Abstract
Free full text

Suppression of inflammation by a synthetic histone mimic
Abstract
Interaction of pathogens with cells of the immune system results in activation of inflammatory gene expression. This response, while vital for immune defence, is frequently deleterious to the host due to the exaggerated production of inflammatory proteins. The scope of inflammatory responses reflects the activation state of signalling proteins upstream of inflammatory genes as well as signal-induced assembly of nuclear chromatin complexes that support mRNA expression1–4. Recognition of post-translationally modified histones by nuclear proteins that initiate mRNA transcription and support mRNA elongation is a critical step in the regulation of gene expression5–10. Here we present a novel pharmacological approach that targets inflammatory gene expression by interfering with the recognition of acetylated histones by the Bromodomain and Extra Terminal domain (BET) family of proteins. We describe a synthetic compound (I-BET) that by “mimicking” acetylated histones disrupts chromatin complexes responsible for the expression of key inflammatory genes in activated macrophages and confers protection against LPS-induced endotoxic shock and bacteria-induced sepsis. Our findings suggest that synthetic compounds specifically targeting proteins that recognize post-translationally modified histones can serve as a new generation of immunomodulatory drugs.
BET proteins BRD2, BRD3 and BRD4 (hereafter defined as BET) govern the assembly of histone acetylation-dependent chromatin complexes that regulate inflammatory gene expression5–8. This function of BET suggests the possibility of intervention with inflammatory gene expression by disrupting chromatin complexes essential for mRNA transcription, elongation and splicing.
The diversity of binding surfaces, created by differences in sequences surrounding the bromodomain acetyl-binding pocket of BET, and other bromodomain-containing proteins, provided a foundation for selective pharmacological targeting of BET9, 11–14. Using an approach that utilises the ability of synthetic compounds to bind selectively to individual proteins in cell lysates (see Supporting Online Material) we identified compounds that interact with BET. One of these compounds, GSK525762A (Fig. 1a), henceforth referred to as I-BET, displayed the highest affinity interaction with BET (Fig. 1). The crystal structure of I-BET bound to BRD4-bromodomain 1 (BD1) showed I-BET positioned at the acetyl-lysine (AcK)–binding pocket (Figure 1b and Supplementary Fig 1a & b). Hydrogen bonding interactions essential for binding of AcK to asparagine-140 and tyrosine-97 within the bromodomain was mimicked by the triazoyl ring of I–BET (Fig 1b). The selectivity of I-BET interaction with BET was determined by the ZA hydrophobic channel and WPF shelf outside of the AcK binding pocket, where a conserved isoleucine or valine impose spatial constraints on the size of molecules that can gain access to the WPF shelf (Fig 1b and Supplementary Fig 1b & c). Indeed, an enantiomer compound of I-BET (GSK525768A) had no activity towards BET (Fig 1c, far right panel). The structural features of I-BET allow two molecules of I-BET to bind to the tandem bromodomains of BET with high affinity (Kd 50.5– 61.3 nM; Fig 1c and Supplementary Fig 1d & e). Moreover, I-BET could successfully compete with AcK within the recognition pocket of BET. Fluorescence resonance energy transfer (FRET) analysis demonstrated that I-BET displaced, with high efficacy (IC50 32.5 – 42.5 nM), a tetra-acetylated H4 peptide that had been pre-bound to tandem bromodomains of BET (Fig 1d and Supplementary Fig 1e). I-BET is highly selective as it did not interact with other bromodomain-containing proteins from each arm of the phylogeny tree (Supplementary Fig 1f) and had no activity towards a panel of 38 unrelated proteins (Supplementary Table 1).
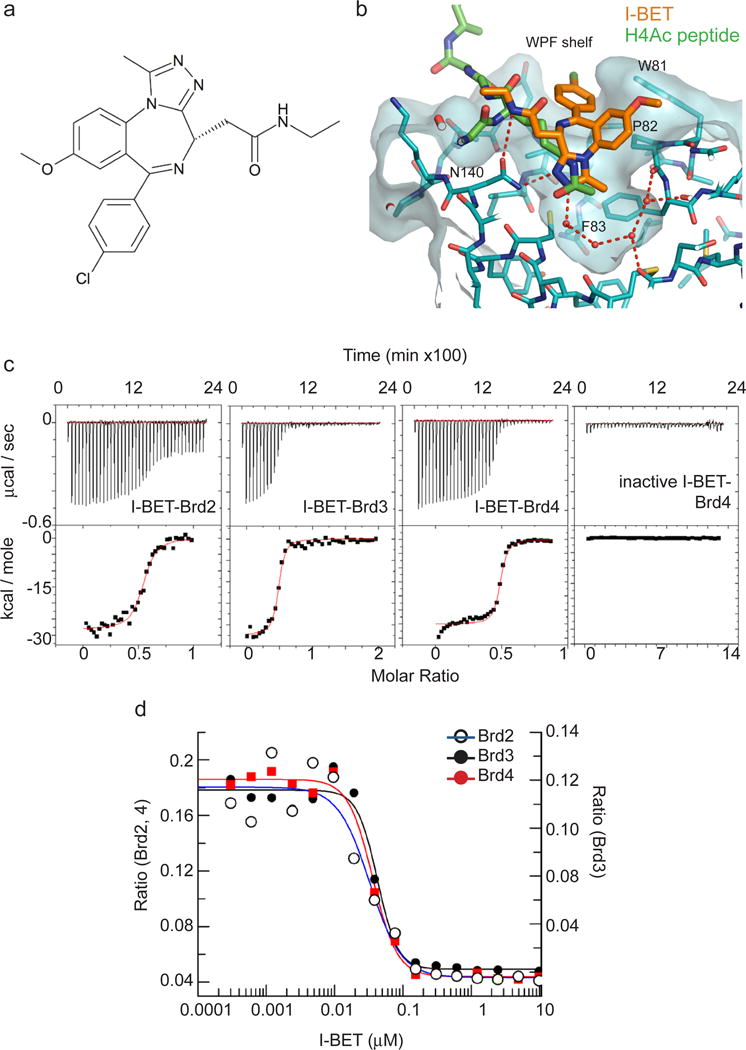
a, Chemical structure of GSK525762A (I-BET). b, Structure of I-BET (orange) bound to the acetyl-binding pocket of BRD4-BD1 overlaid with acetylated histone H4 peptide (H4Ac, green). The ‘WPF shelf’ (W81, P82, F83) as well as the asparagine N140 essential for acetylated lysine (KAc) binding are indicated. c, I-BET binds with high affinity to BET proteins as determined by isothermal titration calorimetry (ITC) of tandem bromodomain fragments of BRD2 (1–473), BRD3 (1–434), BRD4 (1–477) interaction with I-BET or BRD4 (1–477) interaction with an inactive enantiomer of I-BET (inactive I-BET). Time courses of raw injection heats (upper panel) and normalized binding enthalpies, calculated using a single site binding model (Origin software, Microcal, lower panel), are shown. d, I-BET competes with H4Ac peptide for bromodomain binding. Displacement of tetra-acetylated histone H4 peptide from bromodomains of BRD2 (blue), BRD3 (black) and BRD4 (red) by I-BET was determined by FRET analysis.
Stimulation of bone marrow-derived macrophages (BMDMs) with LPS up-regulated numerous inflammatory genes (Fig 2a). Pre-treatment of BMDMs with I-BET shortly before LPS stimulation resulted in the down-regulation of 38 and 151 of the LPS-inducible genes at 1 and 4 hours, respectively (Fig 2a & b and Supplementary Table 2). I-BET suppressed the expression of key LPS-inducible cytokines and chemokines, including il6, ifnb, il1b, il12a, cxcl9 and ccl12. The inhibitory effect of I-BET on the expression of the IL-1β processing enzyme mefv15 underscored the potential of I-BET to control the IL-1β inflammatory circuit. Furthermore, diminished expression of transcription factors rel, irf4 and irf8 point to the ability of I-BET to curtail the initial wave of inflammatory gene expression (Fig 2b, Supplementary Table 2). In the absence of LPS stimulation, treatment of BMDMs with I-BET had a marginal effect on gene transcription (Supplementary Fig 3, Supplementary Table 3) and did not impact the expression of tlrs, myd88, trif, cd14, mapks, ikks and acyloxyacyl hydrolase16 that control LPS sensing and signalling (Supplementary Fig 4). Furthermore, an unaltered pattern of LPS-induced ERK phosphorylation and IκBα degradation in I-BET treated cells excluded the impact of I-BET on gene expression through dysregulation of LPS-induced signalling (Supplementary Fig 5). I-BET also had no effect on the expression of housekeeping genes or the viability of BMDMs (Supplementary Fig 4 & 5). The impact of I-BET on LPS-inducible gene expression is highly selective. The cytokine tnf as well as chemokines ccl2-5, cxcl1/2 were not affected by I-BET (Supplementary Table 2 and Supplementary Fig 6). This specificity and anti-inflammatory potential of I-BET has been validated by the similarity between the effects of I-BET treatment and siRNA-mediated BET knockdown on inflammatory gene expression (Supplementary Fig 7). Notably, knockdown of BET suppressed the expression of tnf that was resistant to I-BET (Supplementary Fig 6 & 7). This result points to the existence of BET-recruiting mechanisms that are independent of BET interaction with acetylated histones. The existence of such a mechanism is supported by findings that show recruitment of BET to acetylated RelA or Mediator complex7, 8, 17, 18. Certain genes were up-regulated by I-BET treatment but none of these have a well-established role in inflammation (Supplementary Table 2). The up-regulation of Brd2 and histone-encoding genes (Supplementary Table 2) may reflect the existence of a positive feedback mechanism where suppression of BET leads to a compensatory increase in the expression of chromatin proteins. The activating effect of I-BET on gene expression may also reflect the ability of BET to function not only as transcriptional co-activators but also as co-repressors19.

a, Venn diagrams display the number of LPS-inducible (>2-fold, red circles) genes that were suppressed (>2 fold, green circles) or up-regulated (>2 fold, yellow circles) by I-BET (1 μM) treatment at 1 or 4 hours after LPS stimulation (left and right panels). b, Heatmap representation of expression levels of genes that were down-regulated by I-BET at 1 hour (left panel) and 4 hours (right panel) after LPS stimulation. Scale ranges from a signal value of 26 (64, green) to 214 (16384, red). Fold change values are listed. Table shows the distribution of down-regulated genes into functional categories.
The genome-wide analysis of the epigenetic states of LPS-inducible genes that were significantly suppressed or not affected by I-BET (sI-BET and naI-BET genes, respectively) provided a clue for the selective effect of I-BET on gene expression. Elevated basal levels of histone H3 and H4 acetylation (H3Ac and H4Ac) at the naI-BET gene promoters suggested that naI-BET genes were already primed or actively involved in transcription (Fig 3 and Supplementary Fig 8). Indeed, the naI-BET gene promoters were associated with higher basal levels of H3K4me3 and RNA Pol II, including the elongation competent RNA Pol II, phosphorylated at Serine 2 (RNA Pol II S2; Fig 3). The important role of the primed/active state in defining the sensitivity to I-BET was underscored by the lack of I-BET effect on expression of housekeeping genes such as gapdh, tubb5 and hprt (Supplementary Fig 4) that are characterized by high levels of H3Ac, H4Ac, H3K4me3 and RNA Pol II at their promoters5. Furthermore, an increase in overall histone acetylation levels caused by BMDM treatment with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) was able to “convert” sI-BET into naI-BET genes (Supplementary Fig 9).

a, Genome-wide epigenetic profiles of sI-BET or naI-BET genes in unstimulated or LPS-stimulated (1 hour) macrophages pre-treated with 5 μM of I-BET or a DMSO control. Analyzed epigenetic marks are indicated. Axes represent the number of reads per million mapped reads. b, Epigenetic profiles of il6 and tnf. The y axes represent the average number of tags per gene per 25 bp per 1,000,000 mapped reads. Scale values are indicated in parentheses. c, The abundance of epigenetic marks on il6 and tnf gene promoters was quantified by ChIP qPCR from four (BRD3, BRD4, Pol II and Pol II S2) or two (BRD2 and P-TEFb) independent experiments performed in triplicate. Error bars are s.e.m. of independent experiments or s.d. of representative experiments, respectively. Asterisks indicate p<0.05 as determined by an unpaired T test.
The primed and/or active transcription state of naI-BET genes prior to LPS stimulation was accomplished without recruitment of significant amounts of BET, thus reducing the likelihood of suppression of these genes by I-BET (Fig 3). Furthermore, treatment with I-BET has less impact on BET association with naI-BET as compared to sI-BET gene promoters in LPS-treated cells (Fig 3). The mechanism of this phenomenon may reflect higher LPS-induced H3Ac and H4Ac levels at naI-BET as compared to sI-BET gene promoters before and after I-BET treatment (Fig 3 and Supplementary Fig 8). Additionally, some of the naI-BET genes may recruit BET via histone acetylation independent mechanisms17.
Treatment of BMDMs with I-BET affected not only the promoter-bound BET but also the levels of H3Ac, H4K5Ac, H4K8Ac, H4K12Ac and total H4Ac on LPS-induced gene promoters (Fig 3 and Supplementary Fig 8). The mechanism of the negative impact of I-BET on histone H3 and H4 acetylation might be twofold. First, by preventing BET from binding to H3Ac/H4Ac, I-BET increases the accessibility of exposed H3Ac/H4Ac to HDACs. This model is supported by a non-enzymatic role of BRD4 in H4Ac preservation in embryonic stem (ES) cells20. It is also possible that I-BET binding to BET prevents the formation of multi-molecular complexes that contain histone acetyltransferases (HATs), other histone modifying enzymes, including lysine H3K4me3 methyltransferases, as well as the positive transcriptional elongation factor b (P-TEFb) and RNA Pol II7, 8, 18, 19. This model is consistent with diminished levels of P-TEFb, that contributes to mRNA elongation by RNA Pol II phosphorylation5, 21, 22, and reduced amounts of H3K4me3 and RNA Pol II at sI-BET genes (Fig 3). The possible direct impact of I-BET on H3Ac/H4Ac through inhibition of bromodomain-containing HATs was excluded by the inability of I-BET to suppress the activity of the most common HATs such as pCAF, p300, GCN5 and CBP (Supplementary Fig 10).
The features of sI-BET and naI-BET genes assessed by the genome-wide analysis were mirrored by the epigenetic states of selected sI-BET and naI-BET genes. Following I-BET treatment, the promoter of the siBET gene il6 displayed a marked reduction in BET recruitment and diminished levels of associated H3K4me3, P-TEFb, RNA Pol II and RNA Pol II S2 (Fig 3b & c). In contrast to il6, the naI-BET gene tnf displayed higher accumulation of BRD2, BRD3, and to a lesser extent BRD4, around the TSS following I-BET treatment (Fig 3b & c). The relatively higher BET levels at the tnf locus were associated with largely unaffected levels of P-TEFb, RNA Pol II and RNA Pol II S2 (Fig. 3b & c). In support of distinct epigenetic states between sI-BET and naI-BET genes, the sI-BET gene il1b had reduced BET accumulation at its TSS that resulted in a drop of P-TEFb, Pol II and Pol II S2 levels. In contrast, the epigenetic landscape of the naI-BET gene nfkbia displayed little change in response to I-BET (Supplementary Fig 11).
The selectivity of gene responses to I-BET correlated inversely with the timing of LPS-induced gene activation. Opposite to early stimulated (primary response) naI-BET genes, the majority of sI-BET genes, with the exception of il1b, belong to the category of secondary response genes (SRG) that become up-regulated at later points of macrophage activation (Supplementary Fig 12 a & c). Most of the sI-BET genes, as well as il1b, were characterized by low basal levels of H3Ac/H4Ac, H3K4me3, RNA Pol II, as well as low CpG content of their promoters (Fig 3, Supplementary Fig 8 & 12b). The latter feature conveys higher stability to promoter-associated nucleosomes that generates a selective barrier for transcriptional activation of secondary response genes23, 24. It is likely that suppression of BET recruitment as well as reduction in H3Ac/H4Ac and H3K4me3 by I-BET aggravates the already non-permissive transcriptional state of the sI-BET genes and reduces the probability of their expression, thus defining the selectivity of I-BET.
The suppression of key inflammatory genes by I-BET suggested a potent ability of the compound to treat inflammatory conditions in vivo. The serum titres of intravenously (i.v.) administered I-BET remain within the effective concentrations for several hours after injection (Supplementary Fig 13). Injection of I-BET in mice before the initiation of LPS- or heat-killed Salmonella Typhimurium-induced endotoxic shock was able to prevent or attenuate death of mice (Fig 4a left and middle panel). Most promisingly for therapeutic applications, a single dose of I-BET applied at 1.5 hours after LPS injection, at the time when mice started to develop symptoms of inflammatory disease, cured the mice (Fig 4a, left panel). Furthermore, in mice that suffer from polymicrobial peritonitis and sepsis caused by cecal ligation and puncture (CLP), twice-daily injections of I-BET for two days protected mice against death caused by sepsis (Fig 4a, right panel).

a, Kaplan-Meier survival curves of: LPS-treated C57BL/6 mice (5 mg/kg, i.p., n=12 per group) that were injected i.v. with a solvent control (black squares) or 30 mg/kg of I-BET 1 hour before (blue triangles) or 1.5 hours after (blue circles) LPS administration (left panel); mice injected i.v. with heat-killed Salmonella typhimurium, strain IR71 (5×109/kg, n=10 per group) (middle panel); or mice subjected to cecal ligation puncture (CLP) procedure that were administered a solvent control or 30 mg/kg of I-BET twice a day for two days (n=8 per group) (right panel) b, Serum titers of indicated cytokines were measured by ELISA (n=10 per group). Mice received a solvent control (black squares) or I-BET (blue triangles) 1 hour before LPS injection and samples were collected at 2 hours after LPS treatment. *** p<0.001, ** p<0.01, *p<0.05 as determined by unpaired T test.
The marked therapeutic effect of I-BET on endotoxic shock and sepsis occurred despite unaltered serum TNF levels (Fig 4b). As TNF is an established mediator of sepsis-associated inflammatory processes, the protective effect of I-BET on sepsis suggests the ability of I-BET to interfere not only with the expression of inflammatory proteins (Fig 4b), but also with TNF-inducible gene expression. Indeed, treatment of BMDMs with I-BET suppressed TNF-inducible key proinflammatory cytokine (il1b, il1a) and chemokine genes (ccl5, cxcl10, cxcl2/3) as well as vasoactive and lipid-related genes (pdgfb, adora2b, fabp3) that contribute to sepsis pathogenesis (Supplementary Fig 14a & b). Notably, similar to the sI-BET genes in LPS-treated BMDMs, the majority of sI-BET genes in TNF-treated cells fit into the secondary response gene category as assessed by epigenetic modifications and CpG content (Supp Fig 14c).
In summary, we show the anti-inflammatory potential of the synthetic compound I-BET that, by interfering with binding of bromodomain-containing BET proteins to acetylated histones, disrupts the formation of the chromatin complexes essential for expression of inflammatory genes. The genes susceptible to I-BET share a common pattern of chromatin modifications at their promoters as well as low promoter CpG content. Suppression of inflammation by I-BET demonstrates the potential of drugs that interfere with protein binding to post-translationally modified histones to achieve a high level of selectivity and potency by exploiting the inherited epigenetic states of genes that contribute to specific physiological and pathological processes.
Methods summary
I-BET is an optimized derivative of benzodiazepine compounds that were identified by high-throughput screening of activators of ApoA1-luciferase reporter in HepG2 cells as described in supplementary information. The chemical synthesis of I-BET is described in supplementary information. The 1.6 Å crystal structure of BRD4-BD1 with I-BET was produced by soaking apo crystals in 2mM I-BET for 4 days. Molecular replacement using 2oss.pdb gave excellent difference density at the acetylated binding site that allowed the ligand binding to be unambiguously modelled. Methods and statistics for data collection and refined coordinates are provided in supplementary information and deposited in the RCSB Protein Data Bank with PDB ID code 3P5O. Bone marrow-derived macrophages (BMDMs) were differentiated from a bone marrow cell suspension obtained from C57BL/6 mice as described in supplementary information. For microarray, qPCR and ChIP analyses, BMDMs were pre-incubated with 1 μM or 5 μM of I-BET, DMSO or an inactive I-BET compound for 30 minutes prior to LPS (100 ng/mL) or TNF (50 ng/mL) stimulation. Microarray experiments were performed using Illumina MouseRef-8 v2.0 expression BeadChip kits (GEO accession code GSE21764). qPCR was performed using SYBR Green (Roche Lightcycler 480). ChIP was performed as described25 and detailed in supplementary information. ChIP sequencing libraries were generated as described26 (GEO accession code GSE21910). For LPS-induced endotoxic shock, 5 mg/kg of LPS was injected intraperitoneally into age-matched C57BL/6 mice. Heat-killed Salmonella typhimurium (IR715; 5×109/kg) was injected intravenously. Cecal Ligation Puncture (CLP) was performed as described27. For in vivo experiments I-BET or a solvent control (20% beta-cyclodextrin, 2% DMSO in 0.9% saline) were given via retro-orbital or tail vein injection (CLP) at a dose of 30 mg/kg.
Acknowledgments
We would like to acknowledge Rachel Grimley and Champa Patel for supplying FRET data and Robert Woodward, Chris Delves, Emma Jones and Paul Holmes for protein production. Jason Witherington, Nick Smithers, Stuart Baddeley, Jon Seal and Leanne Cutler provided compound selectivity and pharmacokinetics data. Gael Krysa, Olivier Mirguet and Romain Gosmini contributed to the discovery, development and characterization of the compound. We thank Robert Anthony and Scott McCleary for assistance with animal models, Ron Gejman for bioinformatics analysis of gene expression kinetics and Angela Santana and Trevor Chapman for technical assistance. We would like to thank Carl Nathan, Ruslan Medzhitov, Sasha Rudensky and Steven Smale for helpful discussions and Srihari Sampath for his contribution to the concept of “histone mimicry”. R.C. is supported by an NIH KL2 Career Development Award and I.M. is supported by the American Italian Cancer Foundation. K.L.J is supported by the National Health and Medical Research Council of Australia and is currently a Rockefeller University Women in Science Fellow.
E.N., S.B., C-W.C, P.W., H.C., J.W., J.K., J.L., R.K.P. and K.L. are employees of GlaxoSmithKline. Research support (excluding salaries to the members of The Rockefeller University), but including protein analysis and compound synthesizing equipment, supplies, and other expense were provided by GlaxoSmithKline.
Footnotes
Author contributions:
E.N. identified, characterized and optimized the compound for in vivo experiments; K.L.J., U.S. and S.B. contributed equally to design, execution and analysis of in vitro and in vivo experiments. C-W.C. performed crystallography, ITC, SPR and thermal shift assays. S.D. performed bioinformatics analysis of ChIP sequencing data; R.C. performed quantitative analysis of epigenetic states of the LPS-inducible genes; I.M. optimized BRD2 and BRD3 profiling of the LPS inducible genes; P.W. performed bioinformatics analysis of gene expression in LPS-stimulated macrophages. H.C., J.W. and J.K. discovered, characterised and optimised the compound for in vivo experiments. J.L., R.K.P. and K.L. contributed to the initiation and development of the studies on pharmacological targeting of proteins that recognize post-translationally modified histones. A.T. conceived and supervised this study, and wrote the final manuscript.Atomic coordinates and structure factors for the BRD4-I-BET crystal structure have been deposited in the RCSB Protein Data Bank under PDB ID code 3P5O. Microarray data has been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE21764. ChIP sequencing data has been deposited under accession number GSE21910. The authors declare competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature09589
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5415086?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101877671
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature09589
Article citations
The BRD4 Inhibitor I-BET-762 Reduces HO-1 Expression in Macrophages and the Pancreas of Mice.
Int J Mol Sci, 25(18):9985, 16 Sep 2024
Cited by: 0 articles | PMID: 39337472 | PMCID: PMC11432103
Cbx4 SUMOylates BRD4 to regulate the expression of inflammatory cytokines in post-traumatic osteoarthritis.
Exp Mol Med, 56(10):2184-2201, 01 Oct 2024
Cited by: 0 articles | PMID: 39349832 | PMCID: PMC11541578
BRD9 promotes the progression of gallbladder cancer via CST1 upregulation and interaction with FOXP1 through the PI3K/AKT pathway and represents a therapeutic target.
Gene Ther, 21 Sep 2024
Cited by: 0 articles | PMID: 39306629
Identification of Novel Bromodomain-Containing Protein 4 (BRD4) Binders through 3D Pharmacophore-Based Repositioning Screening Campaign.
Molecules, 29(17):4025, 26 Aug 2024
Cited by: 0 articles | PMID: 39274873 | PMCID: PMC11397543
RBM25 is required to restrain inflammation via ACLY RNA splicing-dependent metabolism rewiring.
Cell Mol Immunol, 21(11):1231-1250, 09 Sep 2024
Cited by: 1 article | PMID: 39251781
Go to all (919) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (2 citations) GEO - GSE21764
- (2 citations) GEO - GSE21910
Protein structures in PDBe
-
(2 citations)
PDBe - 3P5OView structure
iPPI-DB : An interactive database of protein-protein interactions modulators
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
High-Throughput Screening for CEBPD-Modulating Compounds in THP-1-Derived Reporter Macrophages Identifies Anti-Inflammatory HDAC and BET Inhibitors.
Int J Mol Sci, 22(6):3022, 16 Mar 2021
Cited by: 7 articles | PMID: 33809617 | PMCID: PMC8002291
Nodakenin suppresses lipopolysaccharide-induced inflammatory responses in macrophage cells by inhibiting tumor necrosis factor receptor-associated factor 6 and nuclear factor-κB pathways and protects mice from lethal endotoxin shock.
J Pharmacol Exp Ther, 342(3):654-664, 25 May 2012
Cited by: 37 articles | PMID: 22637723
Anti-inflammatory effects of benzenediamine derivate FC-98 on sepsis injury in mice via suppression of JNK, NF-κB and IRF3 signaling pathways.
Mol Immunol, 67(2 pt b):183-192, 29 May 2015
Cited by: 7 articles | PMID: 26032013
The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins.
Int J Mol Sci, 17(11):E1849, 07 Nov 2016
Cited by: 142 articles | PMID: 27827996 | PMCID: PMC5133849
Review Free full text in Europe PMC




