Abstract
Free full text
Enterotypes of the human gut microbiome
Abstract
Our knowledge on species and function composition of the human gut microbiome is rapidly increasing, but it is still based on very few cohorts and little is known about their variation across the world. Combining 22 newly sequenced fecal metagenomes of individuals from 4 countries with previously published datasets, we identified three robust clusters (enterotypes hereafter) that are not nation or continent-specific. We confirmed the enterotypes also in two published, larger cohorts suggesting that intestinal microbiota variation is generally stratified, not continuous. This further indicates the existence of a limited number of well-balanced host-microbial symbiotic states that might respond differently to diet and drug intake. The enterotypes are mostly driven by species composition, but abundant molecular functions are not necessarily provided by abundant species, highlighting the importance of a functional analysis for a community understanding. While individual host properties such as body mass index, age, or gender cannot explain the observed enterotypes, data-driven marker genes or functional modules can be identified for each of these host properties. For example, twelve genes significantly correlate with age and three functional modules with the body mass index, hinting at a diagnostic potential of microbial markers.
Introduction
Various studies of the human intestinal tract microbiome. based on the 16S ribosomal RNA-encoding gene, reported species diversity within and between individuals1-3 and first metagenomics studies characterized the functional repertoire of the microbiomes of several American4-5 and Japanese6 individuals. Although a general consensus about the phylum level composition in the human gut is emerging1,3,7, the variation in species composition1-2 and gene pools5,8 within the human population is less clear. Furthermore, it is unknown whether inter-individual variation manifests itself as a continuum of different community compositions or whether individual gut microbiota congregate around some preferred, balanced and stable community compositions that can be classified. Studying such questions is complicated by the complexity of sampling, DNA preparation, processing, sequencing and analysis protocols9 as well as by varying physiological, nutritional and environmental conditions. To analyze the feasibility of comparative metagenomics of the human gut across cohorts and protocols and to obtain first insights in commonalities and differences between gut microbiomes across different populations, we Sanger-sequenced 22 European metagenomes from Danish, French, Italian and Spanish individuals that were selected for diversity (Supplementary Notes Section 1), and combined them with existing Sanger (13 Japanese6, 2 American4) and 454 (2 American5) gut datasets – totaling 39 individuals.
Global phylogenetic and functional variation of intestinal metagenomes
The vast majority of sequences in the newly sequenced 22 European samples belong to bacteria – only 0.14% of the reads could be classified as human contamination, all other eukaryotes together only comprised 0.5%, archaea 0.8% and viruses up to 5.8% (see Supplementary Notes Section 2.1 for details).
To investigate the phylogenetic composition of the 39 samples from 6 nationalities, we mapped metagenomic reads, using DNA sequence homology, to 1511 reference genomes (Supplementary Table 3) including 379 publicly available human microbiome genomes generated through the NIH Human Microbiome Project10 and the European MetaHIT consortium11 (Supplementary Methods Section 4.1). To consistently estimate the functional composition of the samples, we annotated the predicted genes from the metagenomes using eggNOG12 orthologous groups (Supplementary Methods Section 6.2). We ensured that comparative analysis using these procedures was not biased by dataset origin, sample preparation, sequencing technology and quality filtering (see Supplementary Notes Section 1). We also investigated whether the relatively low and somewhat arbitrary amounts of sequence per sample (between 53-295 Mb) bias our results: we assigned habitat information to 1368 out of the 1511 reference genomes, distinguished between orthologous groups from gut and non-gut species and conclude that our dataset captures most of the functions from gut species even though functions from ‘non-gut’ species still accumulated with each additional sample (Fig. 1a; see Supplementary Notes Section 1.3).
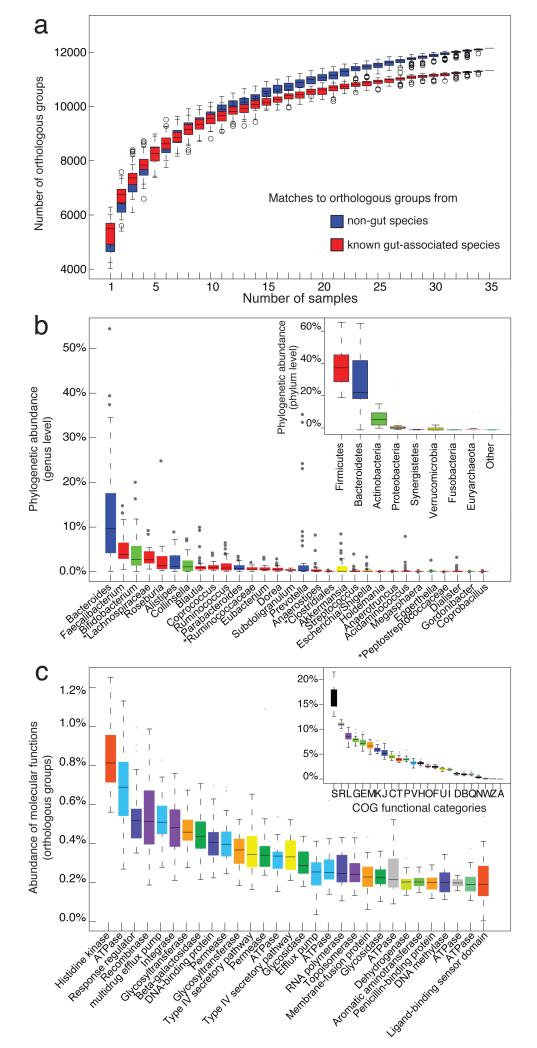
(a) Simulation of the detection of distinct orthologous groups (OGs) when increasing the number of individuals (samples). Complete genomes were classified by habitat-information and the OGs divided into those that occur in known gut-species (red) and those that have not yet associated to gut (blue). The former are close to saturation when sampling 35 individuals (excluding infants) whereas functions from non-gut (probably rare and transient) species are not. (b) Genus abundance variation box plot for the 30 most abundant genera as determined by read abundance. Genera are colored by their respective phylum (see inset for color key). Inset: phylum abundance box plot. Genus and phylum level abundances were measured using reference genome based mapping with 85% and 65% sequence similarity cutoffs. Unclassified genera under a higher rank are marked by asterisks. (c) Orthologous group (OG) abundance variation box plot for the 30 most abundant OGs as determined by assignment to eggNOG12. OGs are colored by their respective functional category (see inset for color key). Inset: abundance box plot of 24 functional categories.
We then characterized the phylogenetic variation across samples at the genus and phylum levels, and functional variation at gene and functional class levels. As infants are known to have very heterogeneous, unstable and distinctive microbiota6,13, we excluded the four respective Japanese samples from the analysis. Using calibrated similarity cutoffs (Supplementary Figure 1), on average, 52.8% of the fragments in each sample could be robustly assigned to a genus in our reference genome set (ranging from 22% to 80.5%), and 80% could be assigned to a phylum (ranging from 64.9% to 91%) implying that the trends observed (Fig. 1b) represent a large fraction of the metagenome.
The phylogenetic composition of the newly sequenced samples confirms that the Firmicutes and Bacteroidetes phyla constitute the vast majority of the dominant human gut microbiota7 (Fig. 1b, inset). Bacteroides was the most abundant but also most variable genus across samples (Fig. 1b; Supplementary Notes Section 2.2), agreeing with previous observations6,14. Our function identification protocol led to a high functional assignment rate: 63.5% of all predicted genes in the Sanger-sequenced samples analyzed (41% of all predicted genes in two samples obtained by pyrosequencing; Supplementary Table 5) can be assigned to orthologous groups (OGs), and OG abundance patterns again agree with previous observations6,15 (e.g. histidine kinases make up the largest group; Fig 1c; Supplementary Notes Section 2.3).
Highly abundant functions from low-abundance microbes
Microbes in the human gut undergo selective pressure from the host as well as from microbial competitors. This typically leads to a homeostasis of the ecosystem in which some species occur in high and many in low abundance16 (the “long-tail” effect, as seen in Fig. 1b), with some low-abundance species, like methanogens17, performing specialized functions beneficial to the host. Metagenomics enables us to study the presence of abundant functions shared by several low-abundance species, which could shed light on their survival strategies in the human gut. In the samples analyzed here, the most abundant molecular functions generally trace back to the most dominant species. However, we identified some abundant orthologous groups that are contributed primarily by low abundance genera (see Supplementary Figure 2, Supplementary Table 6 and Supplementary Notes Section 3). For example, low abundance Escherichia contribute over 90% of two abundant proteins associated with bacterial pilus assembly, FimA (COG3539) and PapC (COG3188), found in one individual (IT-AD-5). Pili enable the microbes to colonize the epithelium of specific host organs; they help microbes to stay longer in the human intestinal tract by binding to the human mucus or mannose sugars present on intestinal surface structures18. They are also key components in the transfer of plasmids between bacteria through conjugation, often leading to exchange of protective functions such as antibiotic resistance18. Pili can thus provide multiple benefits to these low-abundance microbes in their efforts to survive and persist in the human gut. This example illustrates that abundant species or genera cannot reveal the entire functional complexity of the gut microbiota. More reference genomes will facilitate better taxonomic assignments from samples and thus the detection of more low abundance species. However, there is not much room for as yet undetected, abundant genera. Even with our limited genus assignment rate of 52.8% of all reads, we estimate that we miss another 30.7% of the already classified genera due to our strict assignment criteria (Supplementary Figure 1), i.e. only 16.5% of all reads are likely to belong to hitherto unknown genera.
Robust clustering of samples across nations: Identification of enterotypes
To get an overview of the species variation we used phylogenetic profile similarities obtained by mapping metagenomic reads to the 1511 reference genomes (Fig. 2a, see Supplementary Methods Section 4.1). We excluded the two American Sanger-sequenced samples4 from further analysis because of an unusual, very low fraction of Bacteroidetes, and suspected technical artifacts19. Multidimensional cluster analysis and Principal Component Analysis (PCA) revealed that the remaining 33 samples formed three distinct clusters which we designate enterotypes (see Supplementary Notes Section 4.1, Supplementary Figure 3a and Supplementary Table 8). Each of these three enterotypes are identifiable by the variation in the levels of one of three genera: Bacteroides (enterotype 1), Prevotella (enterotype 2) and Ruminococcus (enterotype 3; Fig. 2a and 2d), which was reproduced using independent array-based HITChip20 data in a subset of 22 European samples (Supplementary Figure 4 and Supplementary Notes Section 4.5). The same analysis on two larger published gut microbiome datasets of different origins (16S pyrosequencing data from 154 American individuals5 and Illumina-based metagenomics data from 85 Danish individuals8, Supplementary Methods Section 5) shows that these datasets could also be represented best by three clusters (Supplementary Figure 3b and c, Supplementary Table 9 and Supplementary Table 10). Two of these are also driven by Bacteroides and Prevotella, while the third cluster is mostly driven by related groups of the order Clostridiales, Blautia and unclassified Lachnospiraceae in the 16S rDNA and Illumina data, respectively (Fig. 2b and 2c). This can be explained by a different reference data set in case of 16S rDNA data, different mapping behavior of short reads in case of the Illumina data or current taxonomic uncertainties in the Lachnospiraceae and Ruminococcaceae clades (see Supplementary Notes Section 4.2). The differences might also hint at community subpopulations within this enterotype, which might only be detectable with substantially more samples. Correlation analysis of the Sanger data revealed that abundances of each of the three discriminating genera strongly correlate (that is they co-occur or avoid each other) with those of other genera (Fig. 2d; see Supplementary Methods Section 11), suggesting that the enterotypes are in fact driven by groups of species that together contribute to the preferred community compositions.
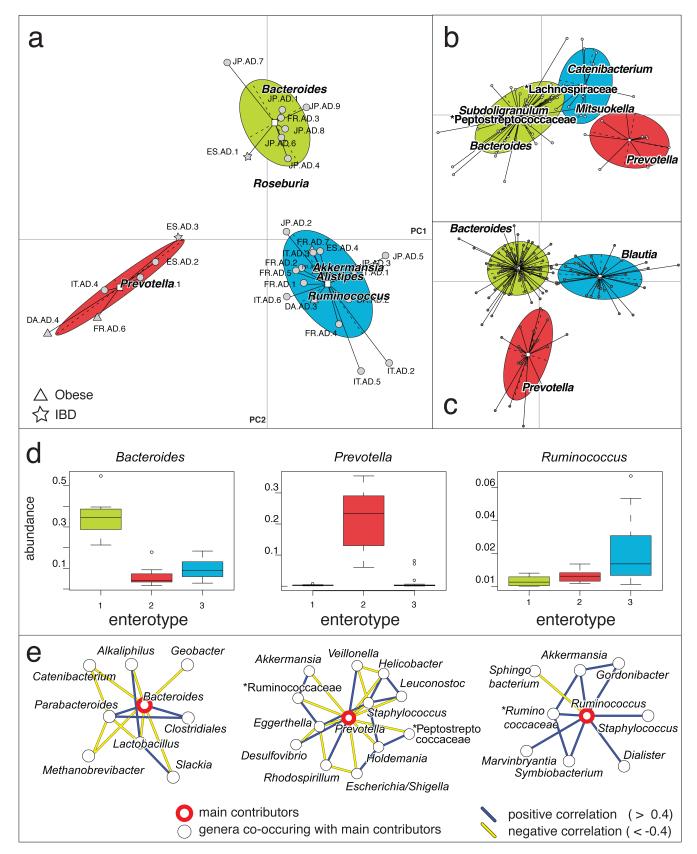
Between class analysis, which visualizes results from Principal Component Analysis and clustering, of the genus compositions of (a) 33 Sanger metagenomes estimated by mapping the metagenome reads to 1511 reference genome sequences using an 85% similarity threshold, (b) Danish subset containing 85 metagenomes from a published Illumina dataset8 and (c) 154 pyrosequencing-based 16S sequences5 reveal three robust clusters that we call enterotypes. Two principal components are plotted using the ade4 package in R with each sample represented by a filled circle. The center of gravity for each cluster is marked by a rectangle and the colored ellipse covers 67% of the samples belonging to the cluster. (d) Abundances of the main contributors of each enterotype from the Sanger metagenomes. (e) Co-occurrence networks of the three enterotypes from the Sanger metagenomes. Unclassified genera under a higher rank are marked by asterisks in (b) and (e).
We further demonstrate the robustness of the enterotypes using two distinct statistical concepts. First we used the silhouette coefficient21 to validate that the three clusters are superior to clusterings obtained from various randomizations of the genus profile data, suggesting a potential role for the interactions between co-occurring genera (see Supplementary Figure 5 and Supplementary Notes Section 4.3). Second we used supervised learning and cross validation to establish that these clusters have non-random characteristics that can be modeled and subsequently used to classify new samples (learning on clusters from randomized genus profiles led to considerably worse classification performance; see Supplementary Figure 6 and Supplementary Notes Section 4.4). These consistent results suggest that enterotypes will be identifiable in human gut metagenomes also from larger cohorts.
We then clustered the 33 samples using a purely functional metric: the abundance of the assigned orthologous groups (Fig. 3a). Remarkably, this clustering also showed a similar grouping of the samples with only minor differences (5 samples placed in different clusters compared to Fig. 2a) indicating that function and species composition roughly coincide with some exceptions such as Spanish sample ES-AD-3 whose genus composition belongs to enterotype 2 while its functional composition is similar to members of enterotype 1. This individual has high levels of phage-related genes compared to the other samples (see Supplementary Figure 7), hinting at partial temporal variability and dynamics of the microbiota, and perhaps indicating phage or virus bursts.
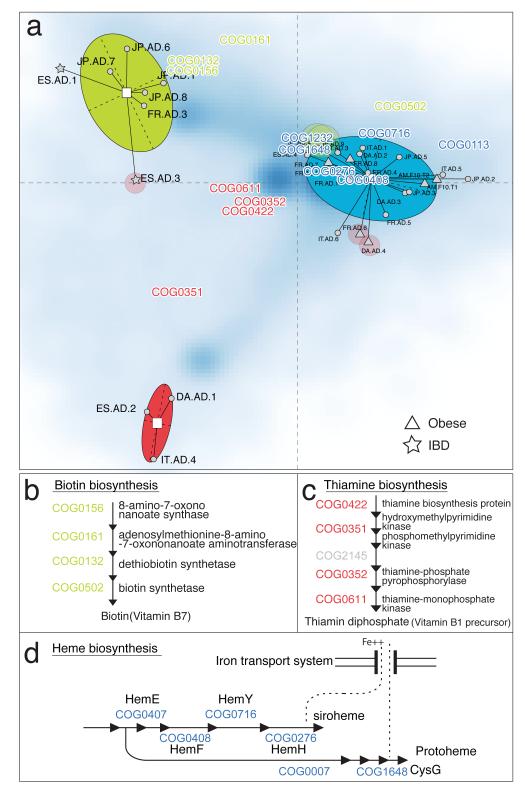
(a) Between class analysis (see Fig. 2) of orthologous group (OG) abundances showing only minor disagreements with enterotypes (transparent circles indicate the differing samples). The blue cloud represents the local density estimated from the coordinates of OGs; positions of selected OGs are highlighted. (b) Four enzymes in the biotin biosynthesis pathway (COG0132, COG0156, COG0161 and COG0502) are overrepresented in enterotype 1. (c) Four enzymes in the thiamine biosynthesis pathway (COG0422, COG0351, COG0352 and COG0611) are overrepresented in enterotype 2. (d) Six enzymes in the heme biosynthesis pathway (COG0007, COG0276, COG407, COG0408, COG0716 and COG1648) are overrepresented in enterotype 3.
The robustness and predictability of the enterotypes in different cohorts and at multiple phylogenetic and functional levels suggests that they are the result of well-balanced, defined microbial community compositions of which only a limited number exist across individuals. These enterotypes are not as sharply delimited as, for example, human blood groups; they are rather densely populated areas in a multidimensional space of community composition. They are nevertheless likely to characterize individuals, in line with previous reports that gut microbiota is rather stable in individuals and can even be restored after perturbation22-25.
Phylogenetic and functional variation between enterotypes
To determine the phylogenetic and functional basis of the enterotypes, we investigated in detail their differences in composition at the phylum, genus, gene and pathway level as well as correlations in abundance of co-occurring genera (Figs. 2 and and3;3; also see Supplementary Methods Sections 10, 11 and 12). Enterotype 1, containing 8 samples, is enriched in Bacteroides (p<0.01; Supplementary Figure 8), which co-occurs, for example, with Parabacteroides (see Supplementary Table 11 for enriched genera and Fig. 2e for correlation networks of co-occurring genera in each enterotype). The drivers of this enterotype seem to derive energy primarily from carbohydrates and proteins through fermentation, since these closely related genera have a very broad saccharolytic potential26 and since genes encoding enzymes involved in the degradation of these substrates (galactosidases, hexosaminidases, proteases) along with glycolysis and pentose phosphate pathways are enriched in this enterotype (see Supplementary Table 12 and Supplementary Table 13). Enterotype 2 contains 6 samples and is enriched in Prevotella (p<0.01; Supplementary Figure 9) and the co-occurring Desulfovibrio, who can act in synergy to degrade mucin glycoproteins present in the mucosal layer of the gut: Prevotella is a known mucin-degrader and Desulfovibrio could enhance the rate-limiting mucin desulfation step by removing the sulfate27. Enterotype 3 is the most frequent one and is enriched in Ruminococcus (p<0.01; Supplementary Figure 10) as well as co-occurring Akkermansia, both known to comprise species able to degrade mucins28. It is also enriched in membrane transporters, mostly of sugars, suggesting the efficient binding of mucin and its subsequent hydrolysis as well as uptake of the resulting simple sugars by these genera. The enriched genera suggest that enterotypes employ different routes to generate energy from fermentable substrates available in the colon reminiscent of a potential specialization in ecological niches or guilds. In addition to the conversion of complex carbohydrates into absorbable substrates, the gut microbiota is also beneficial to the human host by producing vitamins. Although all the vitamin metabolism pathways are represented in all samples, enterotypes 1 and 2 were enriched in biosynthesis of different vitamins: biotin (Fig. 3b), riboflavin, pantothenate and ascorbate in the former, and thiamine (Fig. 3c) and folate in the latter. These phylogenetic and functional differences among enterotypes thus reflect different combinations of microbial trophic chains with a likely impact on the synergistic inter-relations with the human hosts.
Functional biomarkers for host properties
Enterotypes do not seem to differ in functional richness (Supplementary Figure 11), and virtually none of several measured host properties, namely nationality, gender, age or body mass index (BMI), significantly correlates with the enterotypes (with the exception of enterotype 1 which is enriched in Japanese individuals). However, some strong correlations do occur between host properties and particular functions, at the genes or module level (a module is a part of a pathway that is functionally tightly interconnected, see Supplementary Methods Sections 6.3, 13 and Supplementary Notes Section 6). The only significant correlation between a host property and a taxonomic group is a negative one between age and the abundance of an unknown Clostridiales genus (p<0.02) containing three obligate anaerobes (Supplementary Figure 12a; see Supplementary Notes Section 6.2). It should be noted that age is not constant across the nationalities (in our dataset, Italians are relatively old and Japanese young), but that individuals did not stratify by nationality, suggesting that this is not a confounding factor. Our data did not reveal any correlation between BMI and the Firmicutes/Bacteroidetes ratio and we thus cannot contribute to the ongoing debate on the relation between this ratio and obesity29-30.
In contrast to the little phylogenetic signal, we found several significant functional correlations with each of the host properties studied (after correcting for multiple testing to avoid artifacts; see Supplementary Methods Section 13), suggesting that metagenomics-derived functional biomarkers might be more robust than phylogenetic ones. For example, the abundance of 10 orthologous groups (OGs) varies more between than within nationalities (Supplementary Table 14) although overall, the functional composition in total was remarkably similar among the nations (also with respect to the functional core; see Supplementary Figure 13). For gender, we find five functional modules and one OG that significantly correlate (p<0.05; e.g., enriched aspartate biosynthesis modules in males; see Supplementary Table 16). In addition, twelve OGs significantly correlate with age (Supplementary Table 17). For instance, starch degradation enzymes such as glycosidases and glucan phosphorylases increase with age (which could be a reaction to decreased efficiency of host breakdown of dietary carbohydrates with age31) and so does the secA preprotein translocase (Supplementary Figure 14). Conversely, an OG coding for the facultative sigma-24 subunit of RNA polymerase, which drives expression under various stress responses and is linked to intestinal survival32, decreases with age (Fig. 4a). One explanation for this could be the reduced need for stress response in the gut due to the age-associated decline in host immune response33 (immunosenescence). Our analyses also identified three marker modules that correlate strongly with the hosts’ BMI (Supplementary Table 19, Supplementary Figure 14), two of which are ATPase complexes, supporting the link found between the gut microbiota’s capacity for energy harvest and host’s obesity34. Interestingly, functional markers found by a data-driven approach (derived from the metagenomes without previous knowledge) gave much stronger correlations than genes for which a link would be expected (e.g. SusC/SusD, involved in starch utilization26; Fig. 4b). Linear models combining the abundance of only a few functional modules correlate even better with host properties (Fig 4c,d). It should be noted that given the possibility of many confounding variables due to the heterogeneity and size of our cohort, these observations will need to be substantiated using larger, independent cohorts in the future. Furthermore, patterns in metagenomics data can (partly) reflect indirect factors9 such as genome size35 (the smaller the average genome size of a sample, the higher would be the relative fraction of single copy genes therein), which does not matter for diagnostics though.
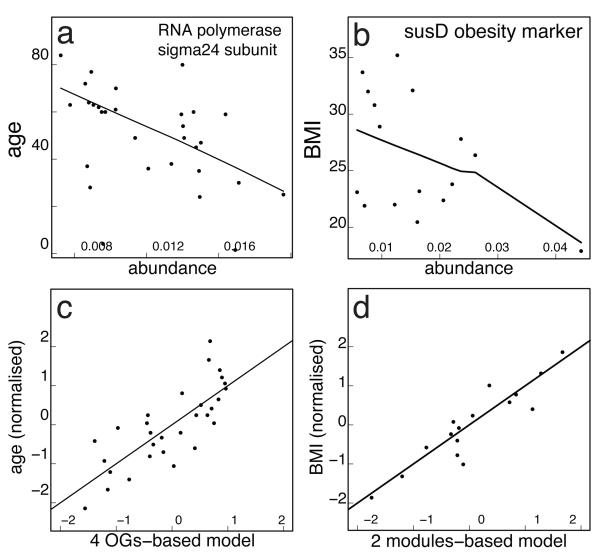
(a) Pairwise correlation of RNA polymerase facultative sigma24 subunit (COG1595) with age (p=0.03, rho=−0.59). (b) Pairwise correlation of SusD, a family of proteins that bind glycan molecules before they are transported into the cell, and body mass index (p=0.27, rho=−0.29, weak correlation). (c) Multiple OGs (COG0085, COG0086, COG0438 and COG0739; see Supplementary Table 18) significantly correlating with age when combined into a linear model (see Supplementary Methods Section 13 and ref. 40 for details; p= 2.75e-05, adjusted R2=0.57). (d) Two modules, ATPase complex and ectoine biosynthesis (M00051), significantly correlating with BMI when combined into a linear model (p= 6.786e-06, adjusted R2=0.82).
While individual host properties don’t explain the enterotypes, the latter might be driven by a complex mixture of functional properties, by host immune modulation or by hitherto unexplored physiological conditions such as transit time or pH of luminal contents. Furthermore, the three major enterotypes could be triggered by the three distinct pathways for hydrogen disposal36 (Supplementary Notes Section 6.4). Indeed, despite their low abundance, Methanobrevibacter (a methanogen) and Desulfovibrio (a known sulfate-reducer) are enriched in enterotypes 3 and 1, respectively.
Taken together, we have demonstrated the existence of enterotypes in the human gut microbiome and have identified three of them that vary in species and functional composition using data that spans several nations and continents. As our current data do not reveal which environmental or even genetic factors are causing the clustering, and as fecal samples are not representative of the entire intestine, we anticipate that the enterotypes introduced here will be refined with deeper and broader analysis of individuals’ microbiomes. Presumably, enterotypes are not limited to humans but also occur in animals. Their future investigations might well reveal novel facets of the human and animal symbiotic biology and lead to the discovery of the microbial properties correlated with the health status of individuals. We anticipate that they might allow classification of human groups that respond differently to diet or drug intake. The enterotypes appear complex, are probably not driven by nutritional habits and cannot simply be explained by host properties such as age or BMI, although there are functional markers such as genes or modules that correlate remarkably well with individual features. The latter might be utilizable for diagnostic and perhaps even prognostic tools for numerous human disorders, for instance colorectal cancer and obesity-linked co-morbidities such as metabolic syndrome, diabetes and cardio-vascular pathologies.
Methods summary
Sample collection
Human fecal samples from European individuals were collected and frozen immediately, and DNA was purified as described previously37.
Sequencing
Random shotgun DNA libraries of 3kb were Sanger-sequenced using standard protocols established at Genoscope.
Sequence processing
Cloning vector, sequencing primers and low quality bases were end-trimmed from raw Sanger reads, and possible human DNA sequences were removed. Reads were processed by the SMASH comparative metagenomics pipeline38 for assembly and gene prediction.
Phylogenetic annotation
Phylogenetic annotation of samples was performed by (1) aligning reads (Sanger/Illumina) against a database of 1511 reference genomes (listed in Supplementary Table 3) or (2) classifying 16S rDNA reads using RDP classifier39. Genus and phylum abundance was estimated after normalizing for genome size for the former, and for 16S gene copy number for the latter.
Functional annotation
Genes were functionally annotated using BLASTP against eggNOG (v2) and KEGG (v50) databases. Protein abundances were estimated after normalizing for protein length. Functional abundance profiles at eggnog-, KEGG orthologous group-, functional module- and pathway-level were created.
Clustering and classification
Samples were clustered using Jensen-Shannon distance and partitioning around medoid (PAM) clustering. Optimal number of clusters was estimated using Calinski-Harabasz (CH) index. We used the silhouette validation technique for assessing the robustness of clusters. Additionally, within a cross-validation scheme, we trained predictive decision tree models on clusters obtained using the same clustering method and evaluated the classification of hold-out samples by accuracy, average precision and average precision gain.
Statistics
Correlations between metadata and feature abundances were computed as described previously40, based on multiple-testing corrected pairwise Spearman correlation analysis and stepwise regression for multi-feature model building. For categorical metadata and enterotype comparisons, samples were pooled into bins (male/female, obese/lean, one enterotype/rest, specific nationality/rest etc.) and significant features were identified using Fisher’s exact test with multiple testing correction of p-values.
Supplementary Material
figures
information
tables
tables 1
Acknowledgements
The authors are grateful to Christopher Creevey, Gwen Falony and members of the Bork group at EMBL for helpful discussions and assistance. We thank the EMBL IT core facility and Yan Yuan for managing the high-performance computing resources. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013): MetaHIT, grant agreement HEALTH-F4-2007-201052 and from EMBL. Obese/non-obese volunteers for MicroObes study were recruited from the SU.VI.MAX cohort study coordinated by S. Hercberg, and metagenome sequencing was funded by ANR; volunteers for MicroAge study were recruited from the CROWNALIFE cohort study coordinated by A. Cresci, and metagenome sequencing was funded by GenoScope. Ciberehd is funded by the Instituto de Salud Carlos III (Spain). The study was supported by grants from the Lundbeck Foundation Centre for Applied Medical Genomics in Personalized Disease Prediction, Prevention and Care (LuCAMP). JR is supported by the IWIOB and the Odysseus programme of the Fund for Scientific Research Flanders (FWO). BGI was partially funded by the International Science and Technology Cooperation Project in China (0806). We are thankful to the Human Microbiome Project for generating the reference genomes from human gut microbes and the International Human Microbiome Consortium for stimulating discussions and the exchange of data.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Information Informed consent was obtained from the 22 European subjects. Sample collection and experiments were approved by the following ethics committees: MetaHIT (Danish) – ethical committee of the Capital Region of Denmark; MetaHIT (Spanish) – CEIC, Hospital Vall d’Hebron; MicroObes – Ethical Committee for Studies with Human Subjects of Cochin Hospital in Paris, France; MicroAge – Joint Ethical Committee of the University of Camerino. Raw Sanger read data from the European fecal metagenomes have been deposited to NCBI Trace Archive with the following project ids: MH6 (33049), MH13 (33053), MH12 (33055), MH30 (33057), CD1 (33059), CD2 (33061), UC4(33113), UC6(33063), NO1 (33305), NO3 (33307), NO4 (33309), NO8 (33311), OB2 (33313), OB1 (38231), OB6 (38233), OB8 (45929), A (63073), B(63075), C (63077), D (63079), E (63081), G (63083). Contigs, genes and annotations are available to download from http://www.bork.embl.de/Docu/Arumugam_et_al_2011/.
The authors declare no competing financial interests.
Additional MetaHIT Consortium members María Antolín1, François Artiguenave2, Hervé M. Blottiere3, Mathieu Almeida3, Carlos Cara4, Christian Chervaux5, Antonella Cultrone3, Christine Delorme3, Gérard Denariaz5, Rozenn Dervyn3, Konrad U. Foerstner6,7, Carsten Friss8, Maarten van de Guchte3, Eric Guedon3, Florence Haimet3, Wolfgang Huber6, Alexandre Jamet3, Catherine Juste3, Ghalia Kaci3, Jan Knol5, Omar Lakhdari3, Severine Layec3, Karine Le Roux3, Emmanuelle Maguin3, Raquel Melo Minardi2, Jean Muller9,10, Raish Oozeer5, Julian Parkhill11, Pierre Renault3, Maria Rescigno12, Nicolas Sanchez3, Shinichi Sunagawa6, Antonio Torrejon1, Keith Turner11, Gaetana Vandemeulebrouck3, Encarna Varela1, Yohanan Winogradsky3, Georg Zeller6
1Digestive System Research Unit, University Hospital Vall d’Hebron, Ciberehd, Barcelona, Spain.
2Commissariat à l’Energie Atomique, Genoscope, 91000 Evry, France.
3Institut National de la Recherche Agronomique, 78350 Jouy en Josas, France.
4UCB Pharma SA, 28046 Madrid, Spain.
5Danone Research, 91120 Palaiseau, France.
6European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
7Darmstadt, Germany.
8Center for Biological Sequence Analysis, Technical University of Denmark, DK-2800 Kongens Lyngby, Denmark.
9Institute of Genetics and Molecular and Cellular Biology, CNRS, INSERM, University of Strasbourg.
10Genetic Diagnostics Laboratory, CHU Strasbourg Nouvel Hôpital Civil, Strasbourg, France
11TheWellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK.
12Istituto Europeo di Oncologia, 20100 Milan, Italy.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature09944
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3728647?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101774303
Article citations
Predictable regulation of gut microbiome in immunotherapeutic efficacy of gastric cancer.
Genes Immun, 12 Nov 2024
Cited by: 0 articles | PMID: 39533019
Gut microbes of a high-value marine fish, Snubnose Pompano (Trachinotus blochii) are resilient to therapeutic dosing of oxytetracycline.
Sci Rep, 14(1):27949, 14 Nov 2024
Cited by: 0 articles | PMID: 39543167 | PMCID: PMC11564560
Efficient Biosynthesis of Theanderose, a Potent Prebiotic, Using Amylosucrase from <i>Deinococcus deserti</i>.
J Agric Food Chem, 72(45):25197-25209, 31 Oct 2024
Cited by: 0 articles | PMID: 39480747 | PMCID: PMC11565756
Utility of dairy microbiome as a tool for authentication and traceability.
Open Life Sci, 19(1):20220983, 30 Oct 2024
Cited by: 0 articles | PMID: 39479351 | PMCID: PMC11524395
Review Free full text in Europe PMC
Osteopontin Depletion in Nonhematopoietic Cells Improves Outcomes in Septic Mice by Enhancing Antimicrobial Peptide Production.
J Infect Dis, 230(5):e1146-e1157, 01 Nov 2024
Cited by: 0 articles | PMID: 38913690 | PMCID: PMC11566238
Go to all (3,701) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Diversity and enterotype in gut bacterial community of adults in Taiwan.
BMC Genomics, 18(suppl 1):932, 25 Jan 2017
Cited by: 44 articles | PMID: 28198673 | PMCID: PMC5310273
A metagenomic approach to dissect the genetic composition of enterotypes in Han Chinese and two Muslim groups.
Syst Appl Microbiol, 41(1):1-12, 10 Nov 2017
Cited by: 14 articles | PMID: 29129355
Enterotypes in the landscape of gut microbial community composition.
Nat Microbiol, 3(1):8-16, 18 Dec 2017
Cited by: 470 articles | PMID: 29255284 | PMCID: PMC5832044
Review Free full text in Europe PMC
Driving gut microbiota enterotypes through host genetics.
Microbiome, 12(1):116, 28 Jun 2024
Cited by: 0 articles | PMID: 38943206
Funding
Funders who supported this work.
European Commission FP7 (1)
Grant ID: FP7_201052
NIDDK NIH HHS (1)
Grant ID: K24 DK002800
Novo Nordisk Fonden (1)
Grant ID: NNF10CC1016517
Wellcome Trust (2)
Grant ID: 076964
Grant ID: 082372




