Abstract
Free full text

Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health
Abstract
The four dengue virus (DENV) serotypes that circulate among humans emerged independently from ancestral sylvatic progenitors that were present in non–human primates, following the establishment of human populations that were large and dense enough to support continuous inter-human transmission by mosquitoes. This ancestral sylvatic–DENV transmission cycle still exists and is maintained in non-human primates and Aedes mosquitoes in the forests of Southeast Asia and West Africa. Here, we provide an overview of the ecology and molecular evolution of sylvatic DENV and its potential for adaptation to human transmission. We also emphasize how the study of sylvatic DENV will improve our ability to understand, predict and, ideally, avert further DENV emergence.
Dengue virus (DENV) is a positive-sense, single-stranded RNA virus of the genus Flavivirus (family Flaviviridae) that uses mosquitoes of the genus Aedes as vectors for transmission among primates (FIG. 1). DENV occurs as four antigenically distinct but genetically related serotypes (DENV-1 to DENV-4) within the dengue antigenic complex1. The molecular biology of DENV is summarized in BOX 1. In recent decades, DENV transmission among humans has intensified, and the virus currently infects 100 million people each year worldwide, being found in over 100 countries2. Most DENV infections are subclinical or result in classical dengue fever, which is characterized by fever, muscle and joint pain, and rash. However, approximately 0.5% of infections result in the most severe manifestation of the disease, dengue haemorrhagic fever (DHF), which can be fatal in as many as 5% of cases. Infection with a given serotype results in lifelong homologous immunity to that serotype but increases the risk of haemorrhagic fever upon infection by a heterologous serotype3. The principal risk factors for developing DHF include the strain of infecting virus4-7, prior infection with a heterologous serotype8-11, and the age12-14 and genetic background of the individual15-19. Other factors that may influence the progression to DHF include gender12-14 and nutrition20,21.
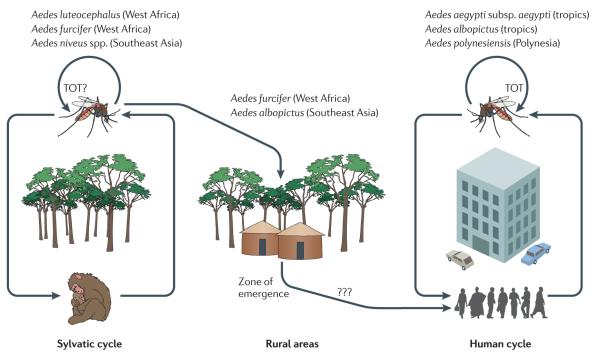
The sylvatic origins of dengue virus, and the ‘zone of emergence’, where sylvatic cycles contact human populations in rural areas of West Africa and Southeast Asia. In addition, dengue virus can persist in mosquito populations by transovarial transmission (TOT), in which virus-infected mosquitoes transfer the virus to their eggs (this has been shown to occur in some species but not in all).
Each of the four DENV serotypes is maintained in two ecologically and evolutionary distinct transmission cycles: a sylvatic cycle and a human cycle. The sylvatic cycle involves non-human primates and arboreal Aedes mosquitoes and has been documented in transmission foci in West Africa and peninsular Malaysia. The human cycle involves the domestic Aedes aegypti subsp. aegypti22 and peridomestic Aedes albopictus mosquitoes and can be found in a diverse range of environments throughout the tropics and subtropics. In the human cycle, humans are the only known reservoir hosts and amplification hosts, a unique host usage pattern among arthropod-borne viruses (arboviruses)23. This Review examines the ecological and evolutionary processes that could have led to the establishment of the four existing serotypes of human DENV, as well as the current disease burden and the future risks that are posed by the continued spillover of sylvatic DENV into human populations. Although the importance of sylvatic-DENV-mediated disease in humans has largely been discounted, we argue that this is premature in light of the paucity of information concerning sylvatic-DENV infection in humans. More generally, it has become increasingly clear that of all the viruses with the potential to shift from an animal reservoir into humans, the most likely to shift are those, like sylvatic DENV, that are carried by our closest relatives, the non-human primates24-41.
Ecology and evolution of sylvatic DENV
The origin of DENV
Humans have undoubtedly been repeatedly exposed to sylvatic DENV, and a process of cross-species transmission (BOX 2) has resulted in four sustained transmission chains in humans, represented by the DENV-1 to DENV-4 lineages that circulate today. Phylogenetic analysis strongly suggests that each of these four lineages jumped from a non-human primate reservoir to humans, as sylvatic strains cluster with human strains in phylogenetic trees of both DENV-2 and DENV-4 (see Supplementary information S1(figure)). This conclusion is also supported by some phylo genetic analyses of DENV-1. Until recently, only a single ‘sylvatic’ isolate of DENV-1 had been sequenced (Malaysia72-P72-1244), but it is possible that this strain in fact represents a spillback from humans. This possibility is supported by the recent isolation of a human DENV-1 from Malaysia (D1.Malaysia.36046/05) that clusters with Malaysia72-P72-1244 on a phylogenetic tree of the envelope (E) gene42, suggesting that both viruses are ultimately of human origin. Finally, although sylvatic DENV-3 strains have not been isolated to date, they are believed to exist in Malaysia, because of the seroconversion of sentinel monkeys43,44.
It is difficult to discern where and when the cross-species transmission events took place to lead to the emergence of DENV infection in humans. Dengue was first documented in the Americas at the end of the eighteenth century, and its arrival on this continent is likely to have been a result of the slave trade. The association between DENV and the slave trade naturally leads to the conclusion that DENV arose in the forests of Central and West Africa, from where slaves were taken and where non-human primates were plentiful. Indeed, just such a scenario seems likely to explain the origin of yellow fever virus (YFV), particularly given the absence of this virus in Asia45 (BOX 3). Because only a few African regions have been intensively surveyed to date, it is also possible that sylvatic DENV may be present but unrecognized in other regions of this continent. Two monkey species that are known to serve as amplification or reservoir hosts for sylvatic DENV in Africa — the African green monkey (Chlorocebus sabaeus) and the Guinea baboon (Papio papio) — are limited to West Africa, but closely related congeners are found across the continent. Moreover, a third primate host of sylvatic DENV, the patas monkey (Erythrocebus patas), has a broad range across central Africa (FIG. 2).
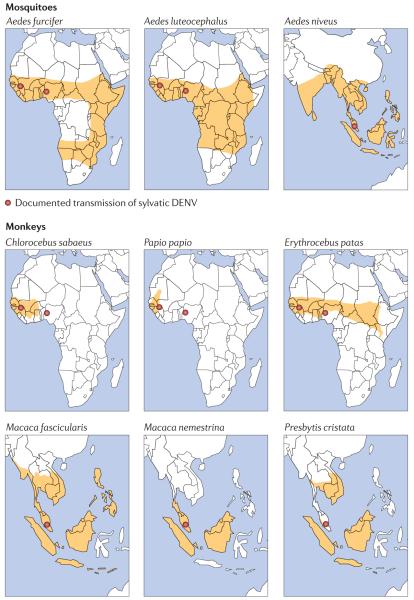
The geographic range of known and putative mosquito vectors and non-human primate hosts for the transmission cycles of sylvatic dengue virus in Africa and Southeast Asia. Although the range of the guinea baboon (Papio papio) is limited to West Africa, closely related species such as Papio anubis, Papio ursinus and Papio cynocephalus are found across the continent and could also be involved in dengue virus transmission.
However, an African origin is inconsistent with the known distribution of DENV diversity in the sylvatic cycle and the prevalence of dengue disease in humans. All four DENV serotypes have been detected in the sylvatic cycle in Southeast Asia43,44,46, whereas only sylvatic DENV-2 has been detected in Africa47-50. In addition, the incidence of dengue fever is very high in Southeast Asia but relatively low in Africa. Epidemics of a disease consistent with dengue were described in Indonesia (then called the Dutch East Indies) during the eighteenth century51 and perhaps even in China 1,000 years earlier52. Recent work also suggests that A. aegypti subsp. formosus, the ancestral progenitor to A. aegypti subsp. aegypti, has only a minor role in the sylvatic-DENV-2 transmission cycle in the forests of West Africa, as it is nearly refractory to infection with sylvatic-DENV33,34,53. By contrast, the Asian tiger mosquito, A. albopictus, is considerably more susceptible to DENV infection than A. aegypti subsp. aegypti54, an observation based on laboratory-based colonies of mosquitoes. Despite the current expanded global distribution of A. albopictus, this species plays a minor role in DENV transmission55. A number of other Aedes species distributed throughout the South Pacific, including Aedes polynesiensis, Aedes pseudoscutellaris, Aedes malayensis and Aedes cooki, also show a higher susceptibility to DENV than A. aegypti54. Indeed, early serological studies suggested that both non-human primates and humans living in the forests of Malaysia had been exposed to DENV, and identified A. albopictus as the likely vector56. Together, these results imply that A. albopictus (and/or other highly susceptible Aedes species) was the original vector of human DENV and that the virus later exploited the highly anthropophilic A. aegypti, which facilitated sustained transmission in humans57. However, a full understanding of the origins and evolutionary history of DENV requires a far larger sample of sylvatic viruses, particularly as we have probably sampled only a small fraction of the total biodiversity of flaviviruses39,58-60.
More puzzling is how DENV became established in monkeys in Asia and Africa. Phylogenetic trees of sylvatic DENV-2 show that viruses derived from Asian and African monkeys form a sister group that is distinct from DENV viruses that infect humans (see Supplementary information S1 (figure)). One explanation is that DENV arose before the separation of African and Asian primates approximately 25 million years ago. However, the low levels of divergence between human and sylvatic DENV strains (<20% across the genome) suggest that such a deep ancestry is unlikely. In addition, all research to date suggests that the mean evolutionary rates for human DENV and sylvatic DENV are approximately the same (between 0.5 × 10−3 and 1 × 10−3 nucleotide substitutions per site per year)25,36,61 and that the timescale of DENV evolution should be measured in hundreds or thousands of years62-64 (see Supplementary information S1 (figure)). It is therefore possible that the African and Asian sylvatic DENV-2 strains diverged within only the past few hundred years and that humans provided the vehicle by which sylvatic viruses moved on a transcontinental scale — either as infected hosts themselves, or as transporters of infected monkeys or mosquito vectors.
The sylvatic transmission cycle in Africa
The earliest report of sylvatic DENV in Africa is from an epidemic in Ibadan, Nigeria, in 1966 (REF. 65), although it was not until 2008 that sylvatic strains were incriminated in this epidemic by phylogenetic analysis29. During the 1970s, five DENV-2 strains were isolated from patients in Senegal47,50,66. Some of these strains, as well as DENV-2 isolates from mosquitoes in other parts of West Africa, are phylogenetically distinct from contemporaneous DENV-2 strains circulating in Asia and the Americas and are therefore likely to constitute a distinct sylvatic cycle41. These strains are discussed in more detail below.
In the 1970s, the isolation of DENV-2 and other arboviruses from humans and mosquitoes in eastern Senegal prompted the initiation of a long-term surveillance programme near the town of Kédougou. Notable findings include the lack of evidence for DENV-2 circulation during most years, despite the surveillance of vast numbers of mosquitoes, punctuated by high mosquito infection rates at intervals of 5–8 years, with most isolations coming at the end of the rainy season32,33,67,68. The principal vectors appear to be Aedes furcifer, Aedes taylori and Aedes luteocephalus, mosquitoes that are typically found in gallery forests. Data from Senegal further indicate that the vector range of sylvatic DENV-2 overlaps extensively with that of YFV, chikungunya virus and Zika virus, although amplifications of these viruses usually occur in different places69. This suggests that factors other than rainfall and vector densities — for example, herd immunity and population turnover in the monkey reservoir species — may determine the amplification periodicity. In addition, a retrospective serological study of monkeys and humans inhabiting the same region in Senegal indicated the presence of successive epizootics in 1974 and 1981 without detectable transmission among the humans residing in outlying forest villages70. However, for the three primate DENV-2-reservoir species that have been identified to date (FIG. 2), their relative roles in viral circulation in eastern Senegal remain unclear. Perhaps the most pertinent point from these studies is that the exposure of humans to sylvatic DENV is not well characterized, and neither is the possible role of humans in the amplification of sylvatic DENV; human infections were only detected serendipitously until a surveillance programme was implemented in 2008.
The sylvatic transmission cycle in Asia
The existence of an enzootic DENV transmission cycle was first suggested in the 1950s, when neutralizing antibodies were found in human populations inhabiting forest and rural areas of peninsular Malaysia that were devoid of A. aegypti subsp. aegypti56,71,72. In a subsequent survey of various mammals, only canopy-dwelling monkeys were positive for DENV-specific antibodies, suggesting that an arboreal vector might be responsible for transmission among non-human primates71. More direct evidence for the existence of an enzootic transmission cycle was provided in the 1960s and 1970s, also in peninsular Malaysia, with the demonstration of high occurrences of DENV-neutralizing antibodies in three species of arboreal primate: the crab-eating macaque (Macaca fascicularis; with antibodies in 68.7% of tested animals), the pig-tailed macaque (Macaca nemestrina; 50.0%) and the silvered leaf monkey (Presbytis cristata; 47.8%). Despite extensive sampling46, attempts to isolate DENV from these monkeys, as well as from over 25,000 mosquitoes (from eight genera and >69 species) were unsuccessful44,46. Isolation of sylvatic DENV from a primate was subsequently achieved when M. fascicularis and Presbytis obscura (dusky leaf monkey) monkeys were placed as sentinels in the canopies of isolated forest reserves.
In 1975, a sylvatic DENV-4 strain was isolated from the mosquito group Aedes niveus s.l., primatophilic arboreal vectors that are abundant in the forest canopy in Malaysia and are also attracted to feeding on humans. In fact, all species of this vector complex (Aedes pseudoniveus, Aedes subniveus, Aedes vanus, Aedes albolateralis, Aedes niveoides and Aedes novoniveus) are primatophilic and known to descend to ground level to feed on humans. This opportunistic feeding behaviour could facilitate the transfer of sylvatic DENV from the forest to peridomestic environments44.
Collectively, these data demonstrate that an enzootic sylvatic-DENV transmission cycle occurs in the forest canopies of Malaysia. This cycle is maintained by species in the A. niveus group, which most probably use monkeys in the genera Macaca and Presbytis as reservoir and amplification hosts. Moreover, the geographical ranges of the primate reservoir hosts and of the arboreal vectors (the mosquitoes) for this zoonotic virus argue that undiscovered transmission foci occur in other locations of tropical Asia as well (FIG. 2). More recently, serological surveys demonstrated the presence of DENV-2-neutralizing antibodies in 28% of wild Bornean orangutans (Pongo pygmaeus)73. However, as it is possible that these orangutans were infected by exposure to mosquitoes that fed on agricultural or timber workers, their role in the sylvatic DENV cycle is unknown.
A sylvatic transmission cycle in the Americas?
The discovery of a sylvatic cycle for YFV in the Neotropic zone, combined with the realization that the virus probably originated in Africa and was introduced into the New World during the slave trade45, begs the question of whether sylvatic DENV might also have been introduced in this manner. DENV seroconversions have been detected among indigenous Ayoreo people in a remote part of Bolivia that is devoid of A. aegypti, suggesting that a sylvatic-DENV cycle may occur there74. However, despite the documented presence of human dengue in the Americas for centuries, no DENV isolates from the Western hemisphere show phylogenetic evidence of a sylvatic lineage. For example, although all four serotypes have reportedly been isolated and sequenced from a number of mammalian species in South America (notably, bats, rodents and marsupials), their close relationship to human viruses on phylogenetic trees75 suggests that they are likely to represent spillbacks from human viruses. These findings are also called into question by the lack of comparable evidence from the many similar studies that have been carried out in other dengue-endemic regions. Serological surveys of primates in regions of Panama that were formerly enzootic for YFV revealed no evidence of enzootic circulation of DENV-1 or DENV-2 (REF. 76), and recent studies employing sentinel monkeys that were maintained in a forest canopy near Iquitos, Peru, also failed to find evidence of DENV circulation (D. M. Watts and G. B. Schoeler, personal communication).
Emergence of sylvatic DENV in humans?
Adaptation of sylvatic DENV to humans
The present danger of the emergence of sylvatic DENV in human populations was highlighted in recent studies that used various laboratory models for the replication of sylvatic DENV in humans. For example, the replication profiles of low-passage DENV-2 strains representing all major genotypes were evaluated in two surrogate models: human monocyte-derived dendritic cells (moDCs) and severe combined immunodeficient (SCID) mice xenografted with human hepatoma (Huh-7) cells24. Despite there being substantial differences among the replication profiles of the strains, there was no consistent difference between sylvatic and human strains. Strikingly, however, the replication of sylvatic DENV-2 in moDCs was similar to that of human DENV-2 from the American genotype24. In addition, the replication of human and sylvatic strains was measured in cultured cells from human hosts (Huh-7 cells), monkey hosts (Vero cells) and mosquito hosts (C6/36 cells from A. albopictus)28. When compared with the sylvatic DENV strains, human strains produced considerably more progeny only in the human cells. Overall, the experimental findings from the surrogate human models of infection and from the cultured cells suggest that there is little or no adaptive barrier for the emergence of sylvatic DENV in human populations, possibly reflecting the evolution of DENV as an opportunistic virus that is capable of infecting a wide range of primate species. The potential for re-emergence of sylvatic DENV from Africa or Asia in the human transmission cycle therefore appears to be high.
Selective forces acting on DENV
Like most arboviruses, the majority of nucleotide sites in the DENV genome are subject to purifying selection. This is reflected in measurements of the relative numbers of non-synonymous (dN) and synonymous (dS) substitutions per site, with mean dN/dS values of <0.1 across the DENV genome (note that a value of 1 equates to selective neutrality). Hence, the vast majority of mutations are deleterious and ultimately purged from the viral population77-80. Such strong purifying selection probably reflects the strong evolutionary trade-offs that are believed to be associated with the replication of arboviruses in cells as different as those of humans and mosquitoes.
It is also striking that there is little difference in dN/dS between human and sylvatic DENV strains. For example, the mean genomic dN/dS values are 0.05 and 0.06 for sylvatic DENV-2 and human DENV-2 (REF. 25). These data imply that the nature of selection pressures is broadly equivalent in human and sylvatic viruses and largely reflects the selective constraints that seem to be inherent to the arbovirus lifestyle. Such a similarity in genome-wide selection pressures again suggests that the successful emergence of a novel sylvatic DENV strain in human populations is unlikely to require a major adaptive challenge.
Ecological forces affecting emergence
Deforestation has been linked to the increased prevalence of several arthropod-borne diseases such as malaria81,82 and Kyasanur forest disease83. Changes in tropical land use, particularly the insertion of human habitations and agriculture into previously undeveloped areas that are enzootic for arboviruses, could promote interactions among DENV vectors and hosts84, increasing the likelihood of sylvatic-DENV re-emergence. Alternatively, deforestation could limit enzootic DENV circulation by reducing the habitat for mosquito vectors and monkey hosts. However, deforested tropical areas typically contain more phytotelmatic habitats (plant-held water bodies) for mosquito larvae than intact forests, making them more suitable for sylvatic-DENV vectors, which lay their eggs in water-filled tree holes85.
Global climate change could also affect sylvatic-DENV circulation by expanding the distributions of sylvatic vectors and tropical primates86. However, changes in rainfall patterns could also reduce forest distributions; for example, the southwards expansion of the Sahara desert is approaching eastern Senegal87. Improved knowledge of sylvatic-DENV transmission cycles is needed before reliable predictions can be made about how deforestation and climate change will affect the re-emergence of sylvatic DENV.
The increasing urbanization of Asian and African regions where sylvatic DENV circulates could lead to larger populations of A. aegypti subsp. aegypti, elevating the risk that persons exposed to sylvatic strains could initiate human–mosquito–human transmission. The recent introduction and expansion of A. albopictus in Africa55,69,88,89 may also increase the risk for DENV emergence from sylvatic foci. Indeed, this vector has already mediated important epidemics of chikungunya virus in Africa in a human–mosquito–human cycle identical to that of DENV90, and the invasion of Italy by this mosquito91 enabled the first chikungunya outbreak in Europe92.
Spillover of sylvatic DENV into human populations
Epidemics and human contact
The frequency and breadth of human interactions with the sylvatic transmission cycle are not well understood. Several obstacles remain to an accurate assessment of human contact with sylvatic DENV and its resulting impact on public health. These include the limited access of researchers to the remote transmission foci and affected individuals, limited health care facilities and diagnostic testing in these regions, co-circulation of several other viruses that are difficult or impossible to distinguish using clinical presentation or existing diagnostic tests, and political and/or social instability. Furthermore, the scarcity of entomological and non-human-primate surveillance prevents the formulation of any meaningful inferences about the degree and routes of human exposure to sylvatic DENV.
Long-term surveillance in Senegal suggests that sylvatic DENV comes into regular contact with humans because the arboreal mosquito A. furcifer acts as a bridge vector between forest and peridomestic habitats32,33. Although this putative mechanism of human exposure is apparently incapable of generating large-scale outbreaks, it probably explains the small numbers of human cases that were documented during the DENV-2 amplifications of 1981–1982 and 1989–1990 (REFS 70,93). However, it is also possible that larger human epidemics go undetected owing to misdiagnosis of dengue as malaria94 and other clinically similar viral diseases (for example, chikungunya disease, o’nyong-nyong fever and Zika fever)95-97. Alternatively, epidemics may go undetected because of a preponderance of subclinical or mild disease, a hypothesis that is supported by serological evidence70,98. Finally, the symptoms associated with infection with sylvatic DENV are indistinguishable from those attributed to infection with human DENV36,50,66. For example, the outbreak of febrile illness that occurred in Ibadan, Nigeria, during the 1960s was initially attributed to human DENV65, although more recent phylogenetic analyses identified the cause as sylvatic DENV29. This outbreak also high-lights the fact that sylvatic epidemics can occur in urban settings with limited spillover.
In Southeast Asia, the earliest evidence of human exposure to sylvatic DENV was in the Philippines, where high frequencies of DENV-neutralizing antibodies were found in isolated aborigine populations that inhabited areas where A. aegypti subsp. aegypti was absent99. Subsequent serological surveys of the Orang Asli, a forest-dwelling people who have little contact with the general population in peninsular Malaysia, indicated high frequencies (82%) of DENV-neutralizing anti bodies43. Indeed, in rural populations inhabiting diverse habitats adjacent to forests and devoid of A. aegypti subsp. aegypti, all of the individuals had been exposed to DENV by the age of 30 (REFS 56, 71). Humans living adjacent to the forest were at higher risk of DENV infection, whereas the lowest incidences of infection were measured among populations with no close contact with the forest and in areas with low densities of A. albopictus43,44.
A variety of mechanisms might explain the limited spread of sylvatic DENV in humans. In West Africa, transmission may be inhibited by the absence of the efficient peridomestic A. aegypti subsp. aegypti and A. albopictus mosquito vectors near sylvatic foci. In Southeast Asia, highly efficient transmission of multiple serotypes of human DENV has generated high and widespread herd immunity100,101 that may restrict the spread of sylvatic DENV. Indeed, sera of convalescent patients with a primary DENV infection strongly neutralize both human and sylvatic DENV strains of the homologous serotype, although they exhibit little hetero-typic (non-homologous) neutralization26. Counteracting the rise of herd immunity, however, are the extensive and ongoing land use changes throughout Southeast Asia102-104, including the spread of plantations and deurbanization drives. These changes have resulted in a shift in agriculture and concomitant human traffic into the forest, which will probably result in increased human contact with the sylvatic cycle and, therefore, an increased re-emergence potential.
Clinical manifestations of sylvatic-DENV infection in humans
Until recently, our understanding of the human illness that is caused by infection with sylvatic DENV derived from a few serendipitous investigations of febrile illness in West Africa for which the associated virus was isolated and characterized. All of these cases coincided with the isolation of virus from arboreal mosquitoes of the genus Aedes, reinforcing the notion of a concurrent sylvatic-DENV amplification cycle in non-human primates and limited spillover events in adjacent ‘zones of emergence’ (REFS 47,50,66). Collectively, these cases indicate that the clinical illness resulting from infection with sylvatic DENV is indistinguishable from classic dengue fever resulting from the human transmission cycle. However, in 2008, a 20-year-old male in Sarawak, Malaysia, developed DHF after returning from a 1-month-long stay in peninsular Malaysia36. Importantly, this individual experienced signs and symptoms that fulfilled the WHO criteria for DHF, including a major drop in his minimum platelet count (to 12,000 platelets per μl), high fever (39.6 °C), mild enlargement of the liver, and a severe drop in blood volume (as indicated by a haematocrit that was elevated by almost 30% above the patient’s baseline) during the acute phase of disease. The cause of this infection was a sylvatic DENV strain, as documented by a phylogenetic analysis36, which revealed that the virus isolated from this patient was closely related to the 1970 sylvatic virus that was isolated from a similar region of peninsular Malaysia, but was different enough to rule out cross-contamination44 (see Supplementary information S1 (figure)). As this patient was identified through a sentinel surveillance programme in Sarawak but was clearly infected in a different part of the country, it was not possible to determine whether this was a primary or secondary exposure. There might be a local community transmission of sylvatic DENV that goes undetected, as virus isolation and sequencing is not commonly performed outside of one or two research laboratories in Malaysia. Not only did this study provide the first unequivocal evidence that sylvatic DENV can cause severe disease, but also the phylogeny strongly suggests that sylvatic DENV has been maintained in non-human primates in Malaysia without detection for nearly four decades.
A similar illness was observed in a 27-year-old male resident of Canchungo, Guinea-Bissau, in 2009, again concomitant with the 2008–2009 sylvatic DENV-2 amplification in West Africa. This patient was probably infected during a trip to Dakar, Senegal, and developed DHF. Virus isolation and phylogenetic analysis based on the complete genome sequence confirmed a sylvatic-DENV-2 infection105. This case is particularly striking given that severe dengue disease is rare in Africa, even during urban transmission of typical human strains.
Prevention of sylvatic-DENV re-emergence
Existing ecological barriers to emergence
To date, no epidemics of sylvatic DENV in humans have been detected, despite the proximity of large urban areas to sylvatic foci. In part, the failure to detect human epidemics may stem from the lack of diagnostic tools to distinguish human-DENV and sylvatic-DENV infections, as discussed in detail below. In addition, the circulation of human DENV in Southeast Asia may constrain the emergence of sylvatic DENV via cross-immunity in human hosts106 and/or competition within peridomestic vectors. Indeed, when cultured cells of A. albopictus were infected with both sylvatic and human DENV strains38, the human strains were competitively superior. Such competitive exclusion would require high levels of human-DENV transmission, and the intense DENV transmission that is currently observed in much of the tropics is a recent phenomenon (since the Second World War). However, in urban areas of Malaysia that are close to sylvatic-DENV foci, infection with human DENV has probably been sustained at high levels since the earliest studies of the 1920s107, suggesting that competitive exclusion of sylvatic DENV could have occurred in this area for at least the last century.
Surveillance
As sylvatic DENV is able to enter human populations and cause disease, including DHF, comprehensive ecological and epidemiological studies are urgently needed to assess the role of non-human primates and possibly other vertebrate hosts in the maintenance of sylvatic DENV, as well as the degree and routes of contact between humans and sylvatic DENV. We contend that this should be a research priority in studies of arbovirus emergence.
To the best of our knowledge, there have been no field investigations of sylvatic DENV in Asia since Rudnick’s studies in the 1960s and 1970s. The situation is better in West Africa, where the Institut Pasteur has conducted systematic collection and screening of sylvatic-DENV vectors for over 40 years67,68. However, there have been far fewer surveys of infection in humans or non-human primates48,50,108. Surveillance of sylvatic DENV is hampered by the difficulties of collecting samples in the forest canopy and by the current lack of diagnostic tests capable of distinguishing the antibodies that are developed in response to sylvatic versus human DENV. However, neither obstacle is insurmountable, and synchronous surveillance of sylvatic DENV in its non-human primate reservoir, in arboreal Aedes spp. vectors and in humans living in areas of potential spillover must be renewed, particularly as sylvatic dengue may prove to be a useful model of human dengue (see below).
Diagnostic tools to distinguish infection by sylvatic and human DENV
Distinguishing between exposure to human DENV and sylvatic DENV on the basis of a clinical disease description or serological assays is impossible at present. To date, most human infections have been defined based on virus isolation or molecular techniques such as reverse transcriptase (RT)-PCR, which offer a limited window of detection because the viraemia phase lasts approximately 2–7 days. A promising diagnostic tool that may be able to distinguish between infections with sylvatic and human viruses would be a blocking enzyme-linked immunosorbent assay (ELISA) specific for DENV. A blocking ELISA, in which a mixture of antibodies compete for binding to the same antigen, is effective for distinguishing seroconversions that result from infections with different subtypes of Venezuelan equine encephalitis virus109. A similar assay has been developed for distinguishing infection with West Nile virus110,111, Murray Valley encephalitis virus112,113 and Japanese encephalitis virus114 from infection with closely related flaviviruses, using recombinant envelope domain III (rEIII) and/or NS1 epitopes as antigens. However, whether the antigenic differences in envelope domain III of DENV strains are sufficient to discriminate between sylvatic and human DENV strains remains to be evaluated.
Sylvatic dengue as a model for human dengue
Despite an urgent need, there are as yet no licensed antiviral therapies and no vaccines available to control human DENV (BOX 4). A major roadblock to the development of DENV control measures is the lack of an animal model that adequately recreates the viral replication or disease which occurs in humans. At present, the best model for the evaluation of therapies or vaccines is efficacy against the replication of human DENV in non-human primates115. Although many primate species can be infected with human DENV, they generally do not develop disease (see REF. 116 for exceptions), and the maximum viraemia titres are several orders of magnitude lower than those observed in humans115. A recent workshop on dengue animal models called for an investigation into the utility of sylvatic-DENV infections in non-human primates as models for human dengue115. Studies of the host range, replication and transmission of sylvatic DENV will be a critical foundation for this effort, particularly for decisions about appropriate non-human primate host species and protocols for infection. To date, there have been no controlled studies of sylvatic-DENV replication or associated disease in monkeys. It will be ironic if sylvatic DENV ultimately represents the best model for understanding and preventing its human counterpart.
Conclusion
Sylvatic DENV strains are ecologically and evolutionarily distinct lineages that circulate in a forest cycle in Southeast Asia and West Africa between monkeys and arboreal canopy-dwelling mosquitoes of the genus Aedes. Although sylvatic-DENV transmission is mainly confined to the forest, there is clear evidence that these viruses come into regular contact with humans in Southeast Asia and Africa and can cause severe disease in isolated cases as well as transient spillover in urban settings. In addition, each of the four serotypes of human DENV represents an independent cross-species transmission of a sylvatic DENV into humans. Furthermore, recent experimental evidence indicates that little or no adaptive barrier exists to the emergence of sylvatic DENV in the human population, implying that re-emergence of sylvatic DENV in the human transmission cycle is a realistic prospect.
However, the ecological contact between sylvatic cycles and human populations is poorly understood, and the range of human infection and disease caused by sylvatic DENV strains is unknown. Understanding the ecology and epidemiology of sylvatic DENV is therefore essential to understanding and predicting viral emergence. Apart from the pioneering studies of Rudnick and colleagues in Malaysia during the 1970s and the long-term surveillance of mosquitoes in Senegal by the Institut Pasteur, this area of research has been largely ignored. For example, nothing is known about the contemporary distribution and ecology of sylvatic DENV in Asia or in other regions in Africa. Therefore, comprehensive prospective epidemiological and ecological studies in enzootic locations of Asia and West Africa are clearly a research priority. The prospect of licensing effective tetravalent vaccines against human DENV raises the potential for the control or even eradication of the human-DENV transmission cycle. This heightens the need not only for an understanding of the ease with which the sylvatic strains can re-emerge in human populations, but also for the development of serological diagnostic tools that are able to distinguish between infections with sylvatic and human viral strains. However, if additional DENV serotypes that have not yet emerged into human transmission cycles already exist in as-yet-undetected sylvatic cycles, there could be major implications for the long-term control of dengue using the vaccines that are currently under development. Finally, as underscored by the major burden of disease that is caused by enzootic YFV in the Americas, the potential for DENV to establish zoonotic transmission cycles in the New World also deserves careful evaluation.
Acknowledgements
N.V. is supported by start-up funds provided by the Department of Pathology, University of Texas Medical Branch. J.C. has been supported in part by operational funds from Universiti Malaysia Sarawak. K.A.H. is supported by US National Institutes of Health (NIH) grants R01AI069145 and P20RR016480. E.C.H. is supported in part by NIH grants R01GM080533 and R01GM83224. S.C.W. is supported for dengue research by NIH grant AI069145 and by the John Sealy Distinguished University Chair in Human Infections and Immunity. Special thanks to S. Rossi for providing the design on which figure 1 is based.
Glossary
| Dengue antigenic complex | An antigenic subgroup within the RNA virus genus Flavivirus that includes all four dengue virus serotypes and is based on the antigenic cross reactivities (cross neutralization) of monoclonal and polyclonal antibodies. |
| Dengue fever | An acute mosquito-borne disease of the tropics, characterized by sudden onset and followed by a benign self-resolving course of transient fever, headache, lymphadenopathy, rash, and severe joint and muscle pain. It is the most common disease manifestation in persons infected with dengue virus, although the majority of infections lack symptoms (an outcome called inapparent infection). |
| Dengue haemorrhagic fever | A syndrome that is caused by infection with dengue virus and characterized by abdominal pain, haemorrhage and circulatory collapse. This syndrome is also referred to as severe dengue disease. |
| Sylvatic | Pertaining to a form of a disease or virus that occurs in wild animals (in the case of dengue virus, in non-human primates) in the forest. |
| Peridomestic | Of a vector or location: in and around human habitats. |
| Amplification hosts | Host in which an infectious agent replicates to high enough concentrations for transmission to additional hosts. For dengue virus, humans and non-human primates are the only known amplification hosts. |
| Arboviruses | Arthropod-borne viruses. In the case of arboviruses that infect humans, mosquitoes are the most common vectors. |
| Spillover | Cross-species virus transmission into small, localized groups of people or other animals, with no sustained onward transmission. |
| Seroconversion | The development of detectable specific antibodies as a result of natural infection or vaccine immunization. |
| Reservoir hosts | Hosts that harbour (usually without major detriment to themselves) microorganisms that are pathogens for a different species. |
| Anthropophilic | Attracted to humans more than to other animals. |
| Herd immunity | Immunity of a proportion of a population to a pathogen, owing to infection or vaccination, that is able to protect the remainder of the population owing to there being insufficient susceptible hosts for efficient pathogen amplification. |
| Epizootics | Amplifications or increased occurrences of a disease or pathogen affecting animals at the same time within a particular region or geographical area. |
| Enzootic | Pertaining to the maintenance of a transmission cycle among non-human vertebrate hosts in a specific geographical region. Also sometimes referring to the disease caused by pathogens that circulate enzootically. |
| Zoonotic | Pertaining to pathogens (and the diseases that they cause) that circulate among wild animals but also have the potential to be transmitted to human populations. |
| Neotropic zone | The ecological region of the New World between the Tropics of Cancer and Capricorn, including southern Mexico, Central and South America, and the West Indies. |
| Purifying selection | The purging of deleterious alleles or mutants as a result of differential reproductive success. |
| of emergence | An ecological transition zone that is adjacent to sylvatic environments; in this zone, prevailing ecological conditions, such as vector abundance and closeness for human contact, enable the cross-species transmission of viruses from animals into humans. |
| Haematocrit | A test to compare the volume of red blood cells to the total blood volume. |
| Viraemia | The presence of a virus in the circulatory system of the host; this is essential for virus transmission by an arthropod vector. |
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Nikos Vasilakis’s homepage: http://www.utmb.edu/pathology/faculty/vasilakis/vasilakis_bio.htm
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrmicro2595
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/nrmicro2595.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101708361
Article citations
Genomic surveillance reveals a dengue 2 virus epidemic lineage with a marked decrease in sensitivity to Mosnodenvir.
Nat Commun, 15(1):8667, 09 Oct 2024
Cited by: 1 article | PMID: 39384752 | PMCID: PMC11464713
Potential Performance of Two New RT-PCR and RT-qPCR Methods for Multiplex Detection of Dengue Virus Serotypes 1-4 and Chikungunya Virus in Mosquitoes.
Curr Issues Mol Biol, 46(10):11048-11056, 30 Sep 2024
Cited by: 0 articles | PMID: 39451536 | PMCID: PMC11505945
A new lineage nomenclature to aid genomic surveillance of dengue virus.
PLoS Biol, 22(9):e3002834, 16 Sep 2024
Cited by: 4 articles | PMID: 39283942 | PMCID: PMC11426435
The Impact of Climate on Human Dengue Infections in the Caribbean.
Pathogens, 13(9):756, 03 Sep 2024
Cited by: 0 articles | PMID: 39338947 | PMCID: PMC11434940
Review Free full text in Europe PMC
Prevalence of Flavivirus and A lphavirus in bats captured in the state of Yucatan, southeastern Mexico.
One Health, 19:100876, 15 Aug 2024
Cited by: 0 articles | PMID: 39258264 | PMCID: PMC11386310
Go to all (185) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Potential role of sylvatic and domestic African mosquito species in dengue emergence.
Am J Trop Med Hyg, 73(2):445-449, 01 Aug 2005
Cited by: 59 articles | PMID: 16103619
Abundance and distribution of sylvatic dengue virus vectors in three different land cover types in Sarawak, Malaysian Borneo.
Parasit Vectors, 10(1):406, 31 Aug 2017
Cited by: 31 articles | PMID: 28859676 | PMCID: PMC5580228
Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains.
Virology, 377(2):296-307, 01 Aug 2008
Cited by: 38 articles | PMID: 18570968 | PMCID: PMC3612928
The history and evolution of human dengue emergence.
Adv Virus Res, 72:1-76, 01 Jan 2008
Cited by: 108 articles | PMID: 19081488
Review
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: P20RR016480
Grant ID: P20 RR016480
NIAID NIH HHS (4)
Grant ID: R01 AI069145-01A2
Grant ID: R01AI069145
Grant ID: R01 AI069145
Grant ID: AI069145
NIGMS NIH HHS (5)
Grant ID: R01 GM083224
Grant ID: R01 GM080533-05
Grant ID: R01GM080533
Grant ID: R01 GM080533
Grant ID: R01GM83224
 ¶ and
¶ and 




