal biopsies (inactive disease state). Tissues or cells were fixed with 4% paraformaldehyde (15 min) and permeabilized PBS-Triton X-100 0.1% (10 min). After washing with PBS, the sections were incubated for 30 min in PBS containing 3M glycine to block the reactive groups of paraformaldehyde. The sections were then incubated for 1 h with a blocking solution containing 10% donkey serum (Rockland, Gilbertsville, PA) and 10% human Fc block reagent (Miltenyi Biotec, Auburn, CA). The sections were then incubated with the primary Abs for 1 h, washed using PBS, incubated with fluorescent secondary Abs (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h, washed with PBS, and incubated with PBS containing 100 μg/ml DABCO (Sigma-Aldrich, St. Louis, MO) as antifading reagent before mounting in Glycergel medium (DakoCytomation, Carpinteria, CA). Fluorescence signals were captured using a laser confocal microscope (model Radiance 2000; Bio-Rad). Image acquisition was performed with LaserSharpScanning software (Bio-Rad).
Flow cytometric analysis
PBMCs were collected and centrifuged at 800 × g for 10 min. Cell pellets were resuspended in PBS and 5% FCS. Surface markers were labeled at 30 min at room temperature using FITC anti-CD3, FITC anti-CD19, and APC anti-CD11b Abs (BD Biosciences, San Jose, CA). After PBS wash, cells were fixed and permeabilized using the Fix-and-Perm permeabilization kit (BD Biosciences). LRRK2 protein was detected using anti-LRRK2 Abs (clone 267) from Novus Biological and Cy3-conjugated secondary Abs from Jackson ImmunoResearch Laboratories. Data were acquired on a FACSCalibur cytometer (BD Biosciences) and analyzed using FlowJo for Mac version 8.2 (Tree Star, Ashland, OR).
Luciferase reporter assays
Cells were transfected using Lipofectamine 2000 Transfection Reagent (Invitrogen) with 1 ng of Renilla luciferase plasmid and with either 10 ng Ig-κ pIV firefly luciferase reporter or 25 ng AP-1 firefly luciferase reporter containing four AP-1 response elements. Activity was measured using the Dual Luciferase reporter assay system (Promega, Madison, WI) in a BD Moonlight 3010 Luminometer (BD Biosciences) in accordance with the manufacturer’s instructions, and normalized to the internal transfection control of Renilla luciferase activity.
Detection of reactive oxygen species
RAW 264.7 cells were collected using cell lifters and maintained on ice and in PBS containing 5 mM glucose, 1 mM MgCl2, 0.5 mM CaCl2, and 100 μM luminol. Cells were split in 96-well plates at 20,000 cells per well. Opsonized zymosan (250 μg/ml) was added in each well, and the plates were centrifuged 200 × g for 5 min at 4°C. Baseline luminescence was measured using a luminescence plate reader. Cells were then incubated at 37°C, and luminescence was measured every 5 min. Each condition was measured in duplicate.
Gentamicin protection assay
RAW 264.7 cells were stimulated with IFN-γ (100 ng/ml) 48 h after siRNA transfection. Cells were infected with Salmonella typhimurium the following day with a multiplicity of infection of 10. Cells were centrifuged at 200 × g for 8 min and incubated at 37°C for 40 min. After two PBS washes, cells were incubated with a high dose of gentamicin (100 μg/ml) for 20 min followed by a 1-h incubation with low-dose gentamicin (15 μg/ml). Cells were then washed twice with PBS and lysed for 10 min at 37°C using PBS with Triton X-100 1%. Cells lysates were diluted in LB medium with 10-fold serial dilutions and 20 μl of each dilution were plated on LB plates. After an overnight incubation at 37°C, the bacteria colonies were counted and the total numbers of intracellular bacteria recovered were calculated for each condition.
Microarray data
The Genomics Institute of the Novartis Research Foundation (GNF) consortium dataset, profiling 79 human tissues in duplicates was obtained from the GNF Symatlas and is available in the genome expression omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE96 (17). Expression profiles from Affymetrix U133A platform and GNF custom probes normalized using MAS5 were used. In addition, present and absent calls were extracted and indicated in shades of gray in Fig. 1A.
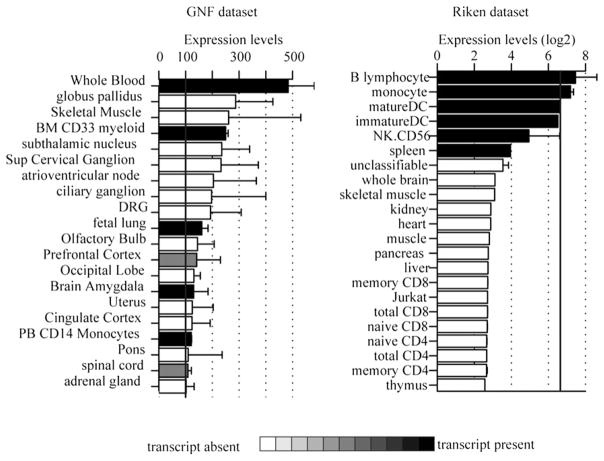
LRRK2 mRNA expression in microarray datasets. Average microarray expression of the highest expressing LRRK2 probe across 79 human tissues obtained from the GNF consortium (left) and 34 tissues obtained from Riken RefDic (right). Expression values were sorted, and only the top 20 are shown. Bars are color coded to reflect the average value of Affymetrix present or absent calls. Black bars indicate that the transcript was identified as present.
The Riken RefDic dataset including 34 human tissues was obtained from the Riken RefDic database (available at http://cibex.nig.ac.jp, under CIBEX accession number CBX19 (18). Raw Affymetrix cell files were normalized using the GCRMA method as described previously (19, 20). Affymetrix MAS5 module of Bioconductor was used to identify present and absent transcripts (19), and probes with no single present call across all tissue or highest expression value below log2(100) were removed.
Results
LRRK2 is expressed in immune cells and is upregulated upon IFN-γ stimulation
To investigate the profile of LRRK2 expression in human cells and tissues, microarray data obtained with LRRK2 probes from GNF (17) and RIKEN datasets (18) were analyzed (Fig. 1). In these datasets, LRRK2 mRNA expression was low to moderate. These findings are consistent with published data in which detection of endogenous LRRK2 protein has been difficult (21, 22). The highest mRNA expression was detected in B cells, monocytes, and dendritic cells. The tissues and cells profiled in these datasets partially overlapped: spleen is absent from the GNF dataset as well as CD56+ NK cells and dendritic cells. Expression in B cells was not detected in the GNF dataset (not shown). Because these data suggested that LRRK2 might be expressed in myeloid-derived cells, we investigated and confirmed by quantitative RT-PCR that LRRK2 expression was higher in CD14-positive and CD19-positive cells (Fig. 2). Among a panel of cell lines, monocyte-derived cell lines (THP-1 and U937) showed the highest expression of LRRK2 mRNA. We were unable to detect LRRK2 in any small intestinal or colonic epithelial cell lines.
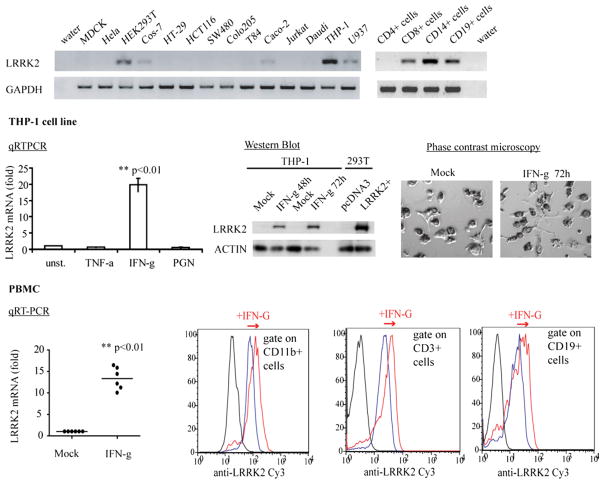
LRRK2 is expressed in immune cells and is upregulated by IFN-γ. Quantitative RT-PCR was performed using cDNA from a Clontech (Mountain View, CA) human cDNA panel or generated from American Type Culture Collection (Manassas, VA) cell lines. Reaction products were loaded into acrylamide gels to check the presence of the unique amplicons corresponding to LRRK2 and GAPDH (left panel). Quantification of the relative amount of LRRK2 mRNA to GAPDH mRNA is shown on the right with normalization to the value obtained from THP-1 cells. THP-1 cells were differentiated into macrophages using PMA 400 nM for 24 h and then treated with TNF-α, IFN-γ (100 ng/ml), or peptidoglycan (10μg/ml). Total mRNA was extracted to quantify LRRK2 mRNA and GAPDH mRNA after 24 h using quantitative RT-PCR, or proteins were extracted to detect LRRK2 and actin by Western blot after 48 or 72 h. Before extractions, cells were observed by phase microscopy to ensure that stimulation did not induce cell death. Blood was collected from healthy donors, and PBMCs were isolated using Ficoll gradients. The following day, PBMCs were treated with IFN-γ (100 ng/ml) or left untreated for 48 h. mRNA was extracted to quantify LRRK2 mRNA and GAPDH mRNA using quantitative RT-PCR (left panel), or PBMCs were analyzed by flow cytometry to detect LRRK2 in CD3-, CD19-, and CD11b-positive cells (right panel). The displayed experiments are representative of three independent experiments. Statistical analyses were performed using Student t test (**p < 0.01).
Intestinal mucosa of patients with CD exhibit a high concentration of proinflammatory cytokines, especially TNF-α and IFN-γ (23–25). To determine whether these cytokines regulate LRRK2 expression, we examined LRRK2 mRNA expression after TNF-α and IFN-γ stimulation. IFN-γ treatment induced a marked increase of LRRK2 mRNA in macrophage-differentiated THP-1 cells (19.7-fold ± 2.1 after 48 h stimulation; p < 0.001), whereas TNF-α had no effect (Fig. 2). Upregulation of LRRK2 mRNA resulted in an increase in LRRK2 protein as determined by Western blot (Fig. 2). Given the importance of the IL-23–Th17 axis in CD, we investigated the effect of an activator of the IL-23 pathway on LRRK2 expression using peptidoglycan stimulation. The IL-23 transcripts were increased 2-fold (not shown), whereas LRRK2 expression remained unchanged (Fig. 2).
To corroborate the results obtained from cell lines, PBMCs from healthy donors were analyzed. LRRK2 mRNA was also upregulated in PBMCs after stimulation for 48 h with IFN-γ (13.3-fold ± 2.6; p < 0.001; Fig. 2). Coimmunostaining of the LRRK2 protein with cell-specific markers in PBMCs and analysis by flow cytometry revealed that IFN-γ induced LRRK2 in CD3-positive T lymphocytes, CD19-positive B lymphocytes, and CD11b-positive monocytes (Fig. 2).
In support of these data indicating IFN-γ regulation of LRRK2 expression, we analyzed the LRRK2 promoter region. Alignment of LRRK2 promoters from 28 different species revealed a highly conserved region containing a consensus sequence of a binding site for IFN response factors (Supplemental Fig. 1), suggesting that LRRK2 upregulation by IFN-γ might be mediated through direct activation via IFN response factors.
LRRK2 is upregulated in the lamina propria of intestinal biopsy specimens from patients with CD
Using intestinal biopsy specimens from patients with CD or UC, we next investigated whether LRRK2 is regulated during in vivo inflammation. For each patient, a pair of biopsy specimens was collected: one from an inflamed area and one from a noninflamed area. The amount of LRRK2 mRNA in each biopsy specimen was normalized to the quantity of GAPDH mRNA. The result from each inflamed biopsy was normalized to the corresponding paired, noninflamed tissue from the same patient to determine the fold induction of LRRK2 expression during CD or UC inflammation. LRRK2 mRNA expression was ~6-fold higher in the biopsy specimens from inflamed tissue compared with a noninflamed area collected in the same patient with CD (p = 0.01; Fig. 3A). Despite a trend, LRRK2 was not statistically significantly increased in inflamed tissues from patients with UC (Fig. 3A), which might reflect that the levels of IFN-γ are higher in CD compared with UC (23, 26, 27).
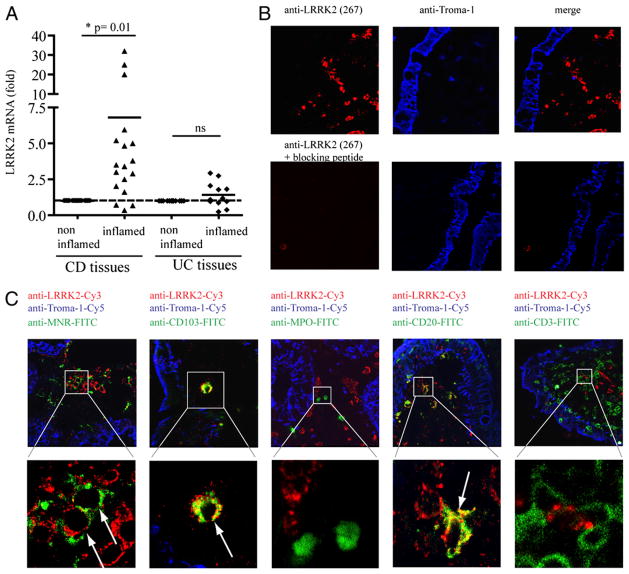
LRRK2 expression in colonic biopsy specimens from patients with CD. A, Paired biopsy specimens from intestinal tissues of patients with IBD (inactive disease state) were collected: one specimen from an inflamed area and one from a control noninflamed area for each patient. Total mRNA was extracted and used for qRT-PCR. The amount of LRRK2 mRNA relative to GAPDH mRNA of each inflamed biopsy specimen was normalized to the quantity measured in the corresponding noninflamed biopsy specimen obtained from the same patient. Statistical analyses were performed using Student t test (*p < 0.05). B and C, Frozen sections (7 μm) were prepared from colonic biopsy specimens obtained from patients with IBD. Tissues were immunolabeled with rabbit anti-LRRK2 (clone 267) and rat anti–troma-1 Abs to detect intestinal epithelial cells. The image gallery in B was performed with anti-LRRK2 Abs preincubated or not with its corresponding blocking peptide. C, Mouse monoclonal anti-CD3, anti-CD19, anti-myeloperoxidase, anti-mannose receptor (MNR), and anti-CD103 Abs were also used to detect T-lymphocytes, B-lymphocytes, neutrophils, macrophages, and dendritic cells, respectively. After immunostaining using secondary FITC-anti–mouse, Cy3-anti–rabbit and Cy5-anti–rat species-specific Abs, fluorescence was detected by confocal microscopy. The image gallery displays single confocal sections (original magnification ×60). The bottom panel arrows indicate the cells containing signals from both LRRK2 and FITC-MNR, CD103, CD20, and CD3 cell specific markers, respectively.
To identify which cell types in intestinal tissue express LRRK2, frozen sections from biopsy specimens of patients with CD were immunostained. LRRK2 was detected in the lamina propria but not in intestinal epithelial cells, which were specifically stained using an anti–Troma-1 Ab (Fig. 3B). We costained LRRK2 with cell-specific markers to detect cell types that microarray datasets had suggested express LRRK2 (Fig. 1), as well as those cell types known to express IFN-γ receptors. Abs directed against cell-specific markers were used to detect T lymphocytes, B lymphocytes, neutrophils, macrophages, and dendritic cells. As shown in Fig. 3C, LRRK2 was detected in a subset of mannose receptor-positive (CD206) macrophages, in CD103-positive dendritic cells, and in CD20-positive B lymphocytes. Mast cells, DC-SIGN–positive dendritic cells, NK cells, and enteric neurons were also localized with a panel of Abs. Although the biopsy specimens contained cells positive for these markers, LRRK2 was not detected in these cell types (not shown).
LRRK2 is able to activate NF-κB pathways
Inflammatory responses are coordinated by many signaling pathways that are controlled by different transcription factors, such as NF-κB and AP-1. We examined whether LRRK2 regulates those signaling pathways using luciferase-based reporters. LRRK2 activated NF-κB–dependent transcription 24 h after transfection (Fig. 4A), but did not activate transcription dependent on AP-1 response elements (Fig. 4B). LRRK2-induced NF-κB activation was dependent on IKK complex, because it was inhibited by a dominant negative form of IKK (Fig. 4C). Moreover, NF-κB activation was independent of LRRK2 kinase activity, because transfection of a dead kinase mutant K1906M (28) still led to NF-κB activation (Fig. 4D). Of note, the main PD-associated LRRK2 mutant (GS2019) induced NF-κB activation in a manner comparable to wild type LRRK2.
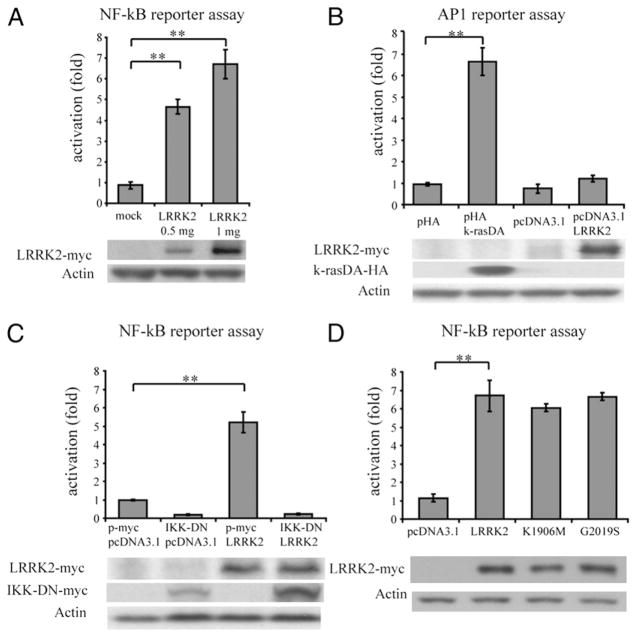
NF-κB activation induced by LRRK2. HEK293T cells were transfected with reporter plasmids encoding firefly luciferase cloned under a promoter containing NF-κB elements (A, C, D) or AP-1 elements (B), and with a plasmid encoding Renilla luciferase as a transfection control at a ratio of 10:1. Transcriptional activation was quantified 24 h after transfection by ratios of firefly luciferase activity to Renilla luciferase activity. A, Luciferase plasmids were cotransfected with empty vector pcDNA3.1 and pcDNA3.1-LRRK2myc. NF-κB activation induced by LRRK2 was normalized to the activation induced by transfection of an empty vector with an equivalent amount. B, Luciferase plasmids for the AP-1 reporter assay were cotransfected with a positive control (k-rasV12), pcDNA-3.1-LRRK2myc, and their corresponding empty backbone vectors (0.5 μg). Activation was normalized to the activation induced by corresponding empty vectors. C, Luciferase plasmids were cotransfected with pc-DNA3.1-LRRK2myc (1 μg) and/or a dominant negative form of IKK-β (0.25 μg). Data were normalized to the activation induced by cotransfection of the corresponding empty backbone vectors. D, Luciferase plasmids were cotransfected with pcDNA3.1 plasmids empty or encoding for LRRK2-myc, kinase-dead mutant LRRK2 K1906M or PD-associated mutant LRRK2 G2019S (1 μg). Data were normalized to the activation induced by the empty vector. Statistical analyses were performed using Student t test (**p < 0.01).
LRRK2 is recruited near pathogens during bacterial infection, and LRRK2 knockdown interferes with reactive oxygen species production during phagocytosis
In contrast to the human THP-1 cell line, LRRK2 can be detected in mouse RAW 264.7 macrophages at baseline and was lower, but significantly induced by IFN-γ (Fig. 5). In unstimulated cells, LRRK2 is distributed in vesicular structures within the cell (Fig. 6). Remarkably, when RAW macrophages were infected with S. typhimurium, a fraction of LRRK2 colocalized with microbes in the cytosol as shown using coimmunostaining of bacteria and LRRK2 (Fig. 6).
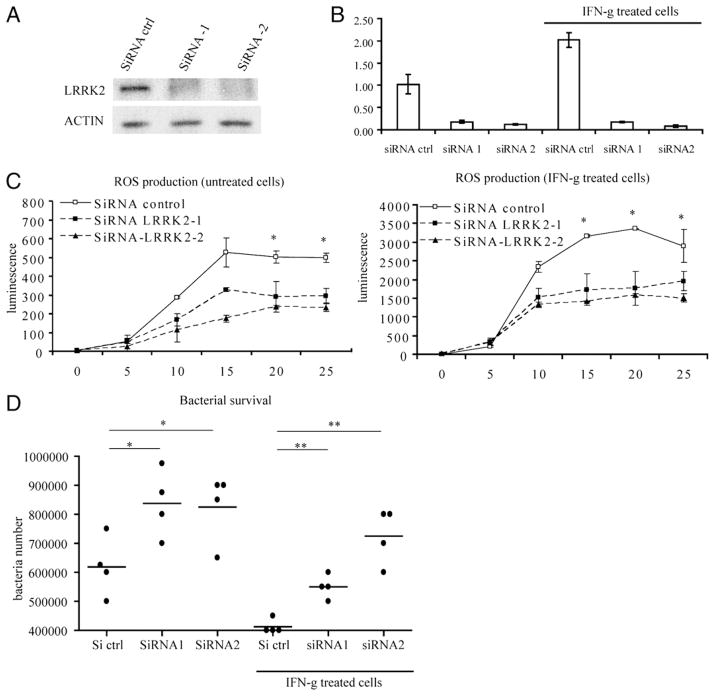
LRRK2 participates in ROS production during phagocytosis and in antibacterial response. Raw 264.7 cells were transfected with siRNA control or two different siRNA-directed against-mouse LRRK2 sequence. After 48 h of transfection, cells were stimulated with IFN-γ or left unstimulated for 24 h. LRRK2 expression was determined by Western blot (A) and quantitative PCR (B) 48 and 72 h after transfection, respectively. Cells were incubated with luminol and with opsonised-zymosan to assess the production of ROS upon phagocytosis (C). Cells were subjected to bacterial infection using S. typhimurium for 40 min and incubated with gentamicin to kill extracellular bacteria. Surviving intracellular bacteria were recovered after a total of 2 h of infection and quantified as detailed in Materials and Methods. Statistical analyses were performed using Student t test (*p < 0.05).
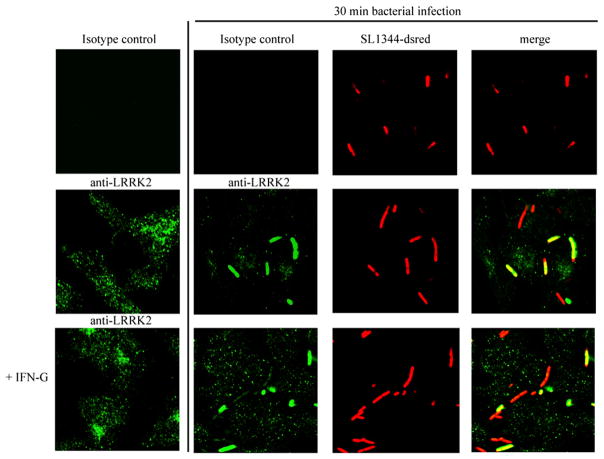
LRRK2 colocalizes with ds-red expressing S. typhimurium during bacterial infection of macrophages. Raw 264.7 cells were treated with IFN-γ (100 ng/ml) for 24 h or left untreated and were then infected using S. typhimurium expressing dsRed for 30 min. Cells were washed using PBS and fixed using paraformaldehyde before LRRK2 immunofluorescent staining using rabbit anti-LRRK2 Ab (clone AL106) and and FITC anti-rabbit Ab. Fluorescence was detected by confocal microscopy (original magnification ×60). Image gallery displays single confocal sections.
Overexpression of LRRK2 has been described to induce a weak but significant increase of reactive oxygen species (ROS) in a neuronal cell line (29). ROS are a key antipathogen response in phagocytic cells through direct action and signaling pathway activation. We next investigated whether LRRK2 might participate in the ROS production during phagocytosis. Mouse macrophage RAW 264.7 cells were transfected with control siRNA or two different siRNA directed against LRRK2 (siRNA transfection was more efficient in RAW 264.7 cells than in THP-1 cells). LRRK2 expression knockdown was assessed by Western blot (Fig. 5A) and the phagocytic burst was measured using opsonized zymosan and luminol in macrophages pretreated with and without IFN-γ. Production of ROS was significantly lower with the knockdown of LRRK2 expression (Fig. 5C). Because ROS play a major role in antibacterial response, we investigated the role of LRRK2 in the killing of intracellular bacteria. The number of surviving S. typhimurium was higher in RAW 264.7 cells with LRRK2 knockdown compared with control cells (Fig. 5D). This finding indicates that LRRK2 contributes to the antibacterial activity of the macrophages. Although recent reports suggested a role for wild-type LRRK2 in autophagy (30), under our experimental conditions knockdown of LRRK2 did not have an effect on autophagy (data not shown). These findings highlight an unanticipated role for LRRK2 in antibacterial host responses and signaling paradigms previously linked to CD.
Discussion
Identification of a number of susceptibility loci for CD in the past several years constitutes significant progress in efforts to understand the pathogenesis of this complex disease. However, functional studies are required to understand the underlying pathophysiologic mechanisms reflected by these many susceptibility loci. IFN-γ is a key cytokine in immune responses. Plasma and mucosal concentrations of IFN-γ are increased in CD (23, 26, 27). Anti–IFN-γ Abs have shown clinical benefit in CD in two independent studies (31, 32). Similarly, anti–IFN-γ showed protective effects in mice with anti-CD40–induced colitis (33). These observations implicate the IFN-γ pathway in the pathogenesis of CD. In this report, we investigated the involvement in the IFN-γ response of LRRK2, whose locus is located downstream of an SNP associated with a higher risk of developing CD. We detected LRRK2 expression in immune cells and tissues, we identified LRRK2 as an IFN-γ target gene upregulated by bacteria-induced or CD inflammation, and we showed that LRRK2 is an activator of the NF-κB pathway. These observations suggest that LRRK2 might be involved in the regulation of mucosal IFN-γ immune responses that are relevant to host responses to pathogens as well as CD.
In nonstimulated conditions, we observed that human monocytes, macrophages, and B lymphocytes express the highest amount of LRRK2 mRNA, although, basal LRRK2 expression is low overall, which is consistent with previous reports (21, 22). However, LRRK2 is highly expressed after IFN-γ stimulation. This striking difference in expression between basal condition and upon IFN-γ stimulation suggests that LRRK2 transcription is tightly regulated in physiologic conditions. Previous reports have already shown LRRK2 expression in extrabrain tissues and in immune-related tissues, such as the spleen, thymus, and lymph nodes (21, 22). A recent study using mice showed that LRRK2 expression is the highest in the spleen and is detected in B cells and macrophages, but not in T cells, which is consistent with our data collected from intestinal human tissues (34). In the context of IBD, the expression in lymphoid organs is especially interesting, because they contain immune cells involved in CD pathogenesis that are responsive to IFN-γ. The fact that LRRK2 is expressed in monocytes, macrophages, dendritic cells, and B cells and can be strongly upregulated by IFN-γ suggests a role for LRRK2 in immune functions. Many of the IFN-γ–induced functions implicated in pathogen resistance require activation of immune-signaling pathways. Our observations show that LRRK2 is able to activate inflammatory signaling pathways, suggesting that LRRK2 is likely involved in the transduction of the IFN-γ signal to downstream effectors. LRRK2 contains two catalytic domains that are functional in vitro and many protein interaction domains whose functions still need to be explored (9–11). Therefore, LRRK2 is thought to be involved in multiprotein-signaling protein complexes. Because LRRK2 is a kinase, we investigated whether the activation of those immune signaling pathways requires the phosphorylation of LRRK2 substrates. Surprisingly, a dead-kinase mutant still induces NF-κB activation, indicating that activation of this pathway is independent of LRRK2 kinase activity. In LRRK2 knockdown experiments, we did not detect any consistent differences in IL-6, MCP-1, and COX-2 transcription activation following Salmonella infection. As the intracellular PAMP receptor Nods, LRRK2 contains a leucin rich repeat domain, can form homodimer, and activates NF-κB in an overexpression setting that favors the homodimerization. We speculate that LRRK2 might have a specific stimulatory ligand (microbial or not) that is not expressed by S. typhimurium and remains to be defined. This ligand will activate LRRK2 dimerization and enhance the effect of LRRK2 on NF-κB signaling.
Mice deficient in IFN-γ or IFN-γ receptor are hypersensitive to bacterial, parasitic, and viral infections (35–37). A GWAS recently demonstrated the association of an SNP located upstream from the LRRK2 locus with a higher susceptibility to Mycobacterium leprae (38). That study also highlighted other genes already known to be associated with CD, such as NOD2, TNFS15, and C13ORF31, suggesting that the pathways involved in the control of M. leprae might be also involved in the susceptibility to CD. Interestingly, previous reports have suggested that M. avium tuberculosis might be involved in CD (39). Our data show that LRRK2 is recruited close to the pathogens during bacterial infection, and LRRK2 participates in the ROS production during the phagocytic burst and the antibacterial activity. These observations suggest that LRRK2 is likely to be involved in the host response to pathogens, particularly in the context of the IFN-mediated immune responses.
Because LRRK2 mutations are the most common genetic alteration associated with PD, it is reasonable to wonder about the potential relevance of the present findings to PD (6). Although our results suggest that LRRK2 polymorphisms might be involved in immune responses, dysregulated in CD or in the host reponse to pathogen, the relevance of our findings regarding PD pathophysiology is uncertain. LRRK2-deficient mice exhibit no obvious phenotype (40), but our findings support the potential relevance of assessing phenotype characterizations in the context of inflammation. Detection of LRRK2 at the protein level has been a challenge that limited the progress in the investigation of LRRK2 functions. For example, although a GWAS identified multiple SNPs associated with a higher risk of developing PD (41), the authors reported their inability to perform quantitative trait analysis to detect differences in LRRK2 expression associated with the disease SNPs because of the insufficient expression at the probe for LRRK2. In this case, our findings indicate the potential relevance of investigating the association of PD-SNPs with LRRK2 expression in the context of IFN stimulation.
Of interest, and in addition to the intestinal macrophages reported in this study, LRRK2 protein has been detected in activated macrophage–microglia in the brain of patients with PD (7). Although the main PD-associated mutant LRRK2-G2019S did not activate NF-κB differently from wild type protein, our data suggest that studies of PD should consider the potential relevance of inflammatory responses mediated by LRRK2 in the pathogenesis of PD. Several studies have suggested that inflammation might be involved in the pathophysiology of PD (42–44), whereas our report provides evidence that LRRK2 has a role in immune responses mediated by IFN-γ.
Acknowledgments
We thank the clinicians, consultants, and nursing staff who recruited subjects and collected samples. We also thank Melissa Cohen for contacting patients and coordinating blood collection, Kathryn Devaney for indexing and processing patient samples for genotyping, Dr. Lisa Westerberg and Dr. Oren Shibolet for helpful suggestions, Susan Davis for manuscript editing, and all individuals, patients, and healthy donors who contributed to this study.
This work was supported by National Institutes of Health Grants AI062773 and DK83756 (to R.J.X.) and DK060049 and DK043351 (to D.K.P.). A.G. was supported by fellowships from La Fondation pour la Recherche Medicale and the Crohn’s and Colitis Foundation of America.
Abbreviations used in this paper
| CD | Crohn’s disease |
| DC-SIGN | dendritic cell-specific intercellular adhesion molecule-3–grabbing nonintegrin |
| GNF | Genomics Institute of the Novartis Research Foundation |
| GWAS | genome-wide association study |
| IBD | inflammatory bowel disease |
| IKK-β | IκB kinase |
| MNR | mannose receptor |
| PD | Parkinson’s disease |
| ROS | reactive oxygen species |
| SNP | single nucleotide polymorphism |
| UC | ulcerative colitis |
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.4049/jimmunol.1000548
Read article for free, from open access legal sources, via Unpaywall:
https://journals.aai.org/jimmunol/article-pdf/185/9/5577/1328965/1000548.pdf
Free after 12 months at www.jimmunol.org
http://www.jimmunol.org/cgi/content/full/185/9/5577
Free after 12 months at www.jimmunol.org
http://www.jimmunol.org/cgi/reprint/185/9/5577
Free to read at www.jimmunol.org
http://www.jimmunol.org/cgi/content/abstract/185/9/5577
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
LRRK2 is not required for lysozyme expression in Paneth cells.
Nat Immunol, 25(11):2037-2039, 08 Oct 2024
Cited by: 0 articles | PMID: 39379660
The immune system in Parkinson's disease: what we know so far.
Brain, 147(10):3306-3324, 01 Oct 2024
Cited by: 0 articles | PMID: 38833182 | PMCID: PMC11449148
Review Free full text in Europe PMC
Exon shuffling and alternative splicing of ROCO genes in brown algae enables a diverse repertoire of candidate immune receptors.
Front Plant Sci, 15:1445022, 23 Aug 2024
Cited by: 0 articles | PMID: 39246816 | PMCID: PMC11378527
Single molecule array measures of LRRK2 kinase activity in serum link Parkinson's disease severity to peripheral inflammation.
Mol Neurodegener, 19(1):47, 11 Jun 2024
Cited by: 2 articles | PMID: 38862989 | PMCID: PMC11167795
Bidirectional communication of the gut-brain axis: new findings in Parkinson's disease and inflammatory bowel disease.
Front Neurol, 15:1407241, 24 May 2024
Cited by: 0 articles | PMID: 38854967
Review
Go to all (251) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE96
SNPs
- (2 citations) dbSNP - rs11175593
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Crohn's and Parkinson's Disease-Associated LRRK2 Mutations Alter Type II Interferon Responses in Human CD14+ Blood Monocytes Ex Vivo.
J Neuroimmune Pharmacol, 15(4):794-800, 16 Mar 2020
Cited by: 14 articles | PMID: 32180132 | PMCID: PMC7718203
Interferon-γ induces leucine-rich repeat kinase LRRK2 via extracellular signal-regulated kinase ERK5 in macrophages.
J Neurochem, 129(6):980-987, 24 Feb 2014
Cited by: 45 articles | PMID: 24479685
An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis.
Sci Transl Med, 10(444):eaan8162, 01 Jun 2018
Cited by: 68 articles | PMID: 29875204 | PMCID: PMC6636639
An emerging role for LRRK2 in the immune system.
Biochem Soc Trans, 40(5):1134-1139, 01 Oct 2012
Cited by: 30 articles | PMID: 22988878
Review
Funding
Funders who supported this work.
NIAID NIH HHS (6)
Grant ID: R01 AI062773-01A1
Grant ID: R01 AI062773-02
Grant ID: R01 AI062773-04
Grant ID: AI062773
Grant ID: R01 AI062773
Grant ID: R01 AI062773-03
NIDDK NIH HHS (21)
Grant ID: DK060049
Grant ID: P30 DK043351-19S1
Grant ID: P30 DK043351-20
Grant ID: P30 DK043351-21
Grant ID: R01 DK060049-09
Grant ID: R01 DK083756-04
Grant ID: DK043351
Grant ID: R01 DK083756-01
Grant ID: P30 DK043351
Grant ID: P30 DK043351-19
Grant ID: R01 DK083756
Grant ID: P30 DK043351-18
Grant ID: P30 DK043351-22
Grant ID: R01 DK083756-02
Grant ID: R01 DK060049-08
Grant ID: DK83756
Grant ID: P30 DK043351-19S2
Grant ID: P30 DK043351-20S1
Grant ID: R01 DK060049
Grant ID: R01 DK060049-10
Grant ID: R01 DK083756-03





