Abstract
Free full text
The columnar and laminar organization of inhibitory connections to neocortical excitatory cells
Abstract
The cytoarchitectonic similarities of different neocortical regions have given rise to the idea of “canonical” connectivity between excitatory neurons of different layers within a column. It is unclear whether similarly general organizational principles also exist for inhibitory neocortical circuits. Here, we delineate and compare local inhibitory-to-excitatory wiring patterns in all principal layers of primary motor (M1), somatosensory (S1), and visual cortex (V1), using genetically targeted photostimulation in a mouse knock-in line that conditionally expresses channelrhodopsin-2 in GABAergic neurons. Inhibitory inputs to excitatory neurons derive largely from the same cortical layer within a three-column diameter. However, subsets of pyramidal cells in layers 2/3 and 5B receive extensive translaminar inhibition. These neurons are prominent in V1, where they might correspond to complex cells, less numerous in barrel cortex, and absent in M1. Although inhibitory connection patterns are stereotypical, the abundance of individual motifs varies between regions and cells, potentially reflecting functional specializations.
The anatomical fine structure of the neocortex is remarkably uniform, suggesting extensive replication of a limited number of circuit motifs1. In support of this view, the excitatory connections of different neocortical areas in different species appear to conform, with minor variations2-5, to the “canonical” laminar organization first described in cat visual cortex6-9: Thalamic afferents arrive in layer 4 (L4), whose neurons project to L2 and L3. Axonal projections of pyramidal cells in these layers terminate in L5 and some of those from L5 in L6.
It has been difficult to determine whether similarly general principles also hold for the organization of inhibitory neocortical circuits10,11. Systematic studies of inhibitory connectivity have been hampered by the relative sparseness of inhibitory neurons and a bewildering diversity of cell types10-14. While the rules governing the interneuron type-specific positioning of inhibitory terminals on post-synaptic target cells are increasingly well understood11,15,16, the connection diagrams between inhibitory and excitatory neurons in different columns and layers, and the extent to which these diagrams generalize across areas, remain largely unknown. Other than a general belief that inhibition is local and largely intralaminar10,14,17-25, only a few wiring principles12,18,21,22,26 of inhibitory neocortical circuits have been formulated, such as disynaptic inhibition of neighboring L5 pyramidal cells by Martinotti cells22,26 and ascending inhibition18,21 from L5 and L4 to L2/3.
The ability to activate inhibitory interneurons remotely with light, using optically gated ion channels that are expressed in genetically defined cell types27-30, removes the difficulties of purely electrophysiological or anatomical searches for synaptic connections. Scanning a stimulating beam across neural tissue31 will generate light-evoked inhibitory currents in postsynaptic partners whenever the focal spot activates a presynaptic interneuron. Depending on which promoter element drives the expression of the light-controlled actuator27-30, and depending on where the recording electrode is placed, the connections of different subclasses of interneurons with different postsynaptic targets can be resolved. Here, we develop this approach to delineate and compare the columnar and laminar origins of inhibitory inputs, without differentiating the interneuron subclasses that emit them, to excitatory neurons in all principal layers of primary motor (M1), somatosensory (S1), and visual cortex (V1) of the mouse.
Results
Conditional expression of ChR2 from a genomic locus
To generate a Cre-responsive actuator27-30,32 allele, the GT(ROSA)26Sor (R26) locus was targeted with a transgene driven by the CMV early enhancer/chicken β-actin (CAG) promoter. The transgene was designed to express a fusion protein of channelrhodopsin-2 (Genbank accession number AF461397) and EGFP (ChR2-EGFP) after Cre-mediated excision of a transcriptional STOP cassette (Fig. 1a). ChR2-EGFP expression in the majority of GABAergic neurons was achieved by crossing this line with a strain carrying a tamoxifen-inducible Cre recombinase (Cre-ERT2) cassette, preceded by an internal ribosome entry site, in the 3′-untranslated region of the Gad2 gene (Fig. 1b). The bicistronic transcript of the targeted Gad2-locus is translated into the 65 kDa isoform of glutamic acid decarboxylase (Gad65) and Cre-ERT2 protein. Because recombination of the R26::ChR2-EGFP locus requires tamoxifen-mediated induction of Cre activity, ChR2-EGFP expression can be timed to the appropriate developmental stage.
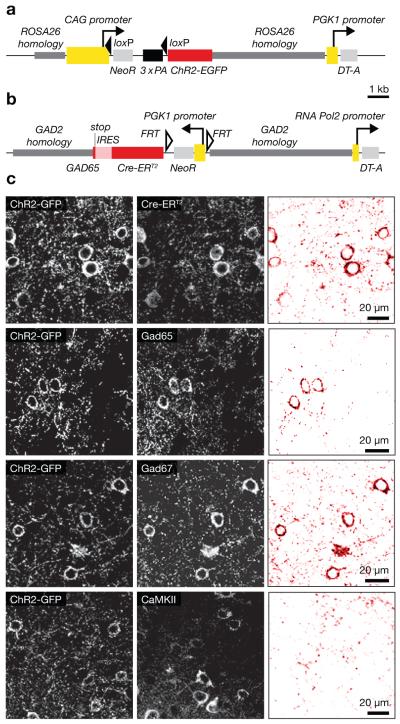
(a, b) Targeting constructs. Homology sequences are indicated in dark grey, promoters in yellow, open reading frames in red, and selection markers in light grey. (a) Construct used to generate the R26::ChR2-EGFP allele. Cre-mediated excision of a triple-polyA transcriptional STOP cassette (3x PA, black) flanked by loxP sites enables ChR2-EGFP expression from the CAG promoter. (b) Construct used to generate the Gad2::Cre-ERT2 allele. An internal ribosome entry sequence (IRES, light red) separates the Gad65 and Cre-ERT2 reading frames. (c) Gad2::CreT2 R26::ChR2-EGFP mice after tamoxifen induction express ChR2-GFP in Cre-positive cells (top), which comprise both Gad65- and Gad67-positive interneurons (middle), but not CamKIIα–positive pyramidal cells (bottom). See Supplementary Fig. 1 for an analysis of interneuron subtypes and Supplementary Table 1 for statistics. The left and center columns show raw confocal images; the right column displays the corresponding colocalization maps, which were produced by multiplying the two fluorescence channels on a pixel-by-pixel basis and normalizing the resulting product image to 8 bits.
Following Cre induction between the fourth and sixth postnatal weeks, ChR2-EGFP was expressed in all major subclasses of GABAergic interneurons11,13,33 (Fig. 1c, Supplementary Fig. 1, Supplementary Table 1) but undetectable in CaMKII-positive pyramidal cells (Fig. 1c).
Genetically targeted photostimulation of interneurons
Focal 473-nm illumination of acute neocortical slices from mice carrying homozygous recombined R26::ChR2-EGFP loci in Gad2-positive cells elicited action potentials in interneurons but not in pyramidal cells (Fig. 2a,b). The comparatively low expression levels of ChR2 from the genomic cassette required longer stimulating light pulses than those used to activate virally transduced or transfected neurons34,35; a 20-ms pulse at 2 mW represented a favorable trade-off between reliable action potential iniation (100 and 91.1 % in cell-attached and whole-cell mode, respectively; n=55 cells) and a ~1:1 ratio of spikes per optical pulse (single action potentials in 8/10 cells in cell-attached mode; occasional doublets in 2/10).
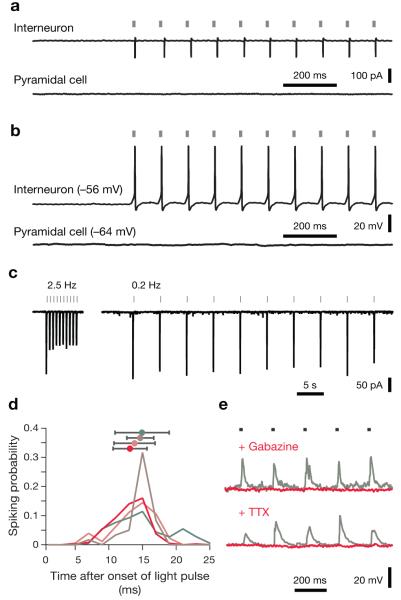
(a, b) Responses to photostimulation in cell-attached (a) and whole-cell current-clamp recordings (b); native resting potentials are indicated in parentheses. Interneurons follow trains of 20-ms optical pulses at 10 Hz with action potentials; pyramidal neurons are unresponsive to light. (c) ChR2-mediated photocurrents desensitize during repeated optical stimulation at frequencies >1 Hz (left) but remain stable at stimulation frequencies ≤0.2 Hz (right). (d) Spiking probabilities as a function of time after stimulus onset were estimated by analyzing 29–198 light-evoked action potentials per cell (n=4 interneurons in cell-attached recordings; colored traces). Spike times are defined as the times at which the upstroke of an action potential reaches half-maximal amplitude. Average spike latencies (± s.d.) are indicated in matching colors. (e) Wide-field optical stimulation at 5 Hz (grey bars) evokes IPSCs in pyramidal cells voltage-clamped at 0 mV (grey traces). IPSCs are abolished after bath application of 10 μM gabazine (red trace, top) or 0.5 μM TTX (red trace, bottom).
Interneurons followed optical pulse trains with maximal frequencies of 0.5–40 Hz in cell-attached (n=10; Fig. 2a) or whole-cell recordings (n=41; Fig. 2b). Individual pulses caused peak photocurrents of −188±106 pA at holding potentials of −70 mV and −145±82 pA at −60 mV (means±s.d.; n=4 cells; Fig. 2c), which evoked action potentials with latencies of 15.8±3.7 ms from the onset of illumination (means±s.d., Fig. 2d). Interneurons in areas M1, S1, and V1 were equally responsive to light, and no significant interregional differences in our ability to control fast- or non-fast-spiking cells were detected (Supplementary Fig. 2).
Driving inhibitory neurons periodically by wide-field illumination caused stimulus-locked IPSCs in pyramidal cells (Fig. 2e). Bath application of the GABAA receptor antagonist SR95531 (gabazine, 10 μM) blocked these IPSCs (Fig. 2e), as did application of the sodium channel blocker tetrodotoxin (TTX, 0.5 μM, Fig. 2e). In contrast to the direct activation of synaptic release commonly seen in virally transduced or transfected neurons34,35, which express ChR2 at sufficiently high levels in axons and synaptic terminals to make vesicle release resistant to TTX, only perisomatically triggered action potentials evoked transmission in our hands. Consistent with this interpretation, axonic stimulation of Purkinje cells in cerebellar slices failed to produce back-propagating action potentials or enhanced somatic depolarizations (Supplementary Fig. 3).
Illuminating the dendritic arbors of neocortical interneurons (Fig. 3a) also generated only insignificant photocurrents: Depolarization amplitudes and spiking probabilities decayed in a roughly radial-symmetric manner as a function of somatic distance, without stimulation hotspots at underlying dendrites (Fig. 3a,b). Our approach thus offers two hitherto unrealized advantages for mapping connectivity: First, it distinguishes synaptic inputs originating locally (that is, from neurons whose somata lie within the illumination cone) from axonal or dendritic projections into the illuminated volume. And second, the optical stimulus selectively addresses inhibitory neurons. Photolysis of caged glutamate, in contrast, activates both inhibitory24,25 and excitatory17,20,25 neurons and is therefore prone to mapping polysynaptic connections, because interneurons at some distance from the stimulation site may be recruited indirectly.
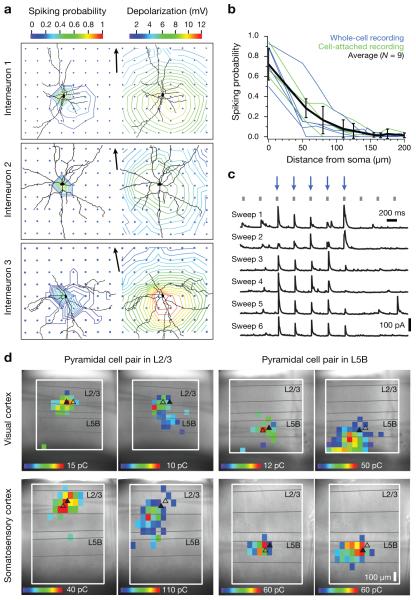
(a) Contour plots depict spiking probabilities (left) and depolarization amplitudes (right) of three interneurons as functions of stimulus location. Blue dots indicate stimulation points; arrowheaded scale bars of 100 μm point to the pial surface. Perisomatic illumination reliably elicits action potentials (left). Positioning the focus of the stimulating beam near dendritic branches does not cause higher spiking probabilities or larger depolarizations than illumination of dendrite-free neuropil equidistant from the soma (right). (b) Spiking probabilities of 9 interneurons as functions of the distance of the stimulation spot from the soma. Cells were recorded in the cell-attached (n=4 cells, green traces) or whole-cell configuration (n=5 cells, blue traces). (c) Sequential illumination of 10 different locations at 2.5 Hz (20 ms, 2 mW, grey tick marks). Illumination of sites marked by blue arrows gives rise to reproducible IPSCs in the recorded pyramidal cell. (d) Maps of inhibitory inputs to pyramidal neurons in V1 (top row) or S1 (bottom row), located in L2/3 (left column) or L5B (right column) at comparable depths (± 7 μm) from the surface of the slice. Two neurons were recorded simultaneously to test whether cells within the same local network could exhibit different connectivity patterns. Cell positions are indicated by triangles; a filled triangle denotes the postsynaptic target for each map. Color on a heat scale symbolizes the average amount of charge flowing during 100 ms following the onset of the IPSC, at a holding voltage of 0 mV.
However, it is formally possible also for an inhibitory input to propagate polysynaptically if the targets of inhibition generate rebound spikes. We tested this scenario and found no evidence in its favor (Supplementary Fig. 4). Pyramidal cells and fast-spiking as well as non-fast-spiking interneurons were hyperpolarized to increasingly negative holding potentials. Rebound spikes appeared in 4/19 pyramidal cells and 3/13 fast-spiking interneurons, but only when these neurons were released from holding potentials <−150 mV (Supplementary Fig. 4a,b). Non-fast-spiking interneurons emitted rebound spikes more frequently (11/24 cells) and readily (Supplementary Fig. 4c), but the spike latencies from the offset of shallow hyperpolarizations were so large (>100 ms) that any IPSCs caused by these spikes would have been excluded in our analysis because the temporal contingency between light pulse and postsynaptic event was broken. To generate short-latency (≤50 ms) rebound spikes, non-fast-spiking interneurons needed to be hyperpolarized below –117 mV (Supplementary Fig. 4c). GABAergic IPSCs could achieve this degree of hyperpolarization only at chloride reversal potentials of ≤−120 mV and, therefore, unphysiologically low36 intracellular chloride concentrations. Our IPSC images thus depict the origins of monosynaptic inhibitory connections.
Optogenetic mapping of inhibitory input distributions
To analyze the laminar and columnar organization of local inhibitory connections, one or two excitatory neurons were voltage-clamped at 0 mV (to maximize chloride currents through GABAA receptors and minimize spontaneous EPSC amplitudes), while a focused laser beam rastered a grid of locations spaced at 55-to-65-μm intervals. The grid spacing reflects the lateral resolution of photostimulation, limited by light scattering in the slice (Fig. 3a,b).
Stimulus-locked IPSCs could be evoked by focal illumination of certain locations but not others, revealing the somatic positions of inhibitory interneurons presynaptic to the recorded excitatory cell(s) (Fig. 3c,d). The distributions of input sources were reproducible for the same postsynaptic neuron during repeated sweeps of the stimulation grid (Fig. 3c) but differed for two simultaneously recorded cells located nearby in the same slice (Fig. 3d).
A cautionary remark, however, is appropriate. Holding the excitatory targets of inhibition at 0 mV could lead to depolarization-induced suppression of inhibition (DSI), which might mask inhibitory inputs37,38. DSI is mediated by retrograde endocannabinoid signals38 that communicate the depolarization of postsynaptic neurons to inhibitory presynaptic terminals, which downregulate GABA release upon activation of cannabinoid receptor-1 (CB-1). To test for a possible effect of DSI, we compared the inhibitory input maps of the same six pyramidal cells when these cells were voltage-clamped at −70 and 0 mV (Supplementary Fig. 5). No differences in the number and distribution of inhibitory inputs were found (P=0.821). In a second set of control experiments, we compared the inhibitory input maps of 17 cells in the presence and absence of the CB-1 antagonist AM251 at 2 μM (Supplementary Fig. 5). Again, no differences in input number or map structure were detected (P=0.379).
Optical raster stimulation allowed us to identify sources of inhibitory inputs to excitatory cells in M1, S1, and V1 and assemble input maps for 6–22 excitatory neurons per layer and cortical area (Supplementary Table 2), yielding a data set of 3,823 inhibitory-to-excitatory connections. The identity and gross integrity of all recorded neurons were confirmed post hoc by visualizing spines on the neurobiotin-filled arbors (Supplementary Fig. 6).
Columnar center-surround structure of inhibitory circuits
Sources of inhibition were confined within horizontal domains spanning at most 550 μm, or 3 somatosensory barrels (179±28 μm per barrel; n=17, Fig. 4), suggesting that inhibitory microcircuits interconnect adjacent columns (Supplementary Table 3). Because the columnar structure is visible in S1, this interpretation could be tested directly by recording simultaneously from pairs of L2/3 pyramidal neurons in the same (n=5 pairs) or adjacent (n=6 pairs) barrel-related columns. While the input maps of L2/3 neurons in the same column overlapped to a large degree (Fig. 4a), those of cells in adjacent columns were shifted by about one barrel diameter (Fig. 4b). Cross-correlation analysis (see Methods) confirmed that the input maps of two neurons in the same barrel-related column were displaced by slightly less than the average distance between the two cells (0.32±0.28 vs. 0.39±0.11 barrel widths, means±s.d.), whereas the maps of cells in different columns were shifted by an average of 0.94±0.09 barrel widths (P=0.002). As would be expected if map displacements occurred in discrete, column-sized steps, they were better described by a step function with a level change at the barrel septum than a linear function of intercell distance (Supplementary Fig. 7). Inhibitory connections thus appear to follow a pericolumnar “center-surround” arrangement, which provides an anatomical substrate by which activity in one column can suppress that of its immediate neighbors1,39.
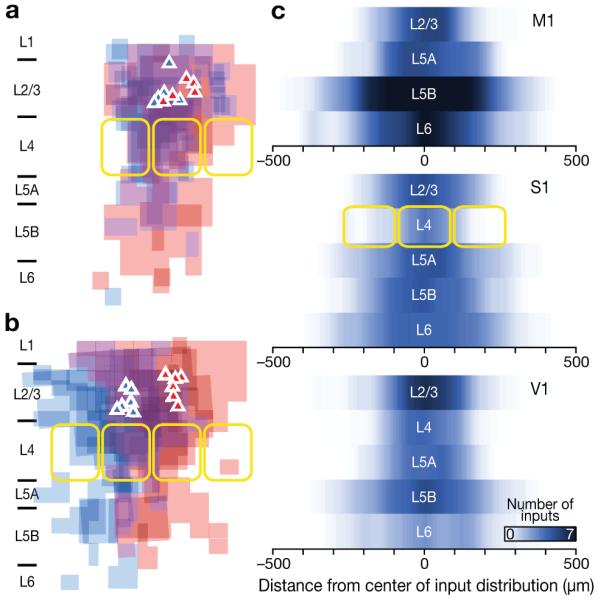
(a, b) Overlay maps of inhibitory inputs to pairs of simultaneously recorded pyramidal neurons in layer 2/3 of S1. The maps depict the locations of inhibitory inputs but not their strength and have been scaled to the size of a standard somatosensory barrel (yellow outlines). Cell positions are marked by triangles. Data from the left cell in each pair are coded in blue, data from the right cell in red. (a) Pairs of pyramidal neurons in the same barrel-related column. (b) Pairs of pyramidal neurons in adjacent barrel-related columns. (c) Horizontal profiles of input distributions show the interpolated number of inhibitory inputs as a function of horizontal distance from the center of the input distribution for each layer, ignoring the laminar (vertical) location of these inputs. Horizontal distances were scaled to the size of a standard somatosensory barrel (yellow outlines) and center-aligned; the number of inhibitory inputs to an excitatory neuron in a given layer at a given distance from the map center was then averaged. The intensity of blue color symbolizes the density of input sources. See Methods and Supplementary Table 3 for statistical detail.
The horizontal reach of inhibitory connections varied from layer to layer, giving rise to hourglass-shaped input profiles with notable constrictions in L4 of sensory cortices (Fig. 4c, Supplementary Table 3). This observation is consistent with the idea that lateral inhibition constrains the flow of information to columnar units in supra- and infragranular layers39, whereas columns in L4 are largely defined by the parcellation of thalamocortical projections.
Area-specific laminar organization of inhibitory circuits
In contrast to a horizontal structure that appeared similar across cortical areas (Fig. 4c, Supplementary Table 3), the vertical organization of inhibitory connections showed clear area-specific variations. With the exception of neurons in L6, excitatory cells in different areas displayed distinct laminar source distributions of inhibitory inputs (Fig. 5, Supplementary Tables 4–7). While the dominant source of inhibition was usually intralaminar—that is, originating from interneurons residing in the same layer as the soma of the excitatory neuron19,24—striking exceptions to this rule were found, particularly in V1. Here, pyramidal cells in L5B were strongly inhibited by interneurons in L6 (Fig. 5d, Supplementary Tables 4–6) and cells in L2/3 and L4 by interneurons in L5, especially L5B (Fig. 5a,b; Supplementary Tables 4–6). Quantitatively, the total inhibitory charge flow (Fig. 5) as well as the normalized numbers of inhibitory inputs (Supplementary Table 5) from the dominant translaminar source (L6 for L5B, and L5B for L2/3 and L4) were at least twice as large in V1 as in M1 and S1 (see also Fig. 6c).
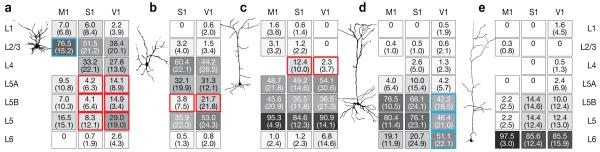
(a–e) Average strength of inhibitory input from the indicated source layers (rows) to excitatory neurons located in L2/3 (a), L4 (b), L5A (c), L5B (d) and L6 (e). An example of a neurobiotin-filled excitatory neuron, recovered after recording, is shown to the left of each panel. The figure summarizes data from 30 neurons in M1, 54 neurons in S1, and 53 neurons in V1. The strength of a connection is expressed as the average percentage of inhibitory charge flow arising from identified inputs in a layer. L5 represents the sum of L5A and L5B. Values are represented numerically (s.d. in parentheses) and by the intensity of grey shading. Colored boxes indicate significant differences (P < 0.05), either between two cortical areas (red) or between one area and the other two (blue), as determined by one-way ANOVA and Tukey-HSD post-hoc tests (Supplementary Tables 2 and 4).

(a) Discriminant analysis of the laminar source distributions of inhibitory inputs to excitatory neurons in different target layers of M1, S1, and V1 (Supplementary Table 8). Neurons are represented as points in the coordinate system spanned by the discriminant functions Y1 and Y2. Borders between colored areas indicate decision boundaries for assigning neurons to M1 (blue), S1 (yellow), and V1 (red). Data points whose fill color matches the background color are classified correctly. Black squares indicate the centroid positions for all cells in each cortical area. (b) Abundance of interneuron subtypes expressing parvalbumin (P), somatostatin (S), and VIP (V) in in the respective cortical layers and areas; pie chart diameters represent overall interneuron densities (Supplementary Table 9). (c) Percentages and absolute numbers of inputs, charge flow, and failure rates (means+s.d.) of the four translaminar motifs exhibiting area-specific differences. Colored bars symbolize data for M1 (blue), S1 (yellow), and V1 (red). Asterisks denote statistically significant differences between cortical areas (P < 0.05), as determined by ANOVA and Tukey-HSD post-hoc tests (Supplementary Tables 5, 6, 7 and 10).
Evidence of putative feedforward inhibition between layers, or of descending inhibition from superficial to deeper laminae, was scant, with two notable exceptions. In S1 and V1, an inhibitory L4-to-L2/3 connection18 ran parallel to the flow of excitatory signals8 from spiny stellate neurons of L4 (Fig. 5a). And in S1, a 5-fold larger amount of L4-derived inhibition than in V1 entered L5A, via marginally faster synaptic connections (Fig. 5c; 20-80 % rise time=5.8±1.1 ms in S1 vs. 7.6±1.5 ms in V1, P=0.06).
Area-specific differences in vertical inhibitory connectivity (Figs. (Figs.55 and and6a)6a) were so characteristic that discriminant analysis could assign >70 % of excitatory neurons of all layers correctly to their cortical areas of origin, based on the laminar distribution of their inhibitory inputs alone (Fig. 6a, Supplementary Table 8). This was true irrespective of whether inhibition was quantified by measuring inhibitory charge flow or the number of synaptic inputs from a layer (Supplementary Table 8). The classification was most accurate in M1 (96 %) and V1 (75 %) but less accurate in S1 (63 %; averages across layers 2/3 to 5B).
Multiple factors could underlie these regional differences in vertical inhibitory connectivity, singly or in combination. The source layers of translaminar inhibition (L4, L5B, and L6) could be populated with interneurons, or particular subtypes of interneurons, at different densities. Alternatively, the translaminar connection probabilities of interneurons and excitatory cells could vary between areas. Finally, regional variations could be due to differences in the properties of these connections, such as their release probabilities, synaptic conductances, and the distribution of synapses along the somatodendritic axis of pyramidal cells. To distinguish between these possibilities, we began by comparing the abundances of the three principal interneuron subtypes defined cytochemically33 in L4, L5B, and L6 of V1, S1, and M1. No consistent covariation between the density of inhibitory neurons or the subtypes expressing parvalbumin, somatostatin, or VIP and the strength of inhibition originating in a layer was detected (Fig. 6b, Supplementary Table 9). We next compared the absolute and normalized numbers of identified translaminar connections (Fig. 6c, Supplementary Tables 5 and 6) as well as three of their functional properties (Fig. 6c, Supplementary Tables 7, 10, and 11): the frequency of transmission failures (an index of release probability, given the uniform light sensitivity of interneurons across areas, as documented in Supplementary Fig. 2); the inhibitory charge flow per IPSC (which is proportional to synaptic conductance); and the 20-80% rise time of IPSCs (which can reflect the electrotonic distance of a synapse from the soma, and is therefore a proxy of anatomical distance). Regional differences in inhibitory connectivity were to a large extent caused by variable connection probabilities; in one instance (the L4-to-L5A motif), there was an additional increase in inhibitory charge flow per IPSC (Fig. 6c). The relative prominence of translaminar inhibition from L6 to L5B in V1, in contrast, was not due to an absolute increase in the number or strength of L6-to-L5B connections but rather a reduction of inputs from other laminar sources (Supplementary Table 5). Differences in transmission failures (Fig. 6c) or the somatodendritic locations of inhibitory synapses, as inferred from IPSC rise times measured in individual trials (Supplementary Table 11), played no discernible roles.
Cell-specific laminar organization of inhibitory circuits
The characteristic differences in average inhibitory connectivity among cortical areas (Supplementary Fig. 8) were not caused by region-specific wiring patterns of individual cells. Rather, an inspection of individual input maps revealed striking cell-to-cell variation of inhibitory connectivity within the same layer and area. For example, some excitatory neurons in L2/3 and L5B of V1 and S1 received strong translaminar inhibition from L5 and L6, respectively, whereas others did not. Cells with dramatically different inhibitory input distributions could be found immediately next to each other, suggesting that cell-to-cell variability is not a slicing artefact (Fig. 3d).
Hierarchical cluster analysis indicated the presence of two distinct subpopulations of pyramidal cells in L2/3 and L5B of V1 and S1 (Fig. 7). Members of one of these populations received the overwhelming majority of inhibitory inputs from their respective home layers; the inhibitory input distribution of these cells therefore resembled that of pyramidal neurons in L2/3 and L5B of M1 (Fig. 5a,d). The other subpopulation appeared specific to sensory cortex; it was found only in V1 and—albeit in smaller numbers—S1 (Fig. 7a,b). This group of neurons is defined by prominent translaminar inhibition, which serves as the dominant source of GABAergic input (Fig. 7a,b). Neurons receiving translaminar inhibition accounted for 26.6 and 4.5 % of cells in L2/3 of V1 and S1, respectively (Fig. 7a), and for 66.6 and 37.5 %, respectively, of neurons in L5B (Fig. 7b). Differences in average inhibitory connectivity among cortical areas thus arise, at least in part, because two subpopulations of excitatory neurons with distinct inhibitory input patterns are present in different proportions.
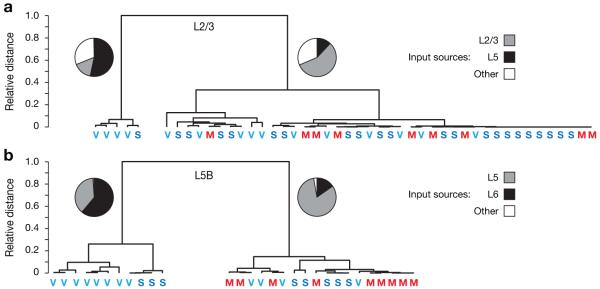
(a, b) Hierarchical clustering of pyramidal cells in layers 2/3 (a) and 5B (b) of M1, S1, and V1. Classification variables were the strengths of intra- and translaminar inhibitory inputs, quantified as normalized inhibitory charge flow. Neurons in both layers fall into two well-separated clusters: a minor population of neurons receiving strong translaminar inhibition (left cluster), and a major population of predominantly home-layer inhibited neurons (right cluster). Bootstrap estimates of cluster distances at the first bifurcation level are 0.64±0.11 and 0.71±0.09 (means±s.d.) for pyramidal cells in layers 2/3 (a) and 5B (b), respectively. Pie charts indicate the average strengths of inhibitory input from the dominant translaminar layer in black (layer 6 for neurons in layer 5B, and layer 5 for neurons in layers 2/3), the home layer in grey, and other layers in white. Colored letters denote the cortical area from which each observation is derived. Note the high frequency of V1 neurons and the absence of M1 neurons in the clusters receiving dominant translaminar inhibition.
Discussion
We have drawn maps of inhibitory-to-excitatory synaptic connections in three neocortical areas (Supplementary Fig. 8). The topographic features depicted in our maps are the somatic locations of presynaptic inhibitory neurons and postsynaptic excitatory cells and the strengths of the connections between them. Our maps do not portray the positions of the synaptic contacts themselves, nor do they differentiate among the large number of interneuron subtypes. We expect these details to be filled in with relative ease as more and better genetic addressing systems29,30 for defined subsets of interneurons become available, simply by repeated application of the optical mapping technique developed here.
Our technique combines the advantages of optogenetic control27-30 and glutamate uncaging3,17,24,25,31: genetic resolution of cell types, and perisomatic restriction of light sensitivity. These qualities are a direct consequence of expressing ChR2 conditionally from an identified genomic locus. Actuator expression in each cell type is perforce comprehensive and uniform, and modest expression levels prevent the accumulation of the actuator in axons, dendrites, and synaptic boutons at densities sufficient to initiate local electrical or release events34,35. Instead, only light pulses directed at the somata of interneurons generate large enough inward currents to elicit action potentials and trigger synaptic release (Fig. 3).
The approach is effective for all broadly defined classes of interneurons but will require fine-tuning of actuator membrane densities to match the biophysical properties of other types of cells. We already know that ChR2 expression from the same homozygous R26::ChR2-EGFP loci, but after Cre-mediated recombination in CaMKIIα-positive excitatory neurons, is insufficient for driving spikes in neocortical pyramidal cells (not shown). This failure is in all likelihood due to an unfavorable constellation of low ChR2 expression levels, tiny ChR2 conductance, low input resistance, and large membrane area in these cells. Parameters for possible adjustment include the strength of the promoter driving the expression of the actuator, the transcriptional landscape of the targeted locus, the number of actuator expression units in the cassette, and the conductance of the actuator itself.
Are inhibitory neocortical microcircuits canonical?
We return to this question, which served as our point of departure, with an ambiguous answer. If the term “canonical” is interpreted in the strictest possible sense, to imply that precisely the same laminar and tangential organization of inhibitory connections is repeated across the entire neocortical mantle, our discovery of characteristic area-specific variations in inhibitory connectivity clearly refutes the notion of canonical wiring. Still, we find that the same distinctly recognizable—perhaps even “canonical”—motifs of inhibitory-to-excitatory connectivity recur in most of the cortical areas we have examined: It is the frequency with which these structural elements are present, not their configuration, that varies between regions.
The most common circuit motif is lateral, intralaminar inhibition of excitatory neurons by interneurons located in either the same column or an immediate neighbor. The abundance of this motif in V1, S1, and M1 supports the traditional view that inhibition is largely local, intralaminar, and uniform across areas14,17-19,21-26. However, there is also evidence that interneuron axons ramify extensively beyond laminar borders16,40 and form synaptic contacts across layers18,21,25. For example, although the ascending axons of somatostatin-positive, non-fastspiking Martinotti cells of L5 are widely thought to connect preferentially with the apical dendrites of L5 pyramidal cells22,26,41, there are now at least three electrophysiologically documented cases of L5 Martinotti cells targeting L2/3 pyramidal neurons21.
The high throughput of our optogenetic mapping technique has facilitated the detection of these rarer connectivity patterns and allowed us to supplant anecdotal evidence from pairwise recordings with quantitative estimates of motif frequencies in a large data set encompassing three cortical areas. Nine translaminar motifs were found, four of which varied between areas (Supplementary Fig. 8). These motifs include inhibitory-to-excitatory connections from L6 to L5B, from L5 (in particular, L5B) to L2/3, from L5B to L4, and from L4 to L5A. The same structural elements are generally present in different cortical regions, but in each region only a subset of excitatory neurons takes part in them (Fig. 7): Like excitatory synapses18,20,42, inhibitory terminals choose postsynaptic targets with high selectivity25.
The most striking examples of translaminar inhibition were seen in V1, where some pyramidal cells in L5B were strongly inhibited by neurons in L6 and cells in L2/3 and L4 by interneurons in L5, especially L5B (Fig. 5). The likely conductors of these inhibitory signals are Martinotti cells, which are thought to provide normalizing dendritic inhibition through axonal arbors extending above their home layers21,22,40,41. Because Martinotti cells are driven intracortically21,22,26,43 rather than by thalamic afferents44, these translaminar inhibitory connections may form part of inhibitory feedback loops from the canonical target layers6-9 of excitatory output: One such loop connects L5 back to L2/3; another loop links L6 back to L5B (Fig. 5, Supplementary Fig. 8).
Inhibitory connectivity and functional specialization
The variable frequencies of inhibitory circuit motifs in different cortical areas (Fig. 5) and the selective participation of pyramidal cells in them (Fig. 7) suggest links between these motifs and the functional specialization of areas and cells. For example, the notable L4-to-L5A connection in S1 may relate to the operation of two thalamocortical input channels in barrel cortex2. These channels, termed the lemniscal and paralemniscal projections, terminate predominantly in L4 and L5A, respectively. Prominent reciprocal inhibitory connections between L4 and L5A may exist in S1 (but not V1) to enable communication between these two input streams (Supplementary Fig. 8).
Translaminar inhibition of pyramidal cells is an especially prominent feature of L2/3 and L5B of V1, suggesting a role in the construction of visual receptive fields. Of the two types of pyramidal cells—those receiving exclusively intralaminar inhibition (“home layer-inhibited neurons”) and those with translaminar inhibitory inputs (“feedback-inhibited neurons”)—home layer-inhibited neurons outnumber feedback-inhibited neurons by ~3:1 in L2/3 (Fig. 7a). In L5B, the ratio inverts and tips by ~1:2 in favor of feedback-inhibited neurons (Fig. 7b). This trend matches closely the proportions of simple to complex cells in L2/3 and L5 (3:1 and 1:3, respectively45), raising the possibility that the two cell types we describe correspond to simple and complex cells in visual cortex.
Assuming this correspondence is genuine (see ref. 46 for a sceptical alternative view), what might be the roles of intra- and translaminar inhibition in the function of simple and complex cells? In L2/3, home layer-inhibited neurons (putative simple cells) collect inhibitory inputs from a more extensive horizontal domain than do feedback-inhibited neurons (putative complex cells) (means±s.d.=471±102 vs. 330±76 μm; P=0.02). This is not entirely surprising if lateral inhibition provides a potential mechanism for generating the push-pull receptive fields of simple cells47 (but see also ref. 46).
Complex cells, in contrast, have been proposed to achieve their characteristic spatial-phase invariance because of strong recurrent input from other simple and complex cells48. However, the high gain produced by recurrent networks can render them unstable and sluggish in their responses to rapidly changing signals49. These problems can, in theory, be solved by adding circuit elements that produce divisive inhibition, or inhibition that scales with the output of the circuit49. It is conceivable that translaminar inhibitory feedback from the target layers of excitatory output subserves precisely this purpose. Distinct patterns of inhibitory connectivity may thus accompany, and perhaps even determine, the functional specialization of cortical pyramidal neurons and areas.
Acknowledgments
We thank P. Chambon, F. Costantini, S. Dymecki, G. Nagel, and P. Soriano for plasmids and T. Ellender, M. Kohl, K. Lamsa, W. Nissen, T. Nottoli, R. Roorda, Y. Tan, and L. Upton for assistance and/or discussions. This work was supported by the Medical Research Council (G.M.), NIH (G.M.), the Dana Foundation (G.M.), the Office of Naval Research (G.M.), the Boehringer Ingelheim Fonds (D.K.), and the Christopher Welch Scholarship Fund (D.K.).
Appendix
Methods
Mouse strain construction
Generation of a Cre-responsive R26::ChR2-EGFP allele
A codon-optimized DNA fragment encoding the 315 N-terminal amino acids of channelrhodopsin-2 (ChR2; GenBank accession number AF461397; ref. 32), including the H134R mutation, was fused via a 10-amino acid linker (EAGAVSGGVY) to EGFP (Clontech). ChR2-EGFP expression was driven by the synthetic CAG promoter after Cre-mediated excision of a loxP-STOP cassette interposed between transcription and translation start sites. Recombination brings the ChR2-EGFP coding sequence in frame with an initiating ATG in the loxP site, resulting in translation of ChR2-EGFP with an 11-amino acid N-terminal “loxP tag” (MYAIRSYELAT). The expression unit was targeted to the GT(ROSA)26Sor (R26) locus after insertion into the ROSA26-PA vector (Fig. 1a).
Generation of a Gad2::CreERT2 allele
To generate the potential for conditional Cre recombinase activity in all GABAergic interneurons, a cassette encoding tamoxifen-inducible Cre-ERT2 was inserted into the Gad2 locus. The GFP coding sequence of plasmid pIRES2-EGFP (Clontech) was replaced with a DNA fragment encoding Cre-ERT2, and this IRES-Cre unit was fused to an FRT-NeoR-FRT selection cassette. The assembly was ligated into a Gad2 targeting vector, which contained a 3168-bp 5′ homology arm (spanning a portion of the last intron and exon of the Gad2 gene, up to and including the stop codon) and a 5571-bp 3′ homology arm (consisting of Gad2 3′ untranslated region immediately following the stop codon). A selection cassette containing the diphteria toxin (DT-A) open reading frame, driven by the RNA polymerase II promoter and terminated by the SV40 polyadenylation signal, was inserted downstream of the 3′ homology arm (Fig. 1b).
R1 embryonic stem (ES) cells (129Sv × 129SvJ F1 hybrid) were electroporated with the linearized targeting vectors, and after G418 selection and expansion, homologous recombinant ES cell clones were identified by PCR and confirmed by Southern blotting. Recombinant ES cells were injected into C57Bl/6J blastocysts to produce germline chimeras. The FRT-NeoR-FRT selection cassette in the Gad2 targeting construct was deleted by breeding founders with the FLPeR strain. Mice are in a mixed (129 × C57Bl/6J) background.
Experimental animals and induction of transgene expression
Most experimental animals were homozygous at both targeted loci. Mice were maintained in top-open cages on a 12 hour light/dark cycle and fed standard diet RM3 (Special Diet Services), containing 19,923 IU/kg vitamin A and 9,577 μg/kg retinol. At 4 to 7 weeks of age, mice were injected i.p. on five consecutive days with 1 mg 4-OH-tamoxifen (Sigma-Aldrich), dissolved in sterile sunflower oil at 10 mg/ml. The mice were used within 7 days after the last injection. All procedures conformed to the UK Animals (Scientific Procedures) Act 1986.
Immunohistochemistry
Animals were perfused with phosphate-buffered saline (PBS, pH 7.4) containing 4 % (w/v) paraformaldehyde (PFA) and 0.2 % (w/v) picric acid under anesthesia (see below). The brain was removed and incubated for 24 h in perfusion solution and subsequently infiltrated with 30 % (w/v) sucrose in PBS for at least 24 h. Coronal sections of 40–60 μm were cut on a Leica SM 2000R sliding microtome. To minimize the PFA exposure of some antigens (particularly calcium binding proteins), 150 μm thick sections were cut acutely in ice-cold aCSF (see below) and incubated in fixation solution (PBS containing 4 % PFA and 0.2 % picric acid) for 2 h.
Sections were rinsed in Tris-buffered saline (TBS, Sigma), TBS containing 3 % (w/v) Triton X-100 (TBS-T), and TBS-T containing 20 % (v/v) horse serum (Vector Labs) and then incubated for 48 h at 4 °C in TBS-T containing 1 % horse serum and combinations of the following primary antibodies: anti-GFP (rabbit, 1:500, Sigma; or chicken, 1:500, AbCam); anti-Cre recombinase (mouse, 1:500, Millipore); anti-Gad65 (rabbit, 1:500, Millipore; mouse, 1:100 Santa Cruz); anti-Gad67 (mouse, 1:1000, Millipore); anti-CaMKIIα (rabbit, 1:400, Epitomics); anti-parvalbumin (mouse, 1:2000, Swant); anti-calretinin (mouse, 1:500, Swant); anti-calbindin (mouse, 1:250, Swant); anti-somatostatin (rabbit, 1:400, Millipore); anti-NPY (rabbit, 1:800, AbCam); and anti-VIP (rabbit, 1:500, ImmunoStar). The sections were rinsed, stained in TBS-T containing 1 % horse serum and Alexa488- and Alexa546-labeled secondary antibodies (Invitrogen), mounted in VectaShield (Vector Labs) and imaged on a Zeiss LSM510 confocal microscope.
Brain slices containing neurobiotin-filled cells were fixed overnight in PBS containing 4 % PFA and 0.2 % picric acid, rinsed in TBS, and stained in TBS-T containing 1 % (v/v) horse serum, 4 μg/ml Alexa-546-labeled avidin (Invitrogen), and 0.0001 % DAPI (Sigma) for 12-24 h. Further processing was as described above.
Electrophysiology and optical stimulation
Experimental animals were anesthetized by injection of 100 μl ketamine (100 mg/ml; Fort Dodge) plus 50 μl medetomidin (1 mg/ml; Pfizer) and perfused cardially with ice-cold solution containing (in mM): 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 10 glucose, 240 sucrose, 0.5 CaCl2, 7 MgCl2, pH 7.4, 320 mOsm. Working under safelight conditions, the brain was recovered into perfusion solution, and coronal neocortical slices of 310 μm were cut on a Leica VT1000S vibratome. To minimize the potential for damage of neuronal processes, the section plane was adjusted to lie perpendicular to the pial surface. Slices were incubated in the dark for one hour at 34 °C and subsequently maintained, shielded from light, at 25 °C in modified aCSF containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 glucose, 1 CaCl2, 2 MgCl2, pH 7.4, 315 mOsm. Recordings were performed at room temperature in aCSF containing (in mM): 125 NaCl, 3.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 glucose, 1.25 CaCl2, 1 MgCl2, pH 7.4, 310 mOsm. All extracellular solutions were bubbled with 95 % O2/5 % CO2.
Patch pipettes had tip resistances of 4-6 MΩ and contained the following internal solutions (in mM): For whole-cell recordings of IPSCs in voltage clamp: 110 CsOH, 110 gluconic acid, 0.2 EGTA, 30 Hepes, 2 MgATP, 0.3 Na2GTP, 4 NaCl, 5 QX-314, 0.2 % neurobiotin. For whole-cell recordings of optically evoked activity in current clamp: 120 K-gluconate, 10 KCl, 10 Hepes, 4 MgATP, 0.3 Na2GTP, 10 phosphocreatine, 0.2 % neurobiotin. For cell-attached recordings: 125 NaCl, 3.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 glucose, 1.5 CaCl2, 1 MgCl2, 0.5 % neurobiotin. All internal solutions were adjusted to pH 7.2-7.25 and an osmotic strength of 270-280 mOsm. Signals were amplified and lowpass-filtered at 2 kHz by a Multiclamp700a amplifier (Molecular Devices) and digitized at 5–10 kHz (Digi-data 1440, Molecular Devices).
Optical stimulation experiments were performed on a Zeiss Axioskop 2FS microscope. A 40 x, 0.8 NA water immersion objective with DIC optics was used for electrode placement and a 10 x, 0.3 NA water immersion objective, without DIC optics, for optical stimulation. The output of a continuous-wave solid-state laser with a maximum power of 325 mW at 473 nm (LRS-473-AH-300-10, Laserglow) was digitally switched and intensity-modulated by an acousto-optic deflector (IntraAction model ASN-802832 with ME-802 driver), positioned by a pair of galvanometric mirrors (GSI Lumonics VM500 with MiniSAX servo controllers), and merged with the epi-illumination path of the microscope via custom-built optics. Light pulses carried 2.0 mW of optical power at the exit pupil of the objective; in a few cases, power was attenuated (to a minimum of 1.5 mW) in order to reduce the intensity of scattered light below the stimulation threshold of nearby neurons. To generate maps of inhibitory inputs, a virtual instrument written in LabVIEW 8.5 delivered focused stimulation light pulses (spot size 3–5 μm, 20 ms duration) at intervals of 400 ms (in mapping experiments) to grids encompassing 14×14 to 22×14 locations.
Data analysis
Data were analyzed in Igor 6 (Wavemetrics), MatLab 7.9 (The Mathworks), and SPSS 17 (SPSS Inc.). Maps of inhibitory inputs were constructed from electrophysiological signals recorded during 8–10 sweeps of the stimulation grid. IPSCs were identified by three criteria. First, the amplitude of the upward deflection in the averaged trace had to exceed 3±0.5 times the average standard deviation of current fluctuations in the absence of an optical stimulus (rms noise). Second, IPSCs had to reach half-maximal amplitude within 5–70 ms after optical stimulus onset. The width of this detection window takes into account the known IPSC latencies of up to 39 ms (ref. 22) and the maximum observed spike latencies of 25 ms after illumination onset (Fig. 2d). Third, IPSCs had to occur in at least three of the 8–10 sweeps and exhibit a temporal jitter of <±10 ms.
Presynaptic sources of IPSCs were allocated to individual cortical layers, which were identified by differences in shading and cell density3,17,20,34,35 (Supplementary Fig. 6). The strength of each synaptic input was measured by integrating the recorded current over a 100-ms interval, beginning at 5 ms before the rising IPSC reached its half-maximal amplitude, to yield the charge flow per IPSC. The contribution of a layer to the total amount of inhibition received by a target cell was quantified as a percentage, which was obtained by calculating the product of the number of IPSCs originating from that layer and their average charge flow and normalizing this value to the total inhibitory charge flow of the cell. Alternatively, inhibition was quantified by counting the number of input sources from a particular layer and dividing by the total number of identified inputs (Supplementary Tables 5 and 6).
Discriminant functions were constructed to examine whether the laminar source distributions of inhibitory inputs differed systematically among cortical areas (Fig. 6a, Supplementary Table 8). These functions incorporated as predictor variables the amount of inhibitory charge flow from all cortical layers but L4 (which we cannot resolve in M1). Layer-by-layer differences were evaluated by one-way ANOVA and nonparametric Kruskal-Wallis test, followed by post-hoc analysis using Tukey’s honestly significant difference (HSD) criterion (Fig. 5, Supplementary Tables 4–7 and 9–11).
To identify subpopulations of pyramidal cells with distinct inhibitory connectivity patterns, data from M1, S1, and V1 were pooled and analyzed by hierarchical clustering. Neurons in L2/3 and L5B were characterized by three parameters: the amount of inhibitory charge flow from the home layer (L2/3 or L5 and L5B, respectively) and from the two most prominent sources of translaminar inhibition (L5B and L5 or L6, respectively). Distances between observations were quantified by the cosine of the angle between the respective data vectors and used to partition the observations iteratively into clusters. At each iteration, the two observations with the smallest distance were combined into one cluster. The distances between the observations joined in each step are represented as branch lengths in the resulting dendrogram (Fig. 7), which was used to estimate the optimal number of clusters.
The horizontal spread of inhibition was quantified by summing the number of inputs in each of the 14 columns of the stimulation grid, i.e., in a direction perpendicular to the pia. The resulting horizontal input profiles for individual target cells were aligned to their medians, interpolated to μm scale, and averaged to produce Fig. 4c. The horizontal distances between the leftmost and the rightmost inhibitory inputs to excitatory neurons in different layers and areas were compared by one-way ANOVA, followed by pair-wise Tukey-HSD post-hoc test and independent-sample t-test (Supplementary Table 3).
To examine the columnar organization of inhibitory inputs in S1, we performed paired recordings from pyramidal cells located in the same or adjacent somatosensory barrels. Three measures were used to quantify the relative tangential displacement of the two inhibitory input maps: the distance between the centers of mass of the normalized charge flow maps; the spatial cross-correlation of the two horizontal input profiles (which were obtained by summing the inhibitory charge flow in each of the 14 columns of the stimulation grid, perpendicular to the pia); and the temporal cross-correlation of the “linearized” IPSC sequences recorded during 6–10 sweeps of the stimulation raster. To generate these linearized sequences, the IPSCs in each recording were re-ordered so that progression in time corresponds to a strictly sequential—as opposed to pseudorandom—sweep of the stimulation grid; a temporal shift of 400 ms (the interval between successive optical stimuli at our standard stimulation frequency of 2.5 Hz) then equals a spatial shift of one grid spacing. Similar trends were reported by all three measures, which were therefore averaged into a single index of map displacement. To enable comparisons between slices, intercell distances (estimated from brightfield images) and map displacements were normalized by dividing measurements on a μm-scale by the horizontal distances between barrel septa.
Footnotes
METHODS
Methods are available in the online version of the paper at http://www.nature.com/natureneuroscience/.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nn.2687
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3011044
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102652927
Article citations
Optogenetic Determination of Dynamic and Cell-Type-Specific Inhibitory Reversal Potentials.
J Neurosci, 44(20):e1392232024, 15 May 2024
Cited by: 1 article | PMID: 38604778 | PMCID: PMC11097265
Agranular frontal cortical microcircuit underlying cognitive control in macaques.
Front Neural Circuits, 18:1389110, 27 Mar 2024
Cited by: 0 articles | PMID: 38601266 | PMCID: PMC11005916
Review Free full text in Europe PMC
Generation and propagation of bursts of activity in the developing basal ganglia.
Cereb Cortex, 33(20):10595-10613, 01 Oct 2023
Cited by: 0 articles | PMID: 37615347 | PMCID: PMC10560579
Multiscale model of primary motor cortex circuits predicts in vivo cell-type-specific, behavioral state-dependent dynamics.
Cell Rep, 42(6):112574, 09 Jun 2023
Cited by: 12 articles | PMID: 37300831 | PMCID: PMC10592234
Foramen lacerum impingement of trigeminal nerve root as a rodent model for trigeminal neuralgia.
JCI Insight, 8(11):e168046, 08 Jun 2023
Cited by: 5 articles | PMID: 37159265 | PMCID: PMC10393239
Go to all (150) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AF461397
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Motor cortex broadly engages excitatory and inhibitory neurons in somatosensory barrel cortex.
Cereb Cortex, 24(8):2237-2248, 31 Mar 2013
Cited by: 61 articles | PMID: 23547136 | PMCID: PMC4148616
Long-range neuronal circuits underlying the interaction between sensory and motor cortex.
Neuron, 72(1):111-123, 01 Oct 2011
Cited by: 314 articles | PMID: 21982373 | PMCID: PMC5047281
An Optogenetic Approach for Investigation of Excitatory and Inhibitory Network GABA Actions in Mice Expressing Channelrhodopsin-2 in GABAergic Neurons.
J Neurosci, 36(22):5961-5973, 01 Jun 2016
Cited by: 65 articles | PMID: 27251618 | PMCID: PMC6601813
Processing in layer 4 of the neocortical circuit: new insights from visual and somatosensory cortex.
Curr Opin Neurobiol, 11(4):488-497, 01 Aug 2001
Cited by: 113 articles | PMID: 11502397
Review
Funding
Funders who supported this work.
Medical Research Council (4)
Function and plasticity of neural circuits in Drosophila
Professor Gero Miesenböck, University of Oxford
Grant ID: G0700888
Professor Miesenboeck - Strategic Appointment
Professor Gero Miesenböck, University of Oxford
Grant ID: G0701225
Function and plasticity of neural circuits in Drosophila
Professor Gero Miesenböck, University of Oxford
Grant ID: G0700888(82521)
Professor Miesenboeck - Strategic Appointment
Professor Gero Miesenböck, University of Oxford
Grant ID: G0701225(84767)
NIDA NIH HHS (5)
Grant ID: R01 DA017297-03
Grant ID: R01 DA017297-01
Grant ID: R01 DA017297-02
Grant ID: R01 DA017297
Grant ID: R01 DA017297-04
Wellcome Trust (3)
Oxford Centre for Neural Circuits and Behaviour.
Professor Gero Miesenböck, University of Oxford
Grant ID: 090309
Grant ID: 090309/Z/09/Z
Grant ID: 84767




