Abstract
Free full text

Turn-on DNA Damage Sensors for the Direct Detection of 8-Oxoguanine and Photoproducts in Native DNA
Associated Data
Abstract
The integrity of the genetic information in all living organisms is constantly threatened by a variety of endogenous and environmental insults. To counter this risk, the DNA-damage response is employed for repairing lesions and maintaining genomic integrity. However, an aberrant DNA-damage response can potentially lead to genetic instability and mutagenesis, carcinogenesis, or cell death. To directly monitor DNA damage events in the context of native DNA, we have designed two new sensors utilizing genetically fragmented firefly luciferase (split luciferase). The sensors are comprised of a methyl-CpG binding domain (MBD) attached to one fragment of split luciferase for localizing the sensor to DNA (50–80% of the CpG dinucleotide sites in the genome are symmetrically methylated at cytosines), while a damage-recognition domain is attached to the complementary fragment of luciferase to probe adjacent nucleotides for lesions. Specifically, we utilized oxoguanine glycosylase 1 (OGG1) to detect 8-oxoguanine caused by exposure to reactive oxygen species and employed the damaged-DNA binding protein 2 (DDB2) for detection of pyrimidine dimer photoproducts induced by UVC light. These two sensors were optimized and validated using oligonucleotides, plasmids, and mammalian genomic DNA, as well as HeLa cells that were systematically exposed to a variety of environmental insults, demonstrating that this methodology utilizing MBD-directed DNA localization provides a simple, sensitive, and potentially general approach for the rapid profiling of specific chemical modifications associated with DNA damage and repair.
Introduction
The human genome has been reported to typically accrue > 104 lesions daily on a per cell basis from a variety of endogenous and environmental insults.1 Direct chemical modifications include bulky DNA adducts,2–4 oxidized or hydrolyzed bases,5 alkylation products,6 and strand breaks.7,8 In order to survive, cells have evolved specific mechanisms to counter DNA damage, collectively termed the DNA-damage response (DDR).9 However, if left to persist, these base adducts and modifications may result in premature aging, carcinogenic mutations, and genetic instability.10,11 Owing to the potentially severe outcomes associated with DNA lesions, methods for detecting these chemical perturbations have been widely pursued. Some commonly utilized methods for evaluating non-specific DNA damage include the Comet assay,12 Ligation-Mediated PCR,13 and the Terminal deoxynucleotidyl transferase-mediated dUTP-biotin Nick End Labeling (TUNEL) assay.14 However, these techniques typically report on the presence of generic DNA damage, rather than quantifying a particular lesion. Methods for the identification of a particular type of DNA lesion most often involve either DNA fragmentation followed by chromatographic separation and mass spectrometry15–17 or immunological methods using damage-specific antibodies.18 While antibodies may have limited specificity for a particular lesion in the context of an excess of undamaged bases,18 mass spectrometric techniques are often burdened by the accrual of DNA damage artifacts (particularly artifactual base oxidation) during processing and the necessity of expensive and specific instrumentation.19 Consequently, it is desirable to design new methods for directly monitoring DNA damage using lesion-specific structure-based or chemically-reactive probes.20–23 In an elegant electrochemical approach, detection of single-base mismatches and DNA lesions was achieved by monitoring attenuation of charge transport through DNA films.24 As a potentially more general alternative, the ongoing elucidation of endogenous repair proteins that directly sense specific types of modified bases as well as proteins that regulate the DDR provides the motivation for a direct protein-based approach for detecting DNA damage and repair, as recently realized in a poly(ADP-ribose) sensor.25 Specifically, the identification and characterization of oxoguanine glycosylase 1 (OOG1) that recognizes 8-oxoguanine (8-oxoG) and damage-DNA binding protein 2 (DDB2) that targets ultraviolet-induced photoproducts provide a protein palette for generating a set of turn-on biosensors for monitoring environmental insults to DNA.26,27
In utilizing a protein-based approach for sensor design, we have previously employed split-protein reassembly (also called protein-fragment complementation), wherein initially non-functional fragments of a split-signaling protein are induced to reassemble through the direct interaction of attached domains.28 Many split proteins have been validated in this regard, including ubiquitin,29 green fluorescent protein (GFP),30 β-lactamase,31 and luciferase.32,33 Using a ternary reassembly approach, this methodology has been applied to sequence-specific DNA detection, in which DNA is targeted by a pair of programmable zinc finger (ZF) domains, and signaling is achieved through the use of split GFP or β-lactamase.34,35 Subsequently, we adapted this methodology for the sequence-specific detection of 5-methylcytosine modifications utilizing a methyl-CpG binding domain (MBD) in conjunction with a sequence-specific ZF attached to the split-protein halves.36–38 Of the many signaling proteins that have been validated for use in split-protein reassembly strategies, we have recently established that the low background and high luminescent signal output of split firefly luciferase makes it particularly attractive for use in a cell-free translation system for biosensor generation.39–42 In order to rapidly probe genome-wide DNA damage utilizing our cell-free split-protein methodology, we reasoned that we would require a generic localization domain for targeting our sensors to any damage-accessible DNA. Transcription factors are generally not suitable for this function, since binding should be largely independent of surrounding sequence. Since cytosine methylation in mammalian genomic DNA is estimated at 50-80% of CpG sites,43 we reasoned that the attachment of MBD1 to one fragment of split luciferase would potentially serve as a generic yet high affinity genome localization domain. To allow our ternary DNA detection technique to report on DNA damage accrued from exposure to reactive oxygen species (ROS), we sought a natural protein domain and attached the relevant domain of OGG1 to the complementary fragment of split luciferase (Figure 1A). Using this same general strategy, we envisioned a second sensor for UV-induced DNA damage by utilizing a domain from DDB2 in conjunction with MBD1 (Figure 1B). Herein we report the design and validation of our OGG1 and DDB2 based split-luciferase biosensors for determining the nature and extent of chemical lesions accrued in oligonucleotides, plasmids, HeLa (mammalian genomic) DNA, as well as live cells, exposed to ROS and UV radiation.
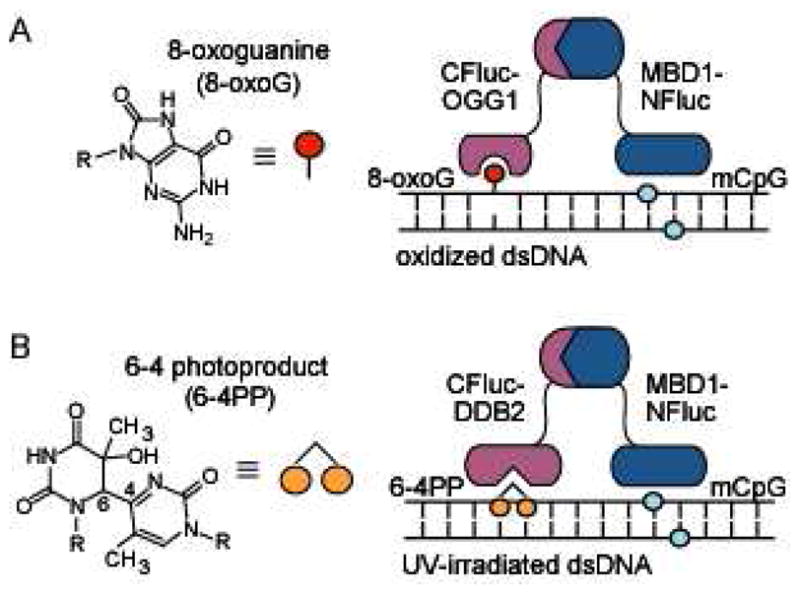
Split-protein DNA damage biosensors. (A) A methyl-CpG binding domain, MBD1, is attached to NLuciferase (NFluc) to localize the construct to methylated DNA, allowing for CLuciferase (CFluc) attached to oxoguanine glycosylase 1 (OGG1) to probe adjacent guanine bases for oxidation. (B) MBD1-NFluc is localized to methylated DNA, allowing for CFluc attached to damaged-DNA binding protein (DDB2) to probe adjacent sites for UV induced damage.
Results and Discussion
Initial design and validation of a split-luciferase sensor for detecting DNA oxidation
Considering the ubiquitous cellular damage associated with oxidation, we began by developing a sensor for detection of 8-oxoG, a common biomarker of oxidative stress. Due to misincorporation events,44 the formation of 8-oxoG is associated with G to T transversions, which represent a common mutational profile in a variety of cancer types.45,46 This lesion is specifically recognized and eliminated in base excision repair (BER) by OGG1.47 OGG1 is quite specific for excision of 8-oxoG and limited structurally related lesions, such as 2,6-diamino-4-hydroxy-5-formaidopyrimidine (FapyG), while excluding oxidized derivatives of adenine, thymine, cytosine, and uracil.48 We therefore utilized the OGG1 domain in a designed split-luciferase reassembly system, in which one half of luciferase is attached to MBD1 in order to localize the constructs to methylated DNA, and the other half is attached to OGG1(K249Q) for the specific interrogation of 8-oxoG lesions (Figure 2A). We utilized this point mutant of OGG1 since it lacks glycosylase/AP lyase activity and has been reported to have high affinity for 8-oxoG (Kd = 11.8 nM).49,50 The specific constructs designed comprised MBD1 (residues 1–69) attached to NLuciferase (residues 2–416) through a 15 amino acid (AA) linker, generating MBD1-NLuciferase, and OGG1(K249Q) (residues 12–325) attached to CLuciferase (residues 398–550) through an 18 AA linker, yielding CLuciferase-OGG1.
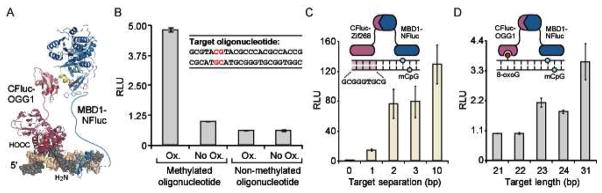
Split-luciferase biosensor for detection of 8-oxoguanine in DNA. (A) Cartoon representation of CFluc-OGG1 and MBD1-NFluc binding to a target oligonucleotide containing an 8-oxoguanine lesion adjacent to a methylated CpG dinucleotide. (B) The sensor pair was incubated with 7.1 nM of the indicated oligonucleotide (inset, site of methylation shown in red) that was exposed to oxidizing conditions (Ox.) of 30 μM CuCl2 and 1 mM H2O2 or 30 μM CuCl2 only (No Ox.). Results are the average of two independent trials, with luminescence readings presented as relative to the No Ox. control. (C) The CFluc-Zif268 and MBD1-NFluc sensors (inset) were incubated with a set of oligonucleotides (10 nM) with a varying separation (0, 1, 2, 3, or 10 bp) between the methyl-CpG site and the zinc finger recognition site. (D) The CFluc-OGG1 and MBD1-NFluc sensors (inset) were incubated with 7.1 nM of the same set of methylated oligonucleotides at described in (C) following exposure to oxidizing conditions.
We initially evaluated our sensor design utilizing a short, 23 base pair (bp) double-stranded DNA oligonucleotide: 5'-GCGTAmCGTACGCCCACGCCACCG, where mC represents 5-methylcytosine. This dsDNA oligonucleotide and the non-methylated equivalent were exposed to oxidizing conditions (1 mM H2O2 and 30 μM CuCl2), which produce hydroxyl radicals through Fenton-type redox chemistry.51 Among other oxidation products, the primary lesion generated under these conditions has been reported to be 8-oxoG.52,53 The complementary oligonucleotides are CG-rich, and contain 17 potentially oxidized guanines at various distances from the methylated-CpG site. We produced the split-protein biosensors, CLuciferase-OGG1 and MBD1-NLuciferase, in a cell-free translation system,39 followed by addition of the oxidized or non-oxidized oligonucleotides. Incubation of 7.1 nM of the oxidized methylated oligonucleotide resulted in a 5-fold signal increase over the non-oxidized equivalent (Figure 2B). Additionally, the non-methylated targets (oxidized or non-oxidized) did not produce significant signal in our system, confirming that the methylation site is essential for biosensor recognition, thus validating the choice of MBD1 as a generic localization domain. This initial validation of the biosensor indicates that oxidation of a methylated oligonucleotide provides accessible binding sites for the MBD1 and OGG1 domains, resulting in split-luciferase reassembly and gain of a luminescent signal.
To further evaluate the constraints that govern split-luciferase reassembly, we evaluated the ability of OGG1 and MBD1 to simultaneous bind to oligonucleotides of varying length. In previous iterations of DNA-templated reassembly of split GFP and split β-lactamase, a significant distance-dependence between protein recognition motifs in the targeted DNA was observed.36,38 To evaluate the effect of target length on split firefly luciferase reassembly, we first examined a previously established system for reporting on site-specific DNA methylation, in which a sequence-specific DNA binding ZF, Zif268, and an MBD are attached to halves of split luciferase.39 Upon incubation of the split-protein sensors with 10 nM DNA targets containing a single methylation site adjacent to a ZF binding site separated by 0, 1, 2, 3, or 10 bp, signal was observed for target lengths up to a 10 bp spacing (Figure 2C). Considering that closer spacing (0–1 bp) may preclude simultaneous binding of the detection domains,36 the wide range of accessible target lengths is presumably attributable to the length and flexible constitution of the linkers connecting the detection domains to split luciferase.42 To confirm this result and to investigate reassembly constraints associated with 8-oxoG detection, we next evaluated our DNA oxidation sensor using the same set of methylated oligonucleotides, which were exposed to oxidizing conditions of 1 mM H2O2 and 30 μM CuCl2. Using this sensor pair, the distance between the single methylation site and the oxidized guanine base is not explicitly known. However, the pattern of reassembly in the presence of 7.1 nM target very closely resembles that of the Zif268/MBD sensor pair, suggesting that reassembly is largely a function of the linkers and split protein utilized (Figure 2D). The reassembly observed for the DNA oxidation sensor utilizing longer DNA targets serves to establish that detection of 8-oxoG at sites distant from the site of methylation are potentially accessible using our biosensor design.
Following initial characterization of our biosensor for DNA oxidation, we next sought to optimize the sensor architecture. The preferred site for attachment of OGG1 to split luciferase may be affected by the accessibility and flexibility of the N- and C-termini and must be determined experimentally. Thus we generated a complementary set of biosensors consisting of MBD1 attached to the C-terminus of CLuciferase (CLuciferase-MBD1) and the OGG1 domain attached to the N-terminus of NLuciferase (OGG1-NLuciferase) (Figure 3A). We produced each complementary set of biosensors in a cell-free translation system, and then added 7.1 nM of oxidized or non-oxidized methylated 23 bp oligonucleotide. We found that each set of biosensors, CLuciferase-OGG1 with MBD1-NLuciferase or CLuciferase-MBD1 with OGG1-NLuciferase, produced similar signal over background (6.8-fold vs. 5.2-fold, respectively), indicating that the orientation of attachment is reasonably flexible for this geometry (Figure 3B). However, since the absolute signal associated with the CLuciferase-OGG1 + MBD1-NLuciferase pair was 4.5-fold higher, this sensor architecture was chosen for further optimization. Because the linker joining the protein fusion may also affect the efficiency of reassembly,42 a third OGG1 fusion construct (CLuciferase-33AA-OGG1) was generated that encodes a 33 AA linker for comparison to the original construct containing an 18 AA linker (Figure 3C). We produced each complementary set of biosensors, CLuciferase-18AA-OGG1 with MBD1-NLuciferase and CLuciferase-33AA-OGG1 with MBD1-NLuciferase, in a cell-free translation system, followed by addition of 7.1 nM of oxidized or non-oxidized methylated oligonucleotide. We found that the 18 AA linker construct contributed a greater relative signal (6.8-fold) than the 33 AA linker construct (3.6-fold) leading us to choose this sensor architecture for further experiments (Figure 3D).

Evaluation of OGG1 sensor architecture. (A) Two sets of oxidation sensors, CFluc-OGG1 with MBD1-NFluc and CFluc-MBD1 with OGG1-NFluc, were compared to evaluate the effect of fusion site on luciferase reassembly. (B) The sensor pairs were reassembled in the presence of oxidized vs. non-oxidized oligonucleotides at 7.1 nM. (C) Two sets of oxidation sensors, CFluc-18AA-OGG1 with MBD1-NFluc and CFluc-33AA-OGG1 with MBD1-NFluc, were compared to evaluate the effect of linker length on luciferase reassembly. (D) The sensor pairs were reassembled in the presence of oxidized vs. non-oxidized oligonucleotides at 7.1 nM.
Evaluation of the sensor for detection of 8-oxoguanine in plasmid DNA
We next sought to evaluate our sensor for detecting 8-oxoG in the context of a much larger DNA target that would better mimic genomic DNA. Towards this goal we chose to utilize a plasmid DNA target containing multiple sites of methylated cytosine. To generate a plasmid target, a high level of CpG methylation must first be achieved since localization of the sensor to DNA is mediated by a domain that recognizes methylation. Because typical laboratory strains of bacteria lack an endogenous methylase specific for generating 5-methylcytosine in CpG dinucleotides, we exogenously methylated a 7667 bp pETDuet-derived plasmid using M.SssI CpG methyltransferase, creating pETm. The degree of methylation was assessed by exposure to a methylation-sensitive restriction enzyme, BstUI, which cleaves only non-methylated 5'-CGCG sites. Only the non-methylated plasmid was digested by BstUI, while the methylated plasmid was fully protected from digestion (Figure 4A, compare lanes 2 and 3). Next, the methylated plasmid was exposed to oxidizing conditions consisting of 1 mM H2O2 and 30 μM CuCl2. We produced the optimized split-protein biosensors, CLuciferase-OGG1 (18 AA linker) and MBD1-NLuciferase, in a cell-free translation system, followed by addition of oxidized or non-oxidized pETm. Upon incubation of 8 ng of oxidized pETm with the split-luciferase constructs, a 6-fold change in luminescent signal over untreated DNA was achieved (Figure 4B). This gain in signal in an oxidation- and methylation-dependent fashion, confirmed that the 8-oxoguanine sensor may potentially be used for detection of DNA oxidation in a genomic target with extensive methylation at CpG sites.
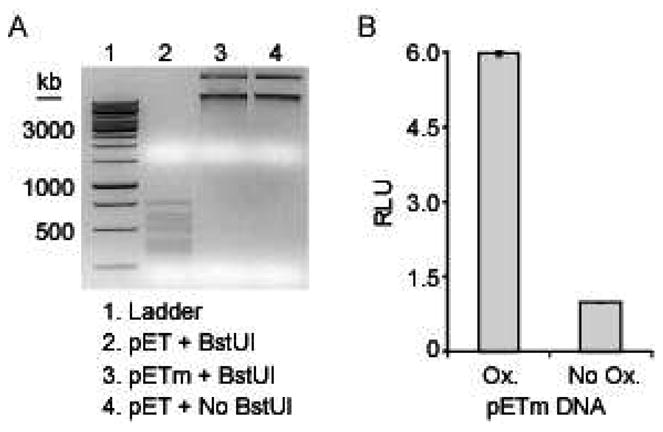
Detection of oxidation in plasmid DNA using the 8-oxoG sensor. (A) Exogenously methylated plasmid DNA (pETm) was compared to the non-methylated plasmid (pET) by restriction enzyme analysis using BstUI, which cleaves non-methylated 5'-CGCG sites. (B) The CLuciferase-OGG1 and MBD1-NLuciferase sensors were incubated with 8 ng of pETm that was exposed to oxidizing conditions (Ox.) of 30 μM CuCl2 and 1 mM H2O2 or non-oxidizing conditions of 30 μM CuCl2 only (No Ox.).
Application of the sensor for detection of 8-oxoguanine in mammalian DNA
We next turned toward confirming the broader utility of our sensor by evaluating its ability to directly detect the oxidation-dependent modification of natively methylated mammalian DNA. We isolated genomic DNA from HeLa cultures and then exposed it to 1 mM H2O2 in the presence of 30 μM CuCl2 to generate 8-oxoG lesions. In the presence of our split proteins, the oxidized HeLa DNA (50 ng) generated a 27-fold signal induction over the non-treated equivalent, thus confirming the utility of our sensor for detection of lesions in mammalian genomic DNA (Figure 5A). Importantly, evaluation of oxidized HeLa DNA using a split-luciferase based methylation sensor did not result in an observable decrease in luminescence, indicating that oxidation at guanines in mCpG sites does not significantly affect MBD1 binding (Supplementary Information).54

Detection of oxidation in HeLa DNA using the 8-oxoG sensor. (A) The CLuciferase-OGG1 and MBD1-NLuciferase sensors were incubated with 50 ng of DNA isolated from HeLa cells that was exposed to oxidizing conditions (Ox.) of 30 μM CuCl2 and 1 mM H2O2 for 30 min or non-oxidizing conditions of 30 μM CuCl2 only (No Ox.). (B) The sensor pair was incubated with 50 ng of HeLa DNA that was treated for 30 min with 1 mM H2O2 in the presence of 15, 30, or 60 μM CuCl2. (C) The sensor pair was incubated with 50 ng of HeLa DNA that was treated with 1 mM H2O2 and 60 μM CuCl2 for 10, 20 or 30 min.
In order to probe oxidation-dependent changes in signal, we next evaluated the extent of DNA damage induced by varying the CuCl2 concentration (15, 30, or 60 μM) present during treatment of HeLa DNA with 1 mM H2O2 for 30 min. The damaged HeLa DNA was exposed to our translated split-protein sensors. A CuCl2 dose-dependent increase in luminescence was observed, wherein the 15, 30, and 60 μM CuCl2 treated oxidized samples provided 13-, 27-, and 32-fold signal over the non-oxidized equivalents (Figure 5B). The effect of varying the H2O2 concentration (0.5 mM, 1 mM, and 1.5 mM) was also investigated, although this did not have a great effect on sensor response at a given concentration of CuCl2 (Supplementary Information). Finally, the exposure time for the 1 mM H2O2 and 60 μM CuCl2 treatment was varied (t = 10, 20, and 30 min) to identify the final optimized conditions. Equivalent signals were observed for the 10 and 20 min treatments (~37-fold), while a decrease in signal to 31-fold was observed as the time of oxidation was increased to 30 min, possibly indicating the formation of alternate oxidation products (for example, further oxidation of 8-oxoguanine55 or increased strand breakage51) that are not recognized by OGG1 (Figure 5C). At times longer than 2 h, the 8-oxoG sensor no longer bound the oxidized target with greater than 2-fold preference over the non-oxidized equivalent (Supplementary Information). Gel electrophoresis of the oxidized DNA revealed a direct correlation between the time of oxidation and increased degradation of the DNA target, possibly preventing OGG1 binding (Supplementary Information). Finally, we chose to use an independent enzyme-linked immunosorbent assay (ELISA), which was more sensitive than LC-MS, for quantifying 8-oxoguanine from hydrolyzed DNA. We directly confirmed the presence of 8-oxoG lesions using ELISA of hydrolyzed nucleosides from both oxidized and non-oxidized HeLa DNA. By comparison to the ELISA standard curve utilizing pure 8-oxoguanosine, it was determined that DNA exposed to oxidizing conditions of 1 mM H2O2 and 60 μM CuCl2 for 10 min yields ~ 1200 8-oxoG lesions in 106 DNA bases (Supplementary Information), which is consistent with results from previous studies using liquid chromatography with electrochemical detection (LC-ECD).56 The equivalent non-oxidized samples did not yield any 8-oxoguanosine detectable in the ELISA. Considering the proportion of 8-oxoG lesions generated using our optimized oxidation conditions, the 50 ng DNA sample evaluated with our split-protein biosensors contains ~ 200 fmol of 8-oxoG. Further confirmation of 8-oxoG in the oxidized HeLa DNA by LC-MS was not feasible due to limitations in instrument sensitivity of ~ 30 pmol 8-oxoguanosine. These results illustrate the ability of our sensors to report on the relative degree of DNA damage generated by modulating the damage-inducing conditions.
Initial design and validation of a split-luciferase biosensor for UV photodamage
To further investigate the generality of our DNA damage sensor platform and to expand the repertoire of detectable lesions, we next designed a biosensor for detection of UV-induced photoproducts including cyclobutane pyrimidine dimers (CPDs) and 6–4 pyrimidine-pyrimidone photoproducts (6-4PPs). If left unrepaired these bulky DNA adducts can cause base misincorporation or stalled DNA replication.57 DNA photoproducts are reported to be initially recognized in the global genome nucleotide excision repair (GG-NER) pathway by the DDB1-DDB2 complex,58–60 which binds 6-4PPs (Kd ~ 63 pM), trans, syn-CPDs (Kd ~ 77 pM), and Dewar thymine dimers (Kd ~ 210 pM),61 as well as other lesions that cause distortions or flexibility in the DNA.62–64 The DDB2 monomer has also recently been reported to be capable of independently binding photoproducts in DNA,65,66 indicating its potential utility in our split-protein reassembly system. For this detection system, we chose to use the Danio rerio DDB2 ortholog of which a high resolution crystal structure has recently been solved.27 The domain utilized herein has 52% identity and 73% similarity to the Homo sapiens protein, and the overlaid structures have a Cα-backbone RMSD of 1.1 Å.
Thus for the design of a UV-induced photoproduct(s) sensor, we attached DDB2 (residues 94–457) to the C-terminus of CLuciferase (residues 398–550) through an 18 AA linker and utilized MBD1-NLuciferase for localization to methylated target DNA (Figure 6A). We initially evaluated our sensors using a 23 bp double-stranded methylated oligonucleotide target 5'-GCGTAmCGTACGCCCACGCCACCG (mC represents 5-methylcytosine), which was exposed to 2 h of UV radiation from a germicidal lamp (peak output at 254 nm). The expected modifications induced by exposure to UVC radiation have been previously described, with 5'-TT and 5'-TC dinucleotides being more photoreactive than 5'-CT and 5'-CC.67 In our oligonucleotide target only 5'-CC dinucleotides are present, which, in contrast to the other dipyrimidine sequences, yield the 6–4 photoproduct (~50 % of products) as the predominant lesion over the cyclobutane dimer and the Dewar valence isomer.67
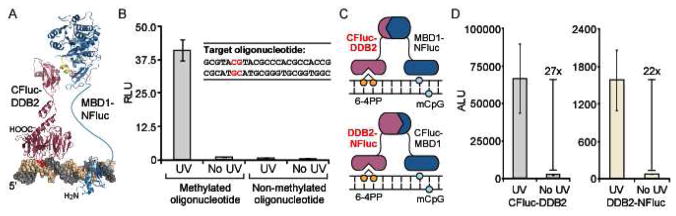
Split-luciferase biosensor for the detection of UV damage in DNA. (A) Cartoon representation showing CFluc-DDB2 and MBD1-NFluc bound to a DNA target containing a 6-4 pyrimidine-pyrimidone photoproduct adjacent to a methylated CpG dinucleotide. (B) The sensor pair was incubated with 7.1 nM of methylated or non-methylated oligonucleotides (inset, site of methylation shown in red) that were exposed to 2 h of UV radiation (254 nm) or not exposed to UV radiation (No UV). Results are the average of two independent trials, with luminescence readings presented as relative to the No UV control. (C) Two sets of UV damage sensors, CFluc-DDB2 with MBD1-NFluc and CFluc-MBD1 with DDB2-NFluc, were compared to evaluate the effect of fusion site on luciferase reassembly. (D) Each set of biosensors shown in (C) was reassembled in the presence of 7.1 nM of methylated oligonucleotide exposed to 2 h of UV radiation.
Therefore, we expect a high yield of cytosine 6–4 adducts in the oligonucleotide target. We generated our split proteins, CLuciferase-DDB2 and MBD1-NLuciferase, in a cell-free translation system, followed by addition of the UV-irradiated oligonucleotide. Incubation with 7.1 nM UV-induced target resulted in a 45-fold enhancement of reassembled luciferase activity, establishing a DDB2-specific interaction with irradiated DNA (Figure 6B). Additionally, signal was not observed for the non-methylated oligonucleotides, confirming the requirement for methylation in the target for MBD1-mediated sensor recognition.
We next pursued optimization of the sensor architecture by altering the sites of attachment of the DDB2 and MBD1 domains, creating CLuciferase-MBD1 and DDB2-NLuciferase (Figure 6C). We produced the two complementary sets of biosensors in a cell-free translation system, and then added 7.1 nM of UV or non-UV irradiated oligonucleotides. Despite similar relative signals for the two pairs (27-fold vs. 22-fold), we found that the original biosensor pair, CLuciferase-DDB2 with MBD1-NLuciferase, conferred a significantly higher absolute signal (42-fold) than the newly generated CLuciferase-MBD1 and DDB2-NLuciferase pair (Figure 6D). Therefore, the site of attachment of the DDB2 domain has a significant effect on reassembly efficiency in this case as compared to OGG1, and the original sensor geometry was utilized in subsequent experiments.
Detection of UV damage in plasmid DNA
We next sought to evaluate our sensors for detection of UV damage in exogenously methylated plasmid DNA. To generate UV-induced lesions, we exposed the exogenously methylated plasmid target, pETm, to 2 h UVC light from a germicidal lamp (peak output at 254 nm). To directly confirm the presence of UV-photoproducts in pETm, the UV-treated and untreated plasmids were incubated with MseI, which cleaves the sequence 5'-TTAA between the adjacent thymines (Supplementary Information), while the presence of thymine dimers prevents MseI endonuclease activity. Cleavage was only observed with the untreated plasmid, while the UV-treated plasmid was protected from nuclease cleavage by the formation of pyrimidine dimers (Figure 7A, compare lanes 2 and 4). In order to test the ability of our sensors to detect this UV-induced damage, we expressed the split proteins, CLuciferase-DDB2 and MBD1-NLuciferase, in a cell-free translation system, followed by addition of the UV-irradiated pETm target. We observed a 200-fold increase in signal when compared to non-irradiated target (8 ng), indicating detection of a significant gain in pyrimidine dimers (Figure 7B). Detection of UV damage in HeLa DNA. We next assessed our UV damage sensor in the context of mammalian DNA. We isolated natively methylated genomic DNA from HeLa cells, followed by a 2 h exposure to UVC light. When 50 ng of the UV-treated HeLa DNA was added to the sensors, we observed a ~100-fold increase in signal when compared to an equivalent amount of non-irradiated target (Figure 8A). It has been previously shown that 5-methylcytosine has a reactivity similar to unmodified cytosine for forming cyclobutane pyrimidine dimers upon exposure to UVC (254 nm) radiation.68 Therefore, sequences such as 5'-TmCG and 5'-CmCG may form cyclobutane dimers. However, incubation of the HeLa DNA targets with our methylation-specific biosensor revealed no change in luminescent signal upon UV-irradiation, indicating that MBD1 binding is not significantly compromised by UV treatment of the DNA target (Supplementary Information). Given the sensitivity of the DDB2 domain, a titration of UV-treated HeLa DNA revealed a linear correlation within the range tested (2.5–12.5 ng), with a detection limit of at least 2.5 ng, which was detectable over the average background signal plus three standard deviations (99% confidence level) (Figure 8B).
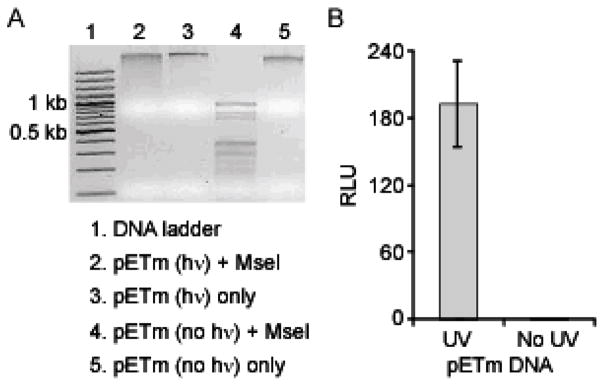
Detection of UV photoproducts in plasmid DNA using the UV damage sensor. (A) Methylated plasmid DNA, pETm, was treated with UVC light (hν) for 2 h or not exposed to UV light (No hν). Irradiated and non-irradiated pETm were incubated with the endonuclease MseI, which is inhibited by the formation of thymine dimers in its recognition sequence (5'-TTAA). (B) The CLuciferase-DDB2 and MBD1-NLuciferase sensor system was incubated with 8 ng of pETm treated with 2 h of UV radiation or no UV.

Detection of UV photoproducts in mammalian DNA using the UV damage sensor. (A) The CLuciferase-DDB2 and MBD1-NLuciferase sensor system was incubated with 50 ng HeLa DNA that was treated with 2 h UVC with a peak output at 254 nm or No UV. (B) The sensor pair was incubated with 12.5, 8.3, 5.6, 3.7, or 2.5 ng of HeLa DNA treated as described in (A), yielding a linear response. (C) (inset) UVC irradiation of HeLa cells results in the formation of DNA photoproducts, such as the 6–4 photoproduct (6–4 PP). The sensor pair was incubated with 50 ng DNA isolated from HeLa cells treated with 10 min of UV exposure or No UV.
Detection of UV damage induced in mammalian cells
Following establishment of our UV damage sensors for the detection of UV-induced photoproducts in exogenously damaged DNA, we proceeded to evaluate our split proteins for detection of DNA damage initiated in live cells. Because endogenous damage may result in cellular toxicity that could prevent further analysis of DNA samples, we first sought to determine an appropriate length of UV exposure time during which cell viability was not greatly compromised. We exposed cells to UV radiation for varying lengths of time and evaluated cell survival using the formazan-based MTT assay. Only ~70% of cells remained viable after a 2 h exposure, which reflects typically observed toxicity associated with longer or more intense UV exposure (Supplementary Information).69,70 However, a 10 min exposure resulted in greater than 95% viability. Therefore, to determine if our sensors were capable of detecting UV damage in DNA directly isolated from cultures, we exposed HeLa cells to UV radiation for only 10 min, followed by immediate harvesting of genomic DNA to limit repair of DNA lesions. Isolated DNA was quantified, and 50 ng was incubated with the UV damage sensor, resulting in a 25-fold signal over background levels of damage (Figure 8C). To further validate the ability of our sensor to detect DNA damage induced in cell populations, various UV exposure times, ranging from 15 min to 1 min, were investigated (Supplementary Information). Thus, these results demonstrate that this split-protein sensor can be utilized for the direct detection of UV lesions in genomic DNA induced in a cellular context using a method that does not require extensive or destructive sample processing following isolation of genomic DNA.
Specificity of the designed DNA damage sensors
Given the highly specific nature of the endogenous domains involved in repair of cellular DNA damage, the designed probes presumably retain this specificity. In an effort to begin validation of the specificity of the designed probes for their intended targets, we evaluated the ability of DDB2 and OGG1 to orthogonally detect damage accrued from exposure to oxidizing conditions (1 mM H2O2 and 60 μM CuCl2) or 2 h UVC radiation. The sensor pair for UV photoproducts, CLuciferase-DDB2 and MBD1-NLuciferase, was generated in a cell-free translation system, followed by incubation with 50 ng HeLa DNA treated with damaging conditions (UV or oxidation) or the corresponding “No Damage” controls. The UV damage sensor detected the induced DNA photoproducts with 152-fold relative signal, while 50-fold relative signal was observed in the cased of oxidized DNA (Figure 9A). Therefore, the UV damage sensor gave a 67% higher relative signal for the UV-irradiated DNA, as compared to that observed for the oxidized target. This off-target reassembly may either be attributable to 1) the ability of DDB2 to bind to lesions other than pyrimidine dimer photoproducts (such as 8-oxoG) or 2) the propensity for Fenton-type oxidation to induce alternate types of damage (such as those bound by DDB2). Although it has previously been established that DDB2 does not bind to 8-oxoG lesions in DNA,62 the observed signal in the presence of the oxidized DNA target may potentially be attributable to the formation of alternate oxidized DNA lesions, such as abasic sites,71 that are bound by DDB2.27,62–64 Next, the sensor pair for 8-oxoG, CLuciferase-OGG1 and MBD1-NLuciferase, was generated in a cell-free translation system, followed by incubation with 50 ng HeLa DNA treated with damaging conditions (UV or oxidation) or the corresponding “No Damage” controls. The 8-oxoG sensor detected the induced oxidation with 37-fold relative signal, while 9-fold relative signal was observed in the case of UV-irradiated DNA (Figure 9B). Therefore, the 8-oxoG sensor gave a 75% higher relative signal in the presence of the oxidized target, as compared to that observed for the UV-irradiated DNA. The observed signal in the presence of the UV-irradiated DNA target may be attributable to the accrual of base oxidation that is recognized by OGG1, considering that UVC light has been demonstrated to indirectly induce DNA oxidation through the generation of singlet oxygen.72,73 Therefore, initial results suggest a significant degree of specificity associated with the designed DNA damage biosensors. However, a more rigorous analysis is required to determine the absolute specificity of the designed biosensors for their intended targets. Additionally, the use of alternate protein domains or optimization of the OGG1 and DDB2 domains may be employed to further improve the specificity for a particular lesion.
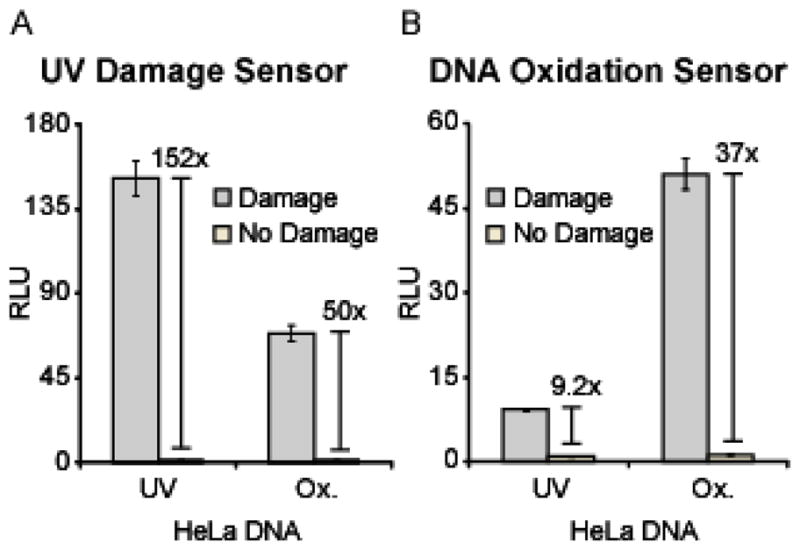
Specificity of detection using the UV damage and DNA oxidation sensors. (A) The CLuciferase-DDB2 and MBD1-NLuciferase sensor pair was incubated in the presence of 50 ng UV-irradiated or oxidized HeLa DNA (“Damage”) or the corresponding “No Damage” controls. (B) The CLuciferase-OGG1 and MBD1-NLuciferase sensor pair was incubated with the same targets as in (A).
Conclusion
The genetic information in our cells is constantly threatened by ubiquitous genotoxins, and cellular viability is predicated on the fidelity of DNA repair mechanisms. For the DNA repair machinery to be successful in consistently eradicating DNA damage, it is likely that mechanisms exist for identifying lesions in a large (> 108-fold) excess of unmodified bases, with current models ranging from protein sliding to extrahelical base-trapping74 and charge transport75 mechanisms. The ongoing study of DNA repair proteins with their exquisite selectivity and sensitivity has allowed us to co-opt them for the development of two new DNA damage sensors.
Building upon our previous ternary sensor architecture for site-specific recognition of methyl-CpG dinucleotides, which utilized an MBD in conjunction with a sequence-specific ZF domain, we have successfully developed a robust and potentially general methodology for reporting on DNA oxidation and UV damage. Specifically, we established that MBD1 can serve as a generic localization domain for targeting our split-protein biosensors to methylated DNA, which occurs frequently in the genome.76,77 To report on chemical modifications to DNA, we utilized the base excision repair glycosylase OGG1 for binding selectively to 8-oxoguanine and the nucleotide excision repair protein DDB2 for reporting on the presence of pyrimidine dimer photoproducts. Based on the modular and general nature of this detection platform, there are a number of DNA-damage recognition modules that have potential utility in this split-protein reassembly system, including additional glycosylases and proteins involved in recognition of mismatched bases.78,79 By employing the genetically encoded split luciferase as the split-signaling domain, we are able to produce our biosensors in vitro in 1.5 h without further purification, providing ease of access to the biosensor. Furthermore, the sensitivity associated with a luminescent readout allows for detection of lesions present in low nanogram DNA samples. However, high-affinity single-chain antibody fragments, which have recently been established for use in the split-luciferase assay, may provide additional sensitivity to the detection platform.80
We envision that the genetically encoded sensors described herein or their analogues25 may potentially be used directly in living cells with appropriate imaging modalities. The ability to control cellular processes, including nucleic acid-templated chemical reactions,81 enzymatic manipulation of the genetic information, or modulation of transcription,82 by pursuing unique DNA targets has been the focus of biomedical and synthetic biology applications. Complementary to the established utility of sequence-specific DNA binding ZFs as fusions for programmed targeting of nucleases,83 methylases,84 and integrases,85 we suggest a potentially parallel approach that targets chemical modifications to DNA. Thus the DNA-damage dependent split-protein reassembly approach described herein demonstrates the ability to generate conditionally activated split proteins that may be redesigned to respond to damage and thus modulate cell fate.86
Experimental Methods
General materials
All reagents were from Sigma-Aldrich unless otherwise noted. Cloning enzymes were purchased from New England Biolabs. Copper (II) chloride and hydrogen peroxide were from J.T. Baker. Tris-HCl and DTT were obtained from Research Products International Corp. The luciferase luminescence reagents were from either Promega or Luceome Biotechnologies.
Cell culture
HeLa cells were maintained at 37 °C and 5% CO2 in 90% DMEM/F12 1:1 media (Lonza) supplemented with 10% FBS (Lonza), penicillin-streptomycin (Mediatech), and ampotericin B (JR Scientific). To obtain mammalian genomic DNA, HeLa DNA was isolated from 106 trypsinized cells using the Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions.
Cloning
The luciferase halves utilized in all biosensors were split as previously described.33,39 DNA encoding the damage detection domains, Homo sapiens OGG1 (residues 12–325) and Danio rerio DDB2 (residues 94–457), was amplified from existing plasmids obtained from Open Biosystems (I.M.A.G.E. clone IDs: 3350168 and 7402966, respectively). The resulting PCR products were introduced into a pcDNA3.1(+) vector (Invitrogen) containing CLuciferase (residues 398–505) followed by an 18 AA linker, creating pcDNA3.1(+)-CLuciferase-OGG1 and pcDNA3.1(+)-CLuciferase-DDB2. Site-directed mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) was carried out on pcDNA3.1(+)-CLuciferase-OGG1 to generate the corresponding plasmid containing the catalytically inactive OGG1(K249Q) mutant. Cloning of the other sensor architectures is described in the Supplementary Information. The pcDNA3.1(+)-MBD1-NLuciferase construct was generated by amplifying MBD1-NLuciferase (MBD1 synthesized by GenScript) from an existing plasmid and inserting it into the pcDNA3.1(+) vector. All sequences were confirmed using dideoxynucleotide sequencing.
Oligonucleotide target preparation
All oligonucleotide targets were obtained HPLC purified from Integrated DNA Technologies and are listed in Table S1 (Supplementary Information). All designed DNA targets were annealed in 1× annealing buffer (10 mM Tris-HCl, pH 7.9, 150 mM NaCl, 10 mM MgCl2, 1mM DTT) using the following procedure: heating to 95 °C for 7 min, cooling to 56 °C at a rate of 0.1 °C sec−1, equilibrating at 56 °C for 5 min, cooling to 25 °C at a rate of 0.1 °C sec−1 and finally equilibrating at 25 °C for 10 min using a Labnet Multi Gene II thermocycler.
Generation of DNA oxidation IN VITRO
A pETDuet-derived plasmid (7667 bp) was fully methylated, generating pETm, by incubation with S-adenosylmethionine and M.SssI CpG methyltransferase (New England Biolabs) according to the manufacturer’s protocol. The extent of protection was determined by exposure to the methylation sensitive endonuclease BstUI. To introduce oxidation, 708 nM annealed oligonucleotide, 8 ng/μL pETm, or 50 ng/μL HeLa DNA was treated with 1 mM H2O2 in the presence of 30 μM CuCl2 for 30 min at RT, followed by quenching with 1 mM EDTA. To assess the effect of Cu2+ concentration on induction of base damage, 50 ng/μL HeLa DNA was treated with 1 mM H2O2 in the presence of 15, 30, or 60 μM CuCl2 for 30 min, followed by quenching with 1 mM EDTA. To assess the effect of oxidation time on induction of base damage, 50 ng/μL HeLa DNA was treated with 60 μM CuCl2 and 1 mM H2O2 for 10, 20, or 30 min, followed by quenching with 1 mM EDTA. In all cases control reactions were carried out with CuCl2 in the absence of H2O2, followed by addition of 1 mM EDTA. The DNA was analyzed without further purification.
Detection of DNA oxidation
Preparation of mRNA is described in the Supplementary Information. The mRNAs encoding CLuciferase-OGG1(K249Q) and MBD1-NLuciferase were translated in the Flexi Rabbit Reticulocyte Lysate System (Promega), consisting of 25 μL reactions prepared according to the manufacturer’s instructions. A typical reaction was performed at 30 °C for 1.5 h and contained 0.2 pmol of each mRNA transcript. Following translation, 1.25 μL of 708 nM annealed oligonucleotide, 8 ng/μL pETm, or 50 ng/μL HeLa DNA target was added to 23.75 μL of the translation and binding was allowed to occur for 1 h at 4 °C. Activity was monitored as a luminescent signal produced upon addition of luminescence reagent, where 20 μL of each translation was added to 80 μL of reagent. Luminescent readings were acquired 1 min after mixing using a Turner TD-20e Luminometer with a 10 s integration time. Results are presented as the relative average of two independent experiments.
Optimization of OGG1 sensor architecture
The mRNAs encoding the three sensor pairs (CLuciferase-18 AA-OGG1 + MBD1-NLuciferase, CLuciferase-MBD1 + OGG1-15 AA-NLuciferase, and CLuciferase-33AA-OGG1 + MBD1-NLuciferase) were translated in the Flexi Rabbit Reticulocyte Lysate System (Promega), consisting of 25 μL reactions prepared according to the manufacturer’s instructions. A typical reaction was performed at 30 °C for 1.5 h and contained 0.2 pmol of a pair of mRNA transcripts. Following translation, 1.25 μL of 708 nM oxidized oligonucleotide (23 bp) was added to 23.75 μL of the translation, and binding was allowed to occur for 1 h at 4 °C. Activity was monitored as a luminescent signal produced upon addition of luminescence reagent, where 20 μL of each translation was added to 80 μL of reagent. Luminescent readings were acquired 1 min after mixing using a Turner TD-20e Luminometer with a 10 s integration time. Results are presented as the average readings for two independent experiments.
Evaluation of target length dependence on split-luciferase reassembly
The mRNAs encoding the CLuciferase-Zif268 and MBD1-NLuciferase were translated in the Flexi Rabbit Reticulocyte Lysate System (Promega), consisting of 25 μL reactions prepared according to the manufacturer’s instructions. A typical reaction was performed at 30 °C for 1.5 h and contained 10 μM ZnCl2, 1 pmol of each mRNA transcript, and 1.25 μL of 708 nM methylated oligonucleotide with 0, 1, 2, 3, or 10 bp spacing between the methylation site and the zinc finger binding site. Activity was monitored as a luminescent signal produced upon addition of luminescence reagent, where 20 μL of each translation was added to 80 μL of reagent. Luminescent readings were acquired 1 min after mixing using a Turner TD-20e Luminometer with a 10 s integration time. Results are presented as the relative average of two independent experiments.
Induction of UV damage
For UV damage induction, 708 nM annealed oligonucleotide, 8 ng/μL pETm, or 362 ng/μL HeLa DNA was introduced into a quartz cuvette and exposed for 2 h to a germicidal UV lamp (LightSources, Inc.) with a UV fluency of 240 μW/cm2 at 254 nm according to the manufacturer. The DNA was collected and analyzed without further preparation. To confirm the presence of thymine dimer formation in the UV-irradiated pETm, a nuclease protection assay was performed. Plasmid was incubated with the MseI restriction endonuclease, which cleaves 5'-TTAA sequences between adjacent thymines, followed by agarose gel electrophoresis. To generate UV-induced lesions in vivo, HeLa cells were plated in complete medium at 106 cells per 60 mm dish 24 h prior to treatment. Cells were exposed for 10 min, followed by immediate trypsinization and isolation of genomic DNA using the Wizard® Genomic DNA Purification Kit according to the manufacturer’s instructions. To determine cellular toxicity associated with excessive UV damage, 2.4 x 104 cells per well in a 96 well plate were assayed with a methylthiazolyl tetrazolium (MTT)-based in vitro Toxicology Assay Kit (Sigma) after UV exposure for 2 h, 1 h, 10 min, or no UV. MTT formazan absorbance was measured at 570 nm using a SynergyTM 2 microplate reader (BioTek® Instruments, Inc.), and results are presented as the average of two independent trials.
Detection of UV damage
The mRNAs encoding the split-proteins CLuciferase-DDB2 and MBD1-NLuciferase were translated in the Flexi Rabbit Reticulocyte Lysate System (Promega), consisting of 25 μL reactions prepared according to the manufacturer’s instructions. A typical reaction was performed at 30 °C for 1.5 h and contained 0.2 pmol of each mRNA transcript. Following translation, 1.25 μL of 708 nM UV-treated oligonucleotide, 8 ng/μL UV-treated pETm, 50 ng/μL UV-treated HeLa DNA, or 50 ng/μL HeLa DNA isolated from UV-treated cells was added to 23.75 μL of the translation and binding was allowed to occur for 1 h at 4 °C. To determine the lowest detectable amount of UV-treated HeLa DNA, 1.25 μL of 12.5, 8.3, 5.6, 3.7, or 2.5 ng/μL dilutions of treated or untreated DNA was added to 23.75 μL of the translation. In all cases activity was monitored as a luminescent signal produced upon addition of luminescence reagent, where 20 μL of each translation was added to 80 μL of reagent. Luminescent readings were acquired 1 min after mixing using a Turner TD-20e Luminometer with a 10 s integration time. Results are presented as the relative average of two independent experiments.
Optimization of DDB2 sensor architecture
The mRNAs encoding the two sensor pairs (CLuciferase-DDB2 + MBD1-NLuciferase and CLuciferase-MBD1 + DDB2-NLuciferase) were translated in the Flexi Rabbit Reticulocyte Lysate System (Promega), consisting of 25 μL reactions prepared according to the manufacturer’s instructions. A typical reaction was performed at 30 °C for 1.5 h and contained 0.2 pmol of an mRNA transcript pair. Following translation, 1.25 μL of 2 h UV-irradiated 708 nM oligonucleotide (23 bp) was added to 23.75 μL of the translation and binding was allowed to occur for 1 h at 4 °C. Activity was monitored as a luminescent signal produced upon addition of luminescence reagent, where 20 μL of each translation was added to 80 μL of reagent. Luminescent readings were acquired 1 min after mixing using a Turner TD-20e Luminometer with a 10 s integration time. Results are presented as the average readings for two independent experiments.
Acknowledgments
We thank D. Piwnica-Worms for the luciferase constructs. This research was supported by R21CA143661 and R01GM077403 (NIH). J. L. F. thanks the Philanthropic Education Organization (P.E.O.) Chapter BB and the University of Arizona TRIF Imaging Fellowship for financial support. Molecular structures were rendered using PyMol; DeLano, W. L. (www.pymol.org).
Footnotes
Supporting Information Available. DNA and protein sequences of the split-luciferase sensors, methylated oligonucleotide sequences, and experimental details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
Full text links
Read article at publisher's site: https://doi.org/10.1021/ja1116606
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3206586?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1021/ja1116606
Article citations
Single Cell Determination of 7,8-dihydro-8-oxo-2'-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling.
Molecules, 28(11):4326, 25 May 2023
Cited by: 3 articles | PMID: 37298802 | PMCID: PMC10254197
Development of Novel Functional Molecules Targeting DNA and RNA.
Chem Pharm Bull (Tokyo), 67(6):505-518, 01 Jan 2019
Cited by: 1 article | PMID: 31155555
Review
In Vitro Fluorogenic Real-Time Assay of the Repair of Oxidative DNA Damage.
Chembiochem, 16(11):1637-1646, 12 Jun 2015
Cited by: 19 articles | PMID: 26073452 | PMCID: PMC4586133
Discrimination between 8-oxo-2'-deoxyguanosine and 2'-deoxyguanosine in DNA by the single nucleotide primer extension reaction with adap triphosphate.
Angew Chem Int Ed Engl, 54(17):5147-5151, 26 Feb 2015
Cited by: 7 articles | PMID: 25727406
Metal-mediated DNA damage and cell death: mechanisms, detection methods, and cellular consequences.
Metallomics, 6(8):1358-1381, 01 Aug 2014
Cited by: 41 articles | PMID: 24788233
Review
Go to all (6) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Repair system of 7, 8-dihydro-8-oxoguanine as a defense line against carcinogenesis.
J Radiat Res, 49(4):329-340, 03 Jul 2008
Cited by: 42 articles | PMID: 18596371
Review
Excision of 8-oxoguanine from methylated CpG dinucleotides by human 8-oxoguanine DNA glycosylase.
FEBS Lett, 587(18):3129-3134, 13 Aug 2013
Cited by: 12 articles | PMID: 23954288
NEIL1 and NEIL2 Are Recruited as Potential Backup for OGG1 upon OGG1 Depletion or Inhibition by TH5487.
Int J Mol Sci, 22(9):4542, 27 Apr 2021
Cited by: 8 articles | PMID: 33925271 | PMCID: PMC8123590
Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases.
Free Radic Biol Med, 107:179-201, 27 Nov 2016
Cited by: 70 articles | PMID: 27903453
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R21 CA143661-01
Grant ID: R21CA141974
Grant ID: R21 CA143661
Grant ID: R21 CA143661-02
NIAID NIH HHS (1)
Grant ID: R01AI068414
NIGMS NIH HHS (1)
Grant ID: R01 GM077403





