Abstract
Free full text

pTransgenesis: a cross-species, modular transgenesis resource
Abstract
As studies aim increasingly to understand key, evolutionarily conserved properties of biological systems, the ability to move transgenesis experiments efficiently between organisms becomes essential. DNA constructions used in transgenesis usually contain four elements, including sequences that facilitate transgene genome integration, a selectable marker and promoter elements driving a coding gene. Linking these four elements in a DNA construction, however, can be a rate-limiting step in the design and creation of transgenic organisms. In order to expedite the construction process and to facilitate cross-species collaborations, we have incorporated the four common elements of transgenesis into a modular, recombination-based cloning system called pTransgenesis. Within this framework, we created a library of useful coding sequences, such as various fluorescent protein, Gal4, Cre-recombinase and dominant-negative receptor constructs, which are designed to be coupled to modular, species-compatible selectable markers, promoters and transgenesis facilitation sequences. Using pTransgenesis in Xenopus, we demonstrate Gal4-UAS binary expression, Cre-loxP-mediated fate-mapping and the establishment of novel, tissue-specific transgenic lines. Importantly, we show that the pTransgenesis resource is also compatible with transgenesis in Drosophila, zebrafish and mammalian cell models. Thus, the pTransgenesis resource fosters a cross-model standardization of commonly used transgenesis elements, streamlines DNA construct creation and facilitates collaboration between researchers working on different model organisms.
INTRODUCTION
The ability to engineer genetically modified organisms is essential for establishing the function of genes during development, disease, homeostasis, repair and regeneration (Gama Sosa et al., 2010; Ristevski, 2005). However, a crucial step in engineering genetically modified organisms is the design and generation of the transgene DNA constructions required for a given experiment. For the past thirty years, DNA constructions have been created primarily through restriction enzyme digestion and ligation. However, cloning with restriction enzymes becomes progressively more cumbersome as the complexity of the engineered constructs increases. For this reason, a site-specific recombination-based DNA cloning method was developed that circumvents the use of restriction enzymes (Hartley et al., 2000). The advent of recombination-based cloning brought a series of diverse and pioneering studies showing the utility of this technology in creating DNA constructions for transgenesis (Fisher et al., 2006; Hope et al., 2004; Ikeya et al., 2005; Kappas et al., 2008; Kwan et al., 2007; Nyabi et al., 2009; Semple et al., 2010; Skarnes et al., 2011). However, none had yet been designed specifically for use in Xenopus, a widely used model organism (Amaya, 2005), and the ability to use them across multiple models was limited. When we began to develop a transgenesis plasmid resource for Xenopus, we sought to design a system that would encapsulate multiple advances demonstrated in previous Multisite Gateway-based cloning projects, but we wished to expand on them to make them more universally useful to the developmental biology community at large. In particular, we wished to decouple the screenable elements from the transgenesis-promoting sequences, thus facilitating the transfer of this plasmid resource across different model systems, such as Xenopus, mammals, fish and flies.
By combining these attributes, we created a new modular, cross-species plasmid resource, which we have named pTransgenesis. The pTransgenesis resource is the first modular cloning system that allows the interchange of DNA elements for transgenesis between Xenopus, zebrafish, Drosophila and mammalian cell culture models. The pTransgenesis design and associated resources will greatly facilitate the efficient generation of transgenic organisms and the transfer of transgenic reagents across various developmental model organisms.
MATERIALS AND METHODS
Plasmid construction
We adapted Invitrogen’s Gateway Multisite Cloning Kit (CA, USA) to create the pTransgenesis vectors (note, this is not the Gateway ‘Pro’). BP reactions were performed using Invitrogen’s BP Recombinase and PCR products were cloned with Invitrogen’s pCR8 GW TOPO kit. LR recombinations were performed using Invitrogen’s LR Clonase II+ (Ishibashi et al., 2012), bacterial transformations with DH5a competent cells (Invitrogen); typically 50-100% of colonies yield correct recombinations.
Transgenesis
Restriction enzyme-mediated integration (REMI) transgenesis was performed as described (Breckenridge et al., 2001; Kroll and Amaya, 1996). I-SceI transgenesis was performed by injecting 2 nl of a 10 pg/nl reaction mixture as described previously (Ishibashi et al., 2012). The Cre mRNA and Tol2 mRNA was made using the SP6 mMessage Machine mRNA Kit (Ambion) from a pCS2-CreNLS or pC2-TP template (Kawakami, 2007). For zebrafish injections, ~1 nl of a 20 pg/nl DNA solution with or without 25 pg Tol2 mRNA was injected at the 1-cell stage. HeLa cell lines were made by plasmid transfection (Lipofectamine 2000, Invitrogen), with a selection of 3 μg/ml of puromycin in DMEM performed at 48 hours post-transfection. Transgenic Drosophila were made using P-element mediated transgenesis (Bestgene, CA, USA), and were crossed with engrailed-Gal4 (Bloomington Stock Center) (Brand and Perrimon, 1993; Millard and Martin, 2008), srp-Gal4 (Bruckner et al., 2004) and eval-Gal4 (Bloomington Stock Center) (Landgraf et al., 1999) driver lines.
Immunohistochemistry and in situ hybridization
Sectioning and immunostaining was performed as described previously (Chalmers et al., 2003) using a primary mouse anti-green fluorescent protein (GFP) antibody (1:500, Roche). For in situ hybridization, digoxygenin (DIG)-labelled probes were generated using the X. tropicalis expressed sequence tag (EST) clones from the TTpA043k14 (Vimentin) and TGas140h10 (Nectin2) clones using 10× DIG labelling mix (Roche) (Gilchrist et al., 2004). DIG probe hybridization and staining was performed according to methods described by Harland (Harland, 1991).
Microscopy
Whole-mount imaging was performed using a Leica MZ FLIII fluorescent stereomicroscope and Northern Eclipse software, HeLa cells were imaged with a Olympus IX70 inverted fluorescent microscope with Northern Eclipse software and confocal imaging was performed with an Olympus Fluoview FV1000 imaging system and accompanying software.
RESULTS AND DISCUSSION
The pTransgenesis framework
The pTransgenesis plasmid resource utilizes four separate, modular plasmid libraries (also referred to as reservoirs or positions, Fig. 1A). We named these plasmid positions with the following terminology: p1, the selection marker, designed to contain all necessary DNA sequences to allow selection of transgenic organisms; p2, the promoter DNA sequence, which is sufficient to drive expression of a coding gene; p3, the coding gene with a poly-adenlyation signal; and p4, containing sequences that facilitate genome integration and/or chromosomal attachment sites. Incubation of a ‘p1 selection marker’, ‘p2 promoter’, ‘p3 coding gene’ and a ‘p4 transgenesis’ vector in the presence of recombinase enzyme in vitro produces a final DNA construction that contains these individual elements in a predictable order (Fig. 1B).
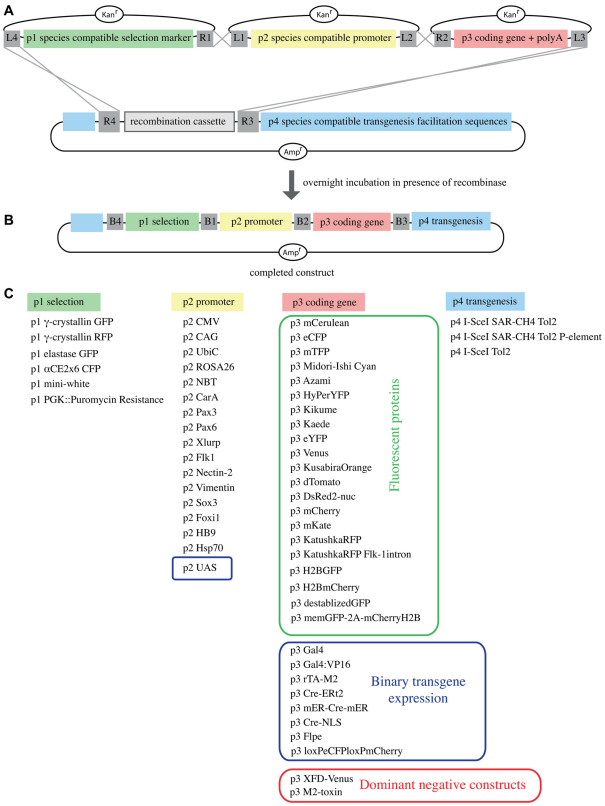
The pTransgenesis framework. (A,B) Four separate vectors (p1, p2, p3 and p4) are recombined without restriction enzymes in a sequential and predictable order. Note: The ‘L’ and ‘R’ elements contained within the grey boxes represent the sequences that facilitate the in vitro recombination between plasmids. B1-B4, attB sequences produced following ‘LR’ recombination. (C) A selection of p1, p2, p3 and p4 constructs available within the pTransgenesis system.
The important outcome of this design is that p3 coding sequences, like GFP, which are used repeatedly in experiments across a wide variety of species, are easily coupled to modular p1, p2 and p4 constructions. Hence, we created a p3 coding sequence library that can be shared and utilized in any compatible species. For this, we cloned into the p3 reservoir 16 different fluorescent proteins, sequences for Gal4-UAS binary transgenic approaches, variations of the Cre-recombinase, and coding sequences that allow genetic manipulation in vivo, such as the dominant-negative Fgf receptor construct (Fig. 1C).
Incorporation of Xenopus compatible elements into pTransgenesis
To demonstrate the utility of the pTransgenesis cloning framework, we used Xenopus, a model which did not possess a recombination-based cloning system. Notably, the Xenopus model allows the generation of non-mosaic, fully transgenic organisms in the F0 generation via REMI transgenesis, thus allowing us to rapidly validate pTransgenesis elements using this method (Kroll and Amaya, 1996).
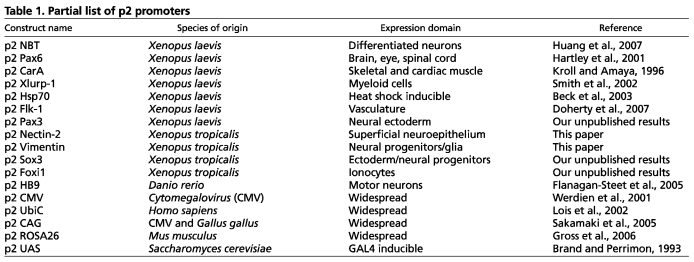
We incorporated three Xenopus-compatible selection markers into the p1 selection marker position: the γ-crystallin promoter driving GFP in the lens (Offield et al., 2000), the elastase promoter driving GFP in the pancreas (Beck and Slack, 1999), and the αCE2x6 promoter driving eCFP (enhanced cyan fluorescent protein) in the lens and pronephros (Matsuo and Yasuda, 1992). Into the p2 position, we inserted five previously characterized tissue-specific promoters, one heat shock-inducible promoter, and four ubiquitous promoters (Fig. 2, Table 1). In addition, we created p4 vectors amenable to I-SceI- and Tol2-mediated transgenesis, some containing SAR-CH4 sequences (human interferon-β scaffold attachment region-chicken β-globin DNase I hypersensitive site 4), reported to protect integrated transgenes from positional effects (Fig. 2A, Fig. 1C) (Allen and Weeks, 2005; Ramezani et al., 2003; Sekkali et al., 2008).
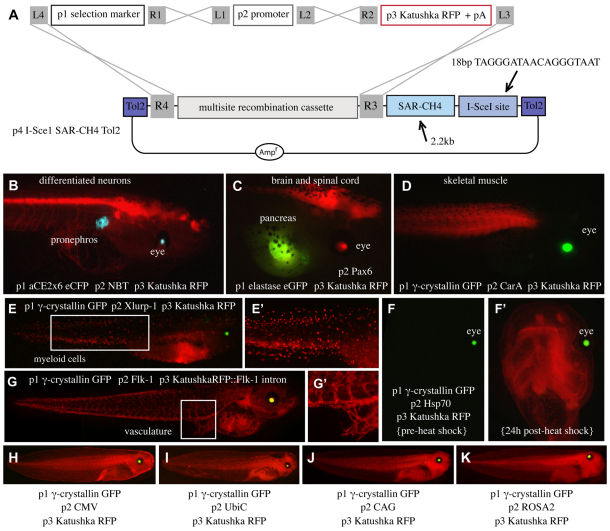
Incorporation of Xenopus-compatible elements into pTransgenesis. (A) Schematic of recombination with a Xenopus-compatible p4 vector containing a single I-SceI site, Tol2 elements, and SAR-CH4 sequences, p3 Katushka RFP and Xenopus-compatible p1 and p2 constructions. (B-K) Transgenic tadpoles from the resulting recombinations are shown. Individual p1, p2, p3 constructions and expression domains are written on each panel in white. Images in E′ and G′ are magnified images of the boxed regions in E and G, respectively. Image in F′ shows the induction of RFP from the heat-shocked tadpole in F.
Table 1.
Partial list of p2 promoters
Following recombination with selectable markers in p1, Katushka red fluorescent protein (RFP) in p3 and transgenesis elements in p4, as shown in Fig. 2A, we tested the functionality of several p2 promoter constructs by REMI transgenesis in F0 X. laevis tadpoles (Fig. 2B-K; Fig. 3D,E, middle panels). In all cases, RFP expression was appropriate to the promoter sequences driving it. We next confirmed the functionality of the p4 I-SceI site by establishing transgenic lines via I-SceI-mediated transgenesis (supplementary material Fig. S1).
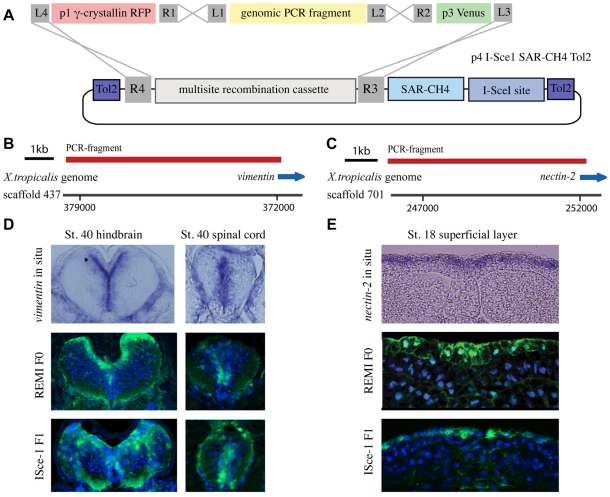
A highly efficient method of testing promoters and creating transgenic lines using pTransgenesis. (A) Genomic PCR fragments are cloned directly into the p2 position, thus allowing the recombination shown. (B,C) PCR products encoding regions 5′ to nectin-2 and vimentin were generated and tested for transcriptional activity in F0 X. laevis and in F1 X. tropicalis. (D) Transverse sections through the hindbrain and spinal cord of embryos (dorsal side up) stained for endogenous vimentin expression (upper panels), or Venus transgene expression (green) in F0 transgenic X. laevis (middle panels) and F1 transgenic X. tropicalis (lower panels). (E) Transverse sections through the neural plate of stage 18 embryos (dorsal side up) stained for endogenous nectin-2 expression (upper panel), or Venus transgene expression (green) in F0 transgenic X. laevis (middle panel) and F1 transgenic X. tropicalis (lower panel). Nuclei are stained with DAPI (blue) in middle and lower panels in D and E.
The pTransgenesis vector framework allows the rapid testing of uncharacterized DNA sequences for promoter activity owing to a commercially available PCR product cloning kit. This method allows cloning of PCR products directly into to the p2 position of the pTransgenesis framework, allowing immediate recombination upstream of a p3 coding gene and downstream of a desired p1 selection marker (Fig. 3A-C). Using this strategy, we tested functionally the promoters of two genes; vimentin, which is highly expressed in glia during Xenopus neural development (Fig. 3D, upper panels) (Yoshida, 2001) and nectin-2, which is expressed in the superficial layer of the neuroepithelium (Fig. 3E, upper panel) (Morita et al., 2010). The new promoters were found to be capable of driving VenusGFP expression in a pattern similar to the endogenous expression patterns of vimentin and nectin-2 in both the F0 and F1 generation, validating the use of these promoters for the study of neural development (Fig. 3D,E, middle and lower panels).
Also, we generated a large repertoire of fluorescent proteins in the p3 coding sequence position, including 15 fluorescent proteins with peak emissions ranging from 475 nm to 630 nm (supplementary material Fig. S2A) (Belousov et al., 2006; Mizuno et al., 2003). Using the p2 CMV promoter, we validated these 15 fluorescent proteins in p3 by linking these sequences with a p1 γ-crystallin RFP or p1 γ-crystallin GFP construction and created F0 transgenic tadpoles using REMI (supplementary material Fig. S2B,C). Taken together, these experiments validated the use of pTransgenesis framework to create a wide variety of transgenic lines in Xenopus.
Conditional transgene expression using pTransgenesis
We adapted the pTransgenesis system to be compatible with two powerful binary transgenic strategies, the Gal4-UAS system, utilized in a variety of model species, including Drosophila, zebrafish and Xenopus (Brand and Perrimon, 1993; Fischer et al., 1988; Hartley et al., 2002; Scheer and Campos-Ortega, 1999) and the Cre-loxP recombination system.
Compatibility of Gal4-UAS binary transgene expression and pTransgenesis was achieved by generating one construct to express p3 Gal4 under the control of a p2 promoter (selectable by RFP in the lens RFP; supplementary material Fig. S3A) and another expressing a p3 transgene of interest downstream of p2 UAS repeats (selectable by GFP in the lens; supplementary material Fig. S3B). Double transgenic lines are easily identified owing to the co-expression of GFP and RFP. We generated a construction expressing Gal4 under the control of the NBT promoter (i.e. expressed in the nervous system; supplementary material Fig. S3C), and VenusGFP or a dominant-negative form of the FGFR1 (XFD) tagged with VenusGFP downstream of p2 UAS (supplementary material Fig. S3D,E). VenusGFP expression was only observed in the nervous system in the double transgenics showing that the binary Gal4-UAS system works (supplementary material Fig. S3F). However, double-transgenic embryos expressing XFD-VenusGFP showed a consistent tail elongation phenotype, suggesting a role for FGF signalling in the nervous system during tail elongation (supplementary material Fig. S3G,H). Importantly, the tail and body length phenotype was also observed in the F1 generation by crossing F0 NBT:Gal4 and UAS:XFD-VenusGFP founder frogs (supplementary material Fig. S3I-K). These data showed the use of pTransgenesis to investigate the role of signalling pathways under strict spatial or temporal control. A more detailed examination of this phenotype is beyond the scope of this report and will be addressed in a separate study.
Another widely used binary transgene approach is via Cre-loxP recombination, in which a floxed transgene is excised by Cre, resulting in the conditional gene activation of another transgene, lying downstream of the distal loxP site (Lakso et al., 1992; Mosimann et al., 2011; Werdien et al., 2001). Using the pTransgenesis resource, we have successfully established via I-SceI transgenesis the first transgenic X. tropicalis line amenable to Cre-loxP recombination. The line expresses cyan fluorescent protein (CFP) ubiquitously in the absence of Cre recombinase (supplementary material Fig. S4A-E), but in the presence of Cre, recombination leads to the replacement of CFP with RFP. We demonstrated the functionality of this transgenic line, by injecting synthetic Cre-recombinase mRNA into one of the two blastomeres at the two-cell stage in the F2 generation of this line, causing the expected hemispheric RFP expression by the neurula stage (supplementary material Fig. S4F-N). This transgenic line will be valuable in long-term fate-mapping studies in X. tropicalis.
Expanding the scope of pTransgenesis
The pTransgenesis resource is designed such that the p3 coding sequence library can be easily adapted to other model species, namely, by including species-compatible selection p1 markers and p2 promoters and, if necessary, species-compatible p4 transgenesis constructs (Fig. 4A). In this section, we outline experiments showing the compatibility of pTransgenesis in Drosophila, zebrafish, and HeLa cells.
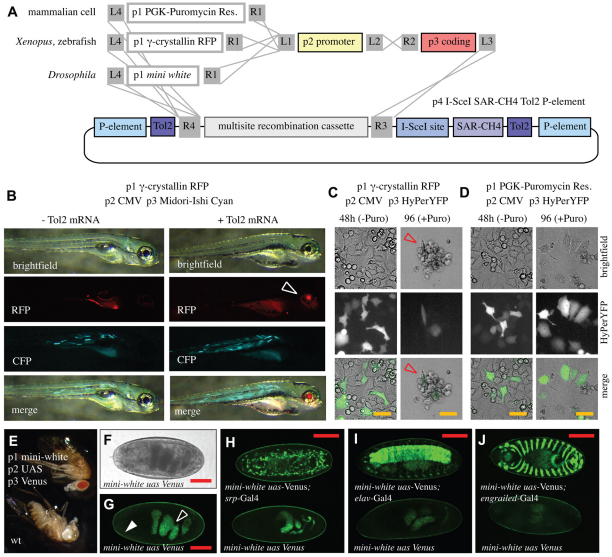
pTransgenesis in various models. (A) Schematic showing plasmid recombinations yielding plasmids compatible with transgenesis in HeLa cells, Xenopus, zebrafish and Drosophila. (B) Images of zebrafish injected with the indicated p1, p2, p3 and p4 elements with or without Tol2 mRNA. Open arrowhead points to activity of the p1 γ-crystallin RFP marker. (C,D) Results from HeLa cell transfections and puromycin (Puro) selection using pTransgenesis-engineered constructions. Red open arrow indicates dying cells. (E) Drosophila melanogaster engineered with the indicated pTransgenesis constructs versus wild type. (F,G) Phase contrast and fluorescence image of pTransgenesis-engineered Drosophila embryo. Open and closed arrowheads indicate gut and cuticle autofluorescence, respectively. (H-J) Confocal images from the indicated Gal4 crosses. (H) srp-Gal4, stage 16 embryo, GFP-labelled hemocytes. (I) eval-Gal4, stage 16 embryo, GFP-labelled CNS. (J) engrailed-Gal4, ventral view of stage15 embryo during dorsal closure process, GFP-labelled engrailed domain segments. Scale bars: 25 μm in C,D; 100 μm in F-J.
For zebrafish, we first confirmed that the p1 γ-crystallin RFP, p2 CMV construction, and a p3 fluorescent protein (Midori-Ishi Cyan) properly express in this model organism. We injected a vector with recombined p1 γ-crystallin RFP, p2 CMV and p3 Midori-Ishi Cyan constructs with or without Tol2 mRNA. We found, like others, that the p1 γ-crystallin RFP screenable marker was functional in the zebrafish eye lens (Fig. 4B, arrow), and the CMV promoter was able to drive widespread Midori-Ishi expression in the fish body (Fig. 4B) (Davidson et al., 2003). Furthermore, we cloned the zebrafish HB9 promoter (Flanagan-Steet et al., 2005) into the p2 position, recombined this with the p1 γ-crystallin RFP screenable marker and p3 VenusGFP (supplementary material Fig. S5A,B) and confirmed its activity in zebrafish using Tol2-mediated transgenesis (supplementary material Fig. S5C-E) and in Xenopus using REMI transgenesis (supplementary material Fig. S5F). Notably, we generated a transgenic zebrafish line with this construct using Tol2-mediated transgenesis, showing the functionality of the p4 Tol2 elements in the pTransgenesis system (supplementary material Fig. S5D,E).
To test the pTransgenesis system in mammalian cells, we created a p1 PGK:Puromycin Resistance cassette, which allows selection following application of puromycin to culture media. We linked this selection marker, or the p1 γ-crystallin RFP construction (negative control), to the p2 CMV and p3 HyPerYFP constructs (Fig. 4C,D). Expression of HyPerYFP was observed in ~50% of cells 48 hours after transfection using either construct (Fig. 4C,D, middle left panels). However, following addition of puromycin in the culture media, only the cells bearing the p1 PGK:PuroR cassette survived and continued to express HyPerYFP (Fig. 4D, middle right panel). Cells were successfully cultured for over a month under selection conditions.
We also tested the pTransgenesis resource in Drosophila. For this purpose, we created a p1 mini-white plasmid, which allows selection for red eyes (Tang and Sun, 2002) and a p4 vector containing sequences enabling P-element mediated transgenesis (Rubin and Spradling, 1982). By linking the p1 mini-white, p2 UAS, p3 VenusGFP and p4 P-element constructions, we created transgenic flies via P-element mediated transgenesis selectable by their red eyes (Fig. 4F). We crossed this pTransgenesis-engineered Drosophila line with previously established transgenic engrailed-Gal4, srp-Gal4 and elav-Gal4 driver lines (Bruckner et al., 2004; Landgraf et al., 1999; Millard and Martin, 2008) (Fig. 4G-K). Control stage 15-16 embryos lacking Gal4 expression showed no fluorescence besides the expected autofluorescence of the gut and cuticle (Fig. 4H, arrowheads) (Bainbridge and Bownes, 1981). By contrast, embryos expressing Gal4 showed the expected Gal4-UAS-driven VenusGFP expression in hemocytes (srp-Gal4, Fig. 4I), motor neurons (elav-Gal4, Fig. 4J) or engrailed domain segments (engrailed-Gal4, Fig. 4K).
Together, these data show that pTransgenesis vectors are compatible with other vertebrate, invertebrate and mammalian cell culture models. Compatibility between pTransgenesis vectors and previously established resources using Multisite Gateway cloning depends on the plasmid ‘RL’-site design of each resource. For example, the previously reported R4-R3 ROSA26 locus targeting (Kappas et al., 2008) and Tol2-transposon-containing vectors (Kwan et al., 2007; Villefranc et al., 2007) function as p4 vectors in the pTransgenesis system. Moreover, the promoters generated in other zebrafish studies are compatible in the p2 promoter position (Fisher et al., 2006). However, unlike earlier projects, pTransgenesis is not designed for the rapid creation of fusion proteins (Akbari et al., 2009; Villefranc et al., 2007) and, thus, these applications are better served by their original resources.
In conclusion, the pTransgenesis resource markedly streamlines the process of creating DNA constructions for engineering transgenic organisms and provides a straightforward framework for the distribution of constructs across various model organisms. An additional salient feature of the pTransgenesis resource is the substantial reservoir of constructs, which readily allows a wide variety of experimental approaches. Finally, the cross-species utility of pTransgenesis will allow researchers to evaluate more easily whether biological processes, which are increasingly being studied with a view to application, are evolutionarily conserved.
Acknowledgments
We thank Sarah Woolner, Tim Mohun, Shane Woods, Matthew Ronshaugen, Jonathan Slack, Caroline Beck, Robert Grainger, Scott Fraser, Igor Samokhvalov, Koichi Kawakami, Makoto Ikeya, Mototsugu Eiraku, Richard Hawley, Ali Ramezani, Paul Krieg, Lyle Zimmerman, Roberto Mayor and John Gurdon for their kind donations of reagents.
Footnotes
Funding
We thank the Wellcome Trust for their support of the European Xenopus Research Centre [M.J.G., E.A.]. This work was funded by the Healing Foundation [N.R.L., R.P., Y.C., E.A.]; the National Science Foundation [N.R.L.]; the Henry Luce Foundation [N.R.L.]; the Winston Churchill Scholarship Foundation [N.R.L.]; and the Wellcome Trust [N.P., E.A.]. K.D. is a Research Councils UK Research Fellow and N.P. is a Wellcome Trust Senior Research Fellow. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/10.1242/dev.066498/-/DC1
References
- Akbari O. S., Oliver D., Eyer K., Pai C. Y. (2009). An Entry/Gateway cloning system for general expression of genes with molecular tags in Drosophila melanogaster. BMC Cell Biol. 10, 8 [Europe PMC free article] [Abstract] [Google Scholar]
- Allen B. G., Weeks D. L. (2005). Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods 2, 975–979 [Europe PMC free article] [Abstract] [Google Scholar]
- Amaya E. (2005). Xenomics. Genome Res. 15, 1683–1691 [Abstract] [Google Scholar]
- Bainbridge S. P., Bownes M. (1981). Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66, 57–80 [Abstract] [Google Scholar]
- Beck C. W., Slack J. M. (1999). Gut specific expression using mammalian promoters in transgenic Xenopus laevis. Mech. Dev. 88, 221–227 [Abstract] [Google Scholar]
- Beck C. W., Christen B., Slack J. M. (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 5, 429–439 [Abstract] [Google Scholar]
- Belousov V. V., Fradkov A. F., Lukyanov K. A., Staroverov D. B., Shakhbazov K. S., Terskikh A. V., Lukyanov S. (2006). Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281–286 [Abstract] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [Abstract] [Google Scholar]
- Breckenridge R. A., Mohun T. J., Amaya E. (2001). A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev. Biol. 232, 191–203 [Abstract] [Google Scholar]
- Bruckner K., Kockel L., Duchek P., Luque C. M., Rorth P., Perrimon N. (2004). The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73–84 [Abstract] [Google Scholar]
- Chalmers A. D., Strauss B., Papalopulu N. (2003). Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development 130, 2657–2668 [Abstract] [Google Scholar]
- Davidson A. E., Balciunas D., Mohn D., Shaffer J., Hermanson S., Sivasubbu S., Cliff M. P., Hackett P. B., Ekker S. C. (2003). Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263, 191–202 [Abstract] [Google Scholar]
- Doherty J. R., Johnson Hamlet M. R., Kuliyev E., Mead P. E. (2007). A flk-1 promoter/enhancer reporter transgenic Xenopus laevis generated using the Sleeping Beauty transposon system: an in vivo model for vascular studies. Dev. Dyn. 236, 2808–2817 [Abstract] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T., Ptashne M. (1988). GAL4 activates transcription in Drosophila. Nature 332, 853–856 [Abstract] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., McCallion A. S. (2006). Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312, 276–279 [Abstract] [Google Scholar]
- Flanagan-Steet H., Fox M. A., Meyer D., Sanes J. R. (2005). Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development 132, 4471–4481 [Abstract] [Google Scholar]
- Gama Sosa M. A., De Gasperi R., Elder G. A. (2010). Animal transgenesis: an overview. Brain Struct. Funct. 214, 91–109 [Abstract] [Google Scholar]
- Gilchrist M. J., Zorn A. M., Voigt J., Smith J. C., Papalopulu N., Amaya E. (2004). Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev. Biol. 271, 498–516 [Abstract] [Google Scholar]
- Gross J. B., Hanken J., Oglesby E., Marsh-Armstrong N. (2006). Use of a ROSA26:GFP transgenic line for long-term Xenopus fate-mapping studies. J. Anat. 209, 401–413 [Abstract] [Google Scholar]
- Harland R. M. (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 [Abstract] [Google Scholar]
- Hartley J. L., Temple G. F., Brasch M. A. (2000). DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 [Europe PMC free article] [Abstract] [Google Scholar]
- Hartley K. O., Hardcastle Z., Friday R. V., Amaya E., Papalopulu N. (2001). Transgenic Xenopus embryos reveal that anterior neural development requires continued suppression of BMP signaling after gastrulation. Dev. Biol. 238, 168–184 [Abstract] [Google Scholar]
- Hartley K. O., Nutt S. L., Amaya E. (2002). Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc. Natl. Acad. Sci. USA 99, 1377–1382 [Europe PMC free article] [Abstract] [Google Scholar]
- Hope I. A., Stevens J., Garner A., Hayes J., Cheo D. L., Brasch M. A., Vidal M. (2004). Feasibility of genome-scale construction of promoter::reporter gene fusions for expression in Caenorhabditis elegans using a multisite gateway recombination system. Genome Res. 14, 2070–2075 [Europe PMC free article] [Abstract] [Google Scholar]
- Huang J. K., Dorey K., Ishibashi S., Amaya E. (2007). BDNF promotes target innervation of Xenopus mandibular trigeminal axons in vivo. BMC Dev. Biol. 7, 59 [Europe PMC free article] [Abstract] [Google Scholar]
- Ikeya M., Kawada M., Nakazawa Y., Sakuragi M., Sasai N., Ueno M., Kiyonari H., Nakao K., Sasai Y. (2005). Gene disruption/knock-in analysis of mONT3: vector construction by employing both in vivo and in vitro recombinations. Int. J. Dev. Biol. 49, 807–823 [Abstract] [Google Scholar]
- Ishibashi S., Love N. R., Amaya E. (2012). A simple method of transgenesis using I-SceI meganuclease in Xenopus. Methods Mol. Biol. (in press) [Abstract] [Google Scholar]
- Kappas N. C., Zeng G., Chappell J. C., Kearney J. B., Hazarika S., Kallianos K. G., Patterson C., Annex B. H., Bautch V. L. (2008). The VEGF receptor Flt-1 spatially modulates Flk-1 signaling and blood vessel branching. J. Cell Biol. 181, 847–858 [Europe PMC free article] [Abstract] [Google Scholar]
- Kawakami K. (2007). Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8, S7 [Europe PMC free article] [Abstract] [Google Scholar]
- Kroll K. L., Amaya E. (1996). Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122, 3173–3183 [Abstract] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 [Abstract] [Google Scholar]
- Lakso M., Sauer B., Mosinger B., Jr, Lee E. J., Manning R. W., Yu S. H., Mulder K. L., Westphal H. (1992). Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl. Acad. Sci. USA 89, 6232–6236 [Europe PMC free article] [Abstract] [Google Scholar]
- Landgraf M., Roy S., Prokop A., VijayRaghavan K., Bate M. (1999). even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron 22, 43–52 [Abstract] [Google Scholar]
- Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D. (2002). Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 [Abstract] [Google Scholar]
- Matsuo I., Yasuda K. (1992). The cooperative interaction between two motifs of an enhancer element of the chicken alpha A-crystallin gene, alpha CE1 and alpha CE2, confers lens-specific expression. Nucleic Acids Res. 20, 3701–3712 [Europe PMC free article] [Abstract] [Google Scholar]
- Millard T. H., Martin P. (2008). Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development 135, 621–626 [Europe PMC free article] [Abstract] [Google Scholar]
- Mizuno H., Mal T. K., Tong K. I., Ando R., Furuta T., Ikura M., Miyawaki A. (2003). Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol. Cell 12, 1051–1058 [Abstract] [Google Scholar]
- Morita H., Nandadasa S., Yamamoto T. S., Terasaka-Iioka C., Wylie C., Ueno N. (2010). Nectin-2 and N-cadherin interact through extracellular domains and induce apical accumulation of F-actin in apical constriction of Xenopus neural tube morphogenesis. Development 137, 1315–1325 [Europe PMC free article] [Abstract] [Google Scholar]
- Mosimann C., Kaufman C. K., Li P., Pugach E. K., Tamplin O. J., Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 [Europe PMC free article] [Abstract] [Google Scholar]
- Nyabi O., Naessens M., Haigh K., Gembarska A., Goossens S., Maetens M., De Clercq S., Drogat B., Haenebalcke L., Bartunkova S., et al. (2009). Efficient mouse transgenesis using Gateway-compatible ROSA26 locus targeting vectors and F1 hybrid ES cells. Nucleic Acids Res. 37, e55 [Europe PMC free article] [Abstract] [Google Scholar]
- Offield M. F., Hirsch N., Grainger R. M. (2000). The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development 127, 1789–1797 [Abstract] [Google Scholar]
- Ramezani A., Hawley T. S., Hawley R. G. (2003). Performance- and safety-enhanced lentiviral vectors containing the human interferon-beta scaffold attachment region and the chicken beta-globin insulator. Blood 101, 4717–4724 [Abstract] [Google Scholar]
- Ristevski S. (2005). Making better transgenic models: conditional, temporal, and spatial approaches. Mol. Biotechnol. 29, 153–163 [Abstract] [Google Scholar]
- Rubin G. M., Spradling A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353 [Abstract] [Google Scholar]
- Sakamaki K., Takagi C., Yoshino J., Yokota H., Nakamura S., Kominami K., Hyodo A., Takamune K., Yuge M., Ueno N. (2005). Transgenic frogs expressing the highly fluorescent protein venus under the control of a strong mammalian promoter suitable for monitoring living cells. Dev. Dyn. 233, 562–569 [Abstract] [Google Scholar]
- Scheer N., Campos-Ortega J. A. (1999). Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 80, 153–158 [Abstract] [Google Scholar]
- Sekkali B., Tran H. T., Crabbe E., De Beule C., Van Roy F., Vleminckx K. (2008). Chicken beta-globin insulator overcomes variegation of transgenes in Xenopus embryos. FASEB J. 22, 2534–2540 [Abstract] [Google Scholar]
- Semple J. I., Garcia-Verdugo R., Lehner B. (2010). Rapid selection of transgenic C. elegans using antibiotic resistance. Nat. Methods 7, 725–727 [Abstract] [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 [Europe PMC free article] [Abstract] [Google Scholar]
- Smith S. J., Kotecha S., Towers N., Latinkic B. V., Mohun T. J. (2002). XPOX2-peroxidase expression and the XLURP-1 promoter reveal the site of embryonic myeloid cell development in Xenopus. Mech. Dev. 117, 173–186 [Abstract] [Google Scholar]
- Tang C. Y., Sun Y. H. (2002). Use of mini-white as a reporter gene to screen for GAL4 insertions with spatially restricted expression pattern in the developing eye in drosophila. Genesis 34, 39–45 [Abstract] [Google Scholar]
- Villefranc J. A., Amigo J., Lawson N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077–3087 [Abstract] [Google Scholar]
- Werdien D., Peiler G., Ryffel G. U. (2001). FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 29, e53 [Europe PMC free article] [Abstract] [Google Scholar]
- Yoshida M. (2001). Glial-defined boundaries in Xenopus CNS. Dev. Neurosci. 23, 299–306 [Abstract] [Google Scholar]
Articles from Development (Cambridge, England) are provided here courtesy of Company of Biologists
Full text links
Read article at publisher's site: https://doi.org/10.1242/dev.066498
Read article for free, from open access legal sources, via Unpaywall:
https://dev.biologists.org/content/develop/138/24/5451.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1242/dev.066498
Article citations
Development of a heat-stable alkaline phosphatase reporter system for cis-regulatory analysis and its application to 3D digital imaging of Xenopus embryonic tissues.
Dev Growth Differ, 66(3):256-265, 04 Mar 2024
Cited by: 1 article | PMID: 38439617 | PMCID: PMC11457516
Duox is the primary NADPH oxidase responsible for ROS production during adult caudal fin regeneration in zebrafish.
iScience, 26(3):106147, 04 Feb 2023
Cited by: 3 articles | PMID: 36843843 | PMCID: PMC9950526
Xenopus Transgenesis Using the pGateway System.
Methods Mol Biol, 2633:97-109, 01 Jan 2023
Cited by: 0 articles | PMID: 36853460
Aerobic glycolysis is important for zebrafish larval wound closure and tail regeneration.
Wound Repair Regen, 30(6):665-680, 05 Oct 2022
Cited by: 12 articles | PMID: 36148505 | PMCID: PMC9828577
A Focal Impact Model of Traumatic Brain Injury in Xenopus Tadpoles Reveals Behavioral Alterations, Neuroinflammation, and an Astroglial Response.
Int J Mol Sci, 23(14):7578, 08 Jul 2022
Cited by: 5 articles | PMID: 35886924 | PMCID: PMC9323330
Go to all (36) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
High-throughput library transgenesis in Caenorhabditis elegans via Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS).
Elife, 12:RP84831, 04 Jul 2023
Cited by: 7 articles | PMID: 37401921 | PMCID: PMC10328503
Serial Recombineering Cloning to Build Selectable and Tagged Genomic P[acman] BAC Clones for Selection Transgenesis and Functional Gene Analysis using Drosophila melanogaster.
Curr Protoc, 3(2):e675, 01 Feb 2023
Cited by: 0 articles | PMID: 36757632 | PMCID: PMC9923880
Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments.
Methods Cell Biol, 135:219-244, 26 Feb 2016
Cited by: 19 articles | PMID: 27443928
Gal4 Driver Transgenic Zebrafish: Powerful Tools to Study Developmental Biology, Organogenesis, and Neuroscience.
Adv Genet, 95:65-87, 13 Jun 2016
Cited by: 35 articles | PMID: 27503354
Review
Funding
Funders who supported this work.
Wellcome Trust (1)
Studying neurogenesis through developmental time.
Professor Nancy Papalopulu, University of Manchester
Grant ID: 090868





