Abstract
Free full text

Distant Metastasis Occurs Late during the Genetic Evolution of Pancreatic Cancer
Summary
Metastasis, the dissemination and growth of neoplastic cells in an organ distinct from that in which they originated 1,2, is the most common cause of death in cancer patients. This is particularly true for pancreatic cancers, where most patients are diagnosed with metastatic disease and few show a sustained response to chemo- or radiation therapy 3. Whether the dismal prognosis of patients with pancreatic cancer compared to patients with other types of cancer is a result of late diagnosis or early dissemination of disease to distant organs is not known. Here we rely on data generated by sequencing the genomes of seven pancreatic cancer metastases to evaluate the clonal relationships among primary and metastatic cancers. We find that clonal populations that give rise to distant metastases are represented within the primary carcinoma, but these clones are genetically evolved from the original parental, non-metastatic clone. Thus, genetic heterogeneity of metastases reflects that within the primary carcinoma. A quantitative analysis of the timing of the genetic evolution of pancreatic cancer was performed, indicating at least a decade between the occurrence of the initiating mutation and the birth of the parental, non-metastatic founder cell. At least five more years are required for the acquisition of metastatic ability and patients die an average of two years thereafter. These data provide novel insights into the genetic features underlying pancreatic cancer progression and define a broad time window of opportunity for early detection to prevent deaths from metastatic disease.
We performed rapid autopsies of seven individuals with end stage pancreatic cancer (Supplementary Table 1). In all patients, metastatic deposits were present within two or more anatomic sites in each patient, most often the liver, lung and peritoneum, as is typical for this form of neoplasia 4.
Low passage cell lines (six patients) or first passage xenografts (one patient) were created from one of the metastases present at each patient’s autopsy. These samples comprised seven of the 24 pancreatic cancers which previously underwent whole exome sequencing and copy number analysis, as described in a mutational survey of the pancreatic cancer genome 5. In this previous study, a total of 426 somatic mutations in 388 different genes were identified among 220,884,033 bp sequenced in the seven index metastatic lesions, corresponding to an average of 61 mutations per index metastatic lesion (range 41-77). In all samples, the vast majority of mutations were represented by missense or silent single base substitutions (Supplementary Fig. 1 and Supplementary Table 2).
For each of the somatic mutations identified in the seven index metastasis lesions, we determined whether the same somatic mutation was present in anatomically distinct metastases harvested at autopsy from the same patients. We also determined whether these mutations were present in the primary pancreatic tumors from which the metastases arose. A small number of these samples of interest were cell lines or xenografts, similar to the index lesions, whereas the majority were fresh-frozen tissues that contained admixed neoplastic, stromal, inflammatory, endothelial and normal epithelial cells (Fig. 1a). Each tissue sample was therefore microdissected to minimize contaminating non-neoplastic elements before purifying DNA.
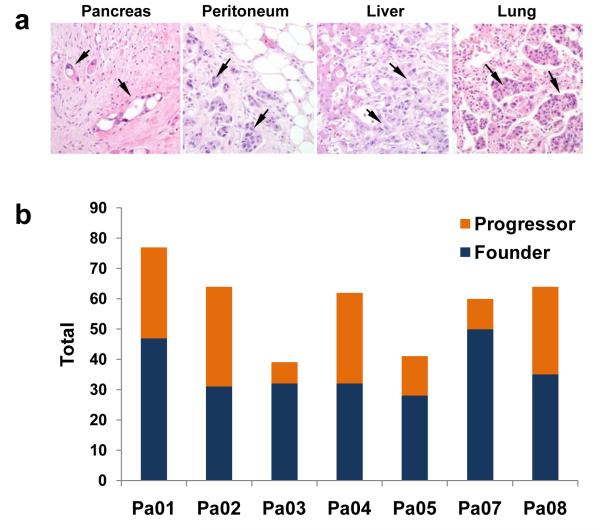
a. Histopathology of primary infiltrating pancreatic cancer and metastatic pancreatic cancer to the peritoneum, liver and lung. In addition to infiltrating cancer cells in each lesion (arrows), non-neoplastic cell types are abundant. b. Total mutations representing parental clones (founder mutations), and clonal evolution (progressor mutations) within the primary carcinoma based on comparative lesion sequencing. Mutations common to all samples analyzed were the most common category identified.
Two categories of mutations were identified (Fig. 1b). The first and largest category corresponded to those mutations present in all samples from a given patient (“founder” mutations, mean of 64%, range 48-83% of all mutations per patient; Fig. 1b, example in Supplementary Fig. 2a). These data indicate that the majority of somatically acquired mutations present in pancreatic cancers occur before the development of metastatic lesions. All other mutations were characterized as “progressor” mutations (mean of 36%, range 17 - 52% of all mutations per patient; Fig. 1b, example in Supplementary Fig. 2b). These mutations were present in one or more of the metastases examined, including the index metastasis, but not the parental clone.
These mutation types were used to classify the lesions that contained them into parental clones (containing only founder mutations) and subclones (containing both founder and progressor mutations). By definition, there could be only one parental clone in a patient, though there could be many different subclones. Parental clones tended to contain more deleterious mutations (nonsense, splice site or frameshift mutations) than subclones (12.6% of the mutations in the parental clones versus 8.1% of the mutations in subclones, Supplementary Table 2). The parental clones had already accumulated mutations in all driver genes (KRAS, TP53 and SMAD4) previously shown to drive pancreatic tumorigenesis 6. Through combined analysis of high density SNP chip data on the index lesion (Supplementary Table 3) plus the sequencing data on all lesions (Supplementary Table 2) we found that the vast majority of homozygous mutations (51 mutations, representing 89% of all homozygous mutations) in the index lesion were already present in the parental clones. Homozygous mutations are characteristic of tumor suppressor genes such as SMAD4 and CDKN2A and often occur in association with chromosomal instability 7. In sum, the parental clones harbored the majority of deleterious genetic alterations and chromosomal instability, upon which were superimposed an accumulation of progressor mutations associated with clonal evolution and metastasis.
Evolutionary maps were constructed for each patient’s carcinoma based on the patterns of somatic mutation and allelic losses and the locations of individual metastatic deposits (Fig. 2, Supplementary Figs. 3 to 8). These maps revealed that, despite the presence of numerous founder mutations within the parental clones, the cells giving rise to the metastatic lesions had a large number of progressor mutations. For example, in Pa01 the parental clone contained 49 founder mutations, yet a clonal expansion marked by the presence of mutations in six additional genes was present in the lung and peritoneal metastases (Supplementary Fig. 3). Moreover, 22 more mutations were found in the liver metastasis. Note that all mutations in the metastatic lesions were clonal, i.e., present in the great majority if not all neoplastic cells of the metastasis, as assessed by Sanger sequencing. Thus, these mutations were present in the cell that clonally expanded to become the metastasis. Similarly, large numbers of progressor mutations were generally observed in the metastases from each of the seven cases examined (Fig. 2 and Supplementary Figs. 3 to 8).
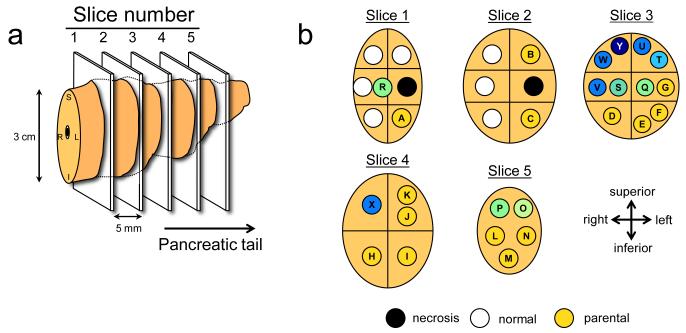
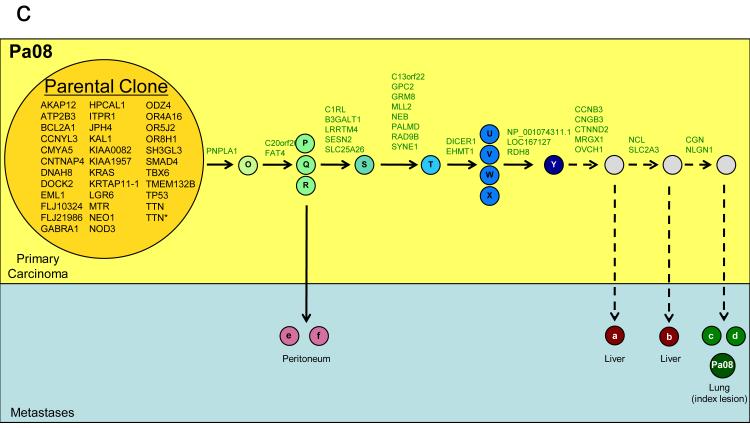
a. Illustration of the pancreatic specimen removed from Pa08 at rapid autopsy, and the planes of sectioning of the specimen. b. Mapping of the parental clone and subclones identified in a by comparative lesion sequencing within serial sections of the infiltrating pancreatic carcinoma. Metastatic subclones giving rise to liver and lung metastases are nonrandomly located within slice 3, indicated by blue circles. These clones are both geographically and genetically distinct from clones giving rise to peritoneal metastases in this same patient, indicated by green. c. Proposed clonal evolution based on the sequencing data. In this model, after development of the parental clone, ongoing clonal evolution continues within the primary carcinoma (yellow rectangle), and these subclones seed metastases in distant sites. * indicates two mutations were found in the TTN gene.
To distinguish between the possibilities that clonal evolution occurred inside the primary cancer versus within secondary sites, we sectioned the primary tumors from two patients into numerous, three-dimensionally organized pieces (Fig. 2a,b) and examined the DNA from each piece for each of the founder and progressor mutations. In Patient Pa08, there were three progressor mutations present in two independent peritoneal metastases (defining one subclone) and 23, 25, or 27 additional progressor mutations present in liver and lung metastases (defining three additional subclones; Fig. 2c). Through the analysis of distinct regions of the primary tumor, it was clear that subclones giving rise to each of these metastases were present in the primary tumor. Moreover, these subclones were not small; from the size of the pieces (Fig. 2a) and the amounts of DNA recovered, each subclone must have contained in excess of 100 million cells. In addition, more than four different subclones, each containing a similarly large numbers of cells, could be identified through the analysis of other pieces of the same tumor. These subclones could be put into an ordered hierarchy establishing an evolutionary path for tumor progression (Fig. 2c). Analysis of multiple primary tumor pieces and metastatic lesions from patient Pa04 revealed a similar clonal evolution, with distinct, large subclones within the primary tumors giving rise to the various metastases (Supplementary Fig. 8).
To further clarify clonal evolution within the primary site, we attempted to correlate the mutation signatures representing the subclones of Pa08 (Fig. 2c) with the geographic location of the pieces of the primary tumor used to define them (Fig. 2a, 2b). Samples representative of the parental clone were located throughout the primary carcinoma. By contrast, samples representing subclones were nonrandomly located in proximity to each other, within which the subclones specifically giving rise to peritoneal versus distant metastases were seen. Thus, we conclude that the genetic heterogeneity of metastases reflects heterogeneity already existing within the primary carcinoma, and that the primary carcinoma is a mixture of numerous subclones, each of which has independently expanded to constitute a large number of cells.
This dataset could also be used to infer the timing of the development of the various stages of pancreatic tumor progression 8. We assume that the tumor is initiated by a genetic event that confers a selective growth advantage to the cell that goes on to become the founder cell of the tumor. To estimate the timing, we first used Ki67 labeling to determine the proliferation rate of seven samples of normal duct epithelium from surgically resected pancreata of individuals without pancreatic cancer as well as of each index metastasis. Ki67-positive nuclei constituted an average of 0.4% of normal ductal cells, while an average of 16.3% of cancer cells within the index metastasis lesions were Ki67-positive, consistent with prior estimates 9-10 (Supplementary Table 4). Based on these data plus that from sequencing of the index lesions, we derived estimates for three critical times in tumor evolution: T1 - the time between tumor initiation and the birth of the cell giving rise to the parental clone; T2 - the subsequent time required for the birth of the cell that gave rise to the index metastasis; and T3 – the time between the dissemination of this cell and the patients’ death (Fig. 3). In other words, there is a time point, t0, when the tumor was initiated, and a time point t1 when a cell is born that has all mutations that exist in the parental clone. Similarly, there is a time point in tumor evolution, t2, when a cell is born that has all the mutations that exist in the index metastasis. T1 is given by t1- t0 and T2 is given by t2- t1. If we denote t3 as the time of patient’s death, then T3 = t3- t2.
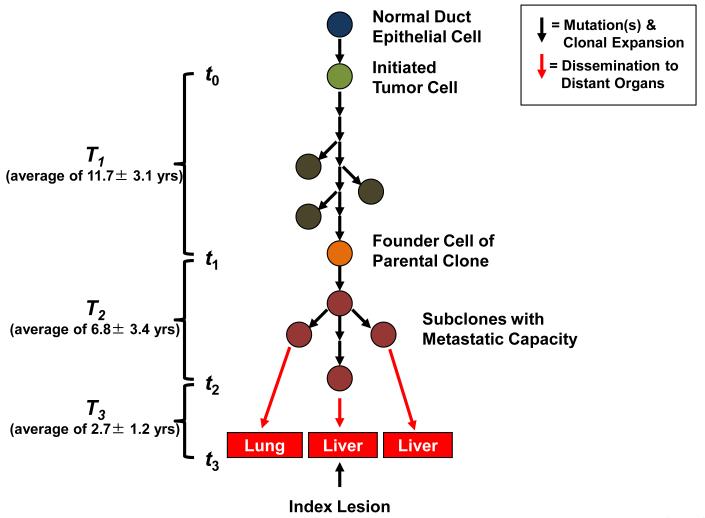
Tumorigenesis begins with an initiating mutation in a normal cell that confers a selective growth advantage. Successive waves of clonal expansion occur in association with the acquisition of additional mutations, corresponding to the progression model of pancreatic intraepithelial neoplasia (PanIN) and time T1. One founder cell within a PanIN lesion will seed the parental clone and hence initiate an infiltrating carcinoma (end of T1 and beginning of T2). Eventually, the cell that will give rise to the index lesion will appear (end of T2 and beginning of T3). Unfortunately, most patients are not diagnosed until well into time interval T3 when cells of these metastatic subclones have already escaped the pancreas and started to grow within distant organs. The average time for intervals T1, T2 and T3 for all seven patients is indicated in the parentheses at left (see also Supplementary Table 6).
Using the mathematical model described in the Methods, we were able to conservatively estimate an average of 11.7 years from the initiation of tumorigenesis until the birth of the cell giving rise to the parental clone, an average of 6.8 years from then until the birth of the cell giving rise to the index lesion, and an average of 2.7 years from then until the patients’ death (see Supplemental Discussion and Supplementary Table 5).
We show, for the first time, that primary pancreatic cancers contain a mix of geographically distinct subclones, each containing large numbers (hundreds of millions) of cells that are present within the primary tumor years before the metastases become clinically evident. The features of these metastatic subclones that promote metastasis formation have yet to be discerned, as no consistent genetic signature of metastatic subclones could be identified. We did identify several genes that were mutated in one or more of the index metastatic lesions from these seven patients with Stage IV disease, but not in the primary pancreatic index lesions from 17 patients with Stage II disease (Supplementary Table 2). These genes include those that may play a role in invasive or metastatic ability through heterotypic cell adhesion (CNTN5), motility (DOCK2), proteolysis (MEP1A) and tyrosine phosphorylation (LMTK2). However, these mutations were not metastasis-specific per se as all but one were present in the matched primary carcinoma of those same seven patients, and there is no evidence that the mutations we observed endowed these genes with metastagenic activity. These data also do not reveal the selective pressures within the primary carcinoma that led to the formation of progressor mutations. In light of recent findings indicating that pancreatic cancers are poorly vascularized 11, one possibility is that intratumoral hypoxia creates a fertile microenvironment for the formation of additional mutations beyond that of the parental clone.
One of the major implications of these data is their implication for screening to prevent pancreatic cancer deaths. Quantitative analysis indicated a large window of opportunity for diagnosis while the disease was still in the curative stage –at least a decade. Our model also predicts an average of 6.8 years between the birth of the cell giving rise to the parental clone and the seeding of the index metastasis. Unfortunately, the great majority of patients are not diagnosed until the last 2 years of the entire tumorigenic process. The challenge is to detect these tumors during time T1, or even after T1 but before seeding of metastases. Advanced imaging methods, as well as blood tests to detect cancer-specific proteins, transcripts, or genes 12, offer hope for such non-invasive early detection.
Methods Summary
Rapid autopsies were performed on seven individuals with Stage IV pancreatic cancer 13. Genomic DNA was extracted from cell lines or xenografts established from one metastasis of each patient and used for exomic sequencing as previously described 5. The Illumina Infinium II Whole Genome Genotyping Assay employing the BeadChip platform was also used to analyze each sample at 1,072,820 (1M) SNP loci as previously described 5. Samples of snap frozen pancreatic cancer tissue were microdissected using a PALM MicroLaser System (Carl Zeiss MicroImaging, Oberkochen, Germany) and DNA extracted using QIAmp DNA Micro Kits (Qiagen, Valencia, CA, USA). Genomic DNA was quantified by calculating long interspersed nuclear elements (LINE) by real-time PCR. Whole genome amplification (WGA) was performed using 10 ng total template DNA and an illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Piscataway, NJ, USA). Ki67 immunolabeling (Clone MIB-1, Dako Cytomation, Glostrup, Denmark) was performed on formalin fixed, paraffin embedded sections of normal pancreatic ducts and metastatic pancreatic cancer tissues for each patient using the Ventana Discovery staining system (Ventana Medical Systems, Tucson, AZ, USA), and this information used to inform computational models of the timing of clonal evolution of each patient’s pancreatic cancer (full details of these models are available in Online Methods).
Methods
Patients and Tissue Samples
Tissue samples from 7 patients with pancreatic ductal adenocarcinoma were collected in association with the Gastrointestinal Cancer Rapid Medical Donation Program (GICRMDP). This program was approved by the Johns Hopkins institutional review board and deemed in accordance with the Health Insurance Portability and Accountability Act. Details of the program have been described in detail previously 13. The tissue harvesting protocol consists of the following; after opening of the body cavity using standard techniques, the whole pancreas including the pancreatic cancer and each grossly identified metastasis were sampled using a sterile blade and forceps. The whole pancreas was sliced into 1 × 1 × 0.4 cm sections for overnight fixation in 10% buffered-formalin, for freezing in Tissue-Tek OCT compound (Sakura Finetechnical, Tokyo, Japan) in liquid nitrogen and for snap-freezing in liquid nitrogen in 1.7 mL cryovials and storage at −80°C. Xenograft enriched or low passage cell lines were generated from the post mortem cancer tissues of these seven patients as previously described 13,14.
Laser Capture Microdissection (LCM)
Frozen tissue sections of autopsy tissues were cut into 7 μm sections using a cryostat and embedded onto UV-treated PALM membrane slides (Carl Zeiss MicroImaging, Oberkochen, Germany) and the slides were stored immediately at −80 °C until subsequent fixation. Tissue sections that underwent LCM were defrosted, fixed in 100% methanol for 3 minutes, stained with toluidine blue before microdissection to remove contaminating stromal elements. Sections were dissected using a PALM MicroLaser System (Carl Zeiss MicroImaging). Dissected tissues were catapulted into adhesive caps. Generally, >20,000 cells were obtained from 5-10 serial sections by LCM in order to have obtain sufficient quantity and quality of genomic DNA for subsequent amplification and sequencing.
Genomic DNA Extraction and Whole Genome Amplification
Genomic DNA from microdissected tissues was extracted using QIAmp DNA Micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Genomic DNA was quantified by calculating long interspersed nuclear elements (LINE) by real-time PCR. The LINE primer set 5′-AAAGCCGCTCAACTACATGG-3′ (forward) and 5′-TGCTTTGAATGCGTCCCAGAG-3′ (reverse) was designed. The real-time PCR condition was 95°C for 10 minutes; 40 cycles of 94°C for 10 seconds, 58°C for 15 seconds and 70°C for 30 seconds. PCR was carried out using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA, USA). To minimize sequencing bias from using low starting templates, only samples for which the measured concentration by LINE assay was ≥3.3 ng/ul (1000 genome equivalents) were used as a starting template for whole genome amplification (WGA). WGA was performed using 10 ng total template DNA and an illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Piscataway, NJ, USA), following the manufacturer’s protocol. WGA products were purified using a Microspin G-50 system (GE Healthcare). The purified WGA products were quantified by NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and diluted to 20 ng/μL for sequencing analysis. Using these methods and quality controls, there was complete concordance in the mutational signatures obtained of cultured cell lines/xenografts versus WGA materials prepared from their matched frozen tissues.
Sanger Sequencing
PCR amplification and sequencing was performed using the conditions and primers described by Jones et al. 5. A small number of sequencing reactions failed (<2% of the total reactions) and these corresponding genes were not included in progression models or quantitative time estimates of clonal evolution.
Genotyping
The Illumina Infinium II Whole Genome Genotyping Assay employing the BeadChip platform was used to analyze tumor samples at 1,072,820 (1M) SNP loci as previously described 5. Briefly, all SNP positions were based on the hg18 (NCBI Build 36, March 2006) version of the human genome reference sequence. The genotyping assay begins with hybridization to a 50 nucleotide oligonucleotide, followed by a two-color fluorescent single base extension. Fluorescence intensity image files were processed using Illumina BeadStation software to provide normalized intensity values and allelic frequency for each SNP position. For each SNP, the normalized experimental intensity value (R) was compared to the intensity values for that SNP from a training set of normal samples and represented as a ratio (called the “Log R Ratio”) of log2 (Rexperimental/Rtraining set). For each SNP, the normalized allele intensity ratio (theta) was used to estimate a quantitative allelic frequency value (called the “B Allele Frequency”) for that SNP 15. Using Illumina BeadStudio software, LogR Ratio and B Allele Frequency values were plotted along chromosomal coordinates and examined visually. Regions of loss of heterozygosity (LOH) were identified as genomic regions >2Mb with consecutive homozygous genotype calls (B Allele Frequency near 0 or 1). Smaller (<2Mb) regions of LOH were identified by requiring co-occurrence of decreased Log R Ratio scores in regions of consecutive homozygous genotype calls (B Allele Frequency near 0 or 1). Visual analysis of these data plotted along chromosomal coordinates was followed by manual analysis of the data for selected genes of interest.
Estimations of Proliferation Rates
To estimate the cell division rate, the Ki-67 labeling index (LI) in the proband lesion for each case was calculated. The Ki-67 LI on the pancreatic ducts in the histologically normal pancreas parenchyma was also calculated. Normal pancreas was collected from 2 autopsied patients who died of causes other than pancreatic cancer and 5 patients who underwent distal pancreatectomy for a serous cystadenoma or a islet cell tumor at The Johns Hopkins Hospital. Paraffin blocks were cut into sections 4 μm thick for Ki-67 immunostaining with all staining processes from deparaffinization to counterstaining with hematoxylin being performed automatically with the Ventana Discovery staining system (Ventana Medical Systems, Tucson, AZ). An anti-human Ki-67 mouse monoclonal antibody (Clone MIB-1, Dako Cytomation, Glostrup, Denmark) was used. At least 12 randomly selected high-power fields containing a minimum of 2000 cells were evaluated for each case, and the labeling index (LI) was calculated as the percentage of positive cell nuclei. Reactive small lymphocytes in each case were regarded as internal positive controls for Ki-67. Equal or more intense nuclear staining in comparison with the internal positive controls was considered to indicate positivity.
Modeling Tumor Evolution
Passenger mutations were defined as those unlikely to drive tumorigenesis. To be conservative, we considered passenger mutations as those not included as candidate cancer genes in a recent study based on whole exome sequencing of 24 pancreatic cancers 5. As the great majority of mutations identified in cancers are believed to be passengers, the results of the model are not highly dependent on the model used to estimate the relatively small number of drivers 16.
Because passenger mutations are neutral and do not affect the evolution in any way, they are accumulated independently in each cell lineage. Following the lineage of the founder cell of the parental clone back in time, we can assume that it acquired a new neutral mutation with rate r at each cell division, with r being the product of the mutation rate per base pair per cell division and the number of base pairs sequenced. The accumulation of neutral mutations in a cell lineage can be well-described by a Poisson process with rate r per cell division. We are interested in the number of cell divisions in the single lineage between tumor initiation and birth of the founder cell of the parental clone during which N1 passenger mutations accumulate. On the other hand, N1 is also the number of mutations that are found in all tumor samples from one patient. Since we sequenced at least one sample from the primary tumor and at least three samples from different metastases from each patient, these specific N1 mutations had to be present in the founder cells of all three metastases and in cells in the primary tumor. Thus there was a cell in the tumor that had these N1 specific mutations for the first time, and that is, by definition, the founder cell of the parental clone. Since we can neglect the accumulation of mutations before the onset of the tumor, these N1 mutations are accumulated along the single lineage from the tumor initiator cell to the founder cell of the parental clone. As the number of cell divisions between two subsequent mutations is distributed according to an exponential distribution with mean 1/r, the required number of cell divisions is the sum of N1 independent exponentially distributed random variables with mean 1/r, and is distributed according to a Gamma distribution with shape parameter N1 and scale parameter 1/r. The mean of this distribution is N1/r and the standard deviation is
Using our measurements of Ki-67 labeling index of the 7 index lesions (average 16.3%), we were able to estimate the S-phase fraction of cells in the 7 index lesions (average LI=9.5%) 17. Assuming a median value for the S-phase duration in human tissues and tumors, Ts, of 10 hours 18, and using the formula for the potential cell doubling time Tpot=λTs/LI, we get an estimate for Tpot of 3.5 days. Here λ is a correction factor for the non-linear age distribution of cells through the cell cycle, which was assumed to be 0.8 19. This estimate is consistent with the average cell doubling time in pancreatic cancer from Amikura et al. 20 of 2.3 days. We use this latter estimate in our analysis, as we believe it is more accurate for pancreatic cancer.
Amikura et al 20 also reported the median doubling time of pancreatic cancer metastases as 56 days. To estimate the age of the index metastasis, we used a two stage model. We estimated the tumor doubling time was equal to the cell doubling time (Tgen) until the tumor size reached 100 microns in diameter at which time angiogenesis is required 21. Thereafter, we used the median doubling time described above.
Our model works very well for estimating the number of cell divisions between discrete events in tumor evolution. In order to go from number of cell divisions to actual time we need to have an estimate for the average rate of cell division. The accuracy of our predictions regarding actual time therefore depends on the accuracy of that estimate. If we let Tgen denote the average time between subsequent cell divisions in a cell lineage, we arrive at the expression for time T1:
We therefore estimate the number of cell divisions, and hence the time T1 between tumor initiation and birth of the founder cell of the parental clone to be proportional to the number of passenger mutations, N1, that the tumor acquired during that time. In our calculations, we use the estimate for cell doubling time in pancreatic cancer from the literature 20 as the value of Tgen.
T2 is determined analogously, with N2 defined as the number of passenger mutations present in the index lesion but not in the parental clone. T3 is determined from literature-based estimates of the tumor and cell doubling times, and the size of the index lesions at autopsy 20.
Acknowledgements
This work was supported by National Institutes of Health grants CA106610 (CID), CA62924 (CID, MAN), CA43460 (BV), CA57345 (KWK and VEV), CA121113 (VEV and KWK), GM078986 (MAN), the Bill and Melinda Gates Foundation Grand Challenges Grant 37874, the Uehara Memorial Foundation (SY), the AACR-Barletta Foundation (CID), the John Templeton Foundation, Pilot Funding from the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Pancreatic Cancer Foundation, the George Rubis Endowment for Pancreatic Cancer Research, the Joseph C. Monastra Foundation for Pancreatic Cancer Research, the Alfredo Scatena Memorial Fund, the Virginia and the D.K. Ludwig Fund for Cancer Research, The Joint Program in Mathematical Biology and J. Epstein. We would like to acknowledge Drs. Toby C. Cornish, Clark Henderson, Noriyuki Omura and Seung-Mo Hong for their technical assistance in selected aspects of this work.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature09515
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3148940
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101860437
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature09515
Article citations
Construction and validation of senescence risk score signature as a novel biomarker in liver hepatocellular carcinoma: a bioinformatic analysis.
Transl Cancer Res, 13(9):4786-4799, 12 Sep 2024
Cited by: 0 articles | PMID: 39430830 | PMCID: PMC11483424
Systemic Therapy for Metastatic Pancreatic Cancer-Current Landscape and Future Directions.
Curr Oncol, 31(9):5206-5223, 04 Sep 2024
Cited by: 0 articles | PMID: 39330013 | PMCID: PMC11430697
Review Free full text in Europe PMC
Clinical Perspectives on the Histomolecular Features of the Pancreatic Precursor Lesions: A Narrative Review.
Middle East J Dig Dis, 16(3):136-146, 31 Jul 2024
Cited by: 0 articles | PMID: 39386334 | PMCID: PMC11459284
Review Free full text in Europe PMC
Panoramic tumor microenvironment in pancreatic ductal adenocarcinoma.
Cell Oncol (Dordr), 47(5):1561-1578, 15 Jul 2024
Cited by: 0 articles | PMID: 39008192
Review
Pancreatic stellate cells and the interleukin family: Linking fibrosis and immunity to pancreatic ductal adenocarcinoma (Review).
Mol Med Rep, 30(3):159, 12 Jul 2024
Cited by: 0 articles | PMID: 38994764 | PMCID: PMC11258612
Review Free full text in Europe PMC
Go to all (1,558) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The patterns and dynamics of genomic instability in metastatic pancreatic cancer.
Nature, 467(7319):1109-1113, 01 Oct 2010
Cited by: 859 articles | PMID: 20981101 | PMCID: PMC3137369
Cancer: Genomic evolution of metastasis.
Nature, 467(7319):1053-1055, 01 Oct 2010
Cited by: 19 articles | PMID: 20981088
Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy.
Cancer Biol Ther, 4(5):548-554, 12 May 2005
Cited by: 96 articles | PMID: 15846069 | PMCID: PMC2771924
Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer.
Mol Oncol, 10(3):481-493, 22 Jan 2016
Cited by: 40 articles | PMID: 26856794 | PMCID: PMC5528974
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (25)
Grant ID: K08 CA106610-04
Grant ID: R01 CA121113-04
Grant ID: R37 CA043460-25
Grant ID: R37 CA057345
Grant ID: R01 CA057345
Grant ID: P50 CA062924-10
Grant ID: R01 CA121113-03
Grant ID: CA106610
Grant ID: CA121113
Grant ID: K08 CA106610
Grant ID: R01 CA121113
Grant ID: R37 CA043460
Grant ID: K08 CA106610-05
Grant ID: R01 CA057345-08
Grant ID: CA43460
Grant ID: P50 CA062924
Grant ID: P50 CA062924-11
Grant ID: R01 CA057345-09
Grant ID: R01 CA140599
Grant ID: P50 CA062924-12
Grant ID: R01 CA057345-10
Grant ID: R01 CA121113-05
Grant ID: R37 CA043460-24
Grant ID: CA57345
Grant ID: R37 CA043460-26
NIGMS NIH HHS (5)
Grant ID: GM078986
Grant ID: R01 GM078986-02
Grant ID: R01 GM078986-03
Grant ID: R01 GM078986
Grant ID: R01 GM078986-04
PHS HHS (1)
Grant ID: A62924




