Abstract
Free full text

Artemis interacts with the Cul4A-DDB1DDB2 ubiquitin E3 ligase and regulates degradation of the CDK inhibitor p27
Abstract
Artemis, a member of the SNM1 gene family, is a multifunctional phospho-protein that has been shown to have important roles in V(D)J recombination, DNA double-strand break repair and stress-induced cell cycle checkpoint regulation. We show here that Artemis interacts with the Cul4A-DDB1 E3 ubiquitin ligase via a direct interaction with the substrate-specificity receptor DDB2. Furthermore, Artemis also interacts with the CDK inhibitor and tumor suppressor p27, a substrate of the Cul4A-DDB1 ligase, and both DDB2 and Artemis are required for the degradation of p27 mediated by this complex. We also show that the regulation of p27 by Artemis and DDB2 is important for cell cycle progression in normally proliferating cells and in response to serum deprivation. These findings thus define a function for Artemis as an effector of Cullin-based E3 ligase-mediated ubiquitylation, demonstrate a novel pathway for the regulation of p27 and show that Cul4A-DDB1DDB2-Artemis regulates G1-phase cell cycle progression in mammalian cells.
Introduction
Artemis is a member of the mammalian SNM1/PSO2 gene family, which also includes SNM1A and SNM1B/Apollo, all of which have proven to have diverse roles in DNA metabolism and cell cycle regulation.1–5 Members of this family share the SNM1 conserved domain, characterized by a metallo-β-lactamase fold and an appended β-CASP (CPSF-Artemis-Snm1-Pso2) motif, the latter of which is unique to this family.6,7 The SNM1 domain encompasses a nuclease activity in the SNM1/PSO2 proteins, which accounts for their function in DNA metabolism, such as that of SNM1B/Apollo in telomere resection8,9 and that of Artemis in V(D)J recombination and the repair of DSBs by nonhomologous end-joining (NHEJ).10,11 Mutations of Artemis thus lead to the development of radiosensitive severe combined immunodeficiency syndrome (RS-SCID), which is characterized by failure in the maturation of both B and T cells and cellular sensitivity to IR.7,11–13 However, in addition to their functions as nucleases SNM1/PSO2 proteins have nonnucleolytic roles in the regulation of the mammalian cell cycle. For instance, both SNM1A and SNM1B/Apollo have roles in the regulation of an early mitotic or prophase checkpoint in response to spindle poisons.14,15 Artemis itself is a phospho-protein that is phosphorylated at multiple sites by PIKKs (phosphoinositide-3-kinase related protein kinases) such as ATM and ATR upon DNA damage.4 These phosphorylations of Artemis are involved in regulating recovery of cells from DNA damage-induced G2/M and S-phase cell cycle checkpoints via regulation of cyclin B-CDK1 and cyclin E, respectively.16–18 Interestingly, the regulation of cyclin E by Artemis was shown to be mediated by its interaction with the SCFFbw7 E3 ubiquitin ligase complex. In addition, Artemis has been shown to be a negative regulator of p53 in response to oxidative or oncogenic stress.19 Thus, Artemis has pleiotropic roles in regulating the mammalian cell cycle in response to various forms of cellular stress, apparently independent of its nuclease activity.
The mammalian cell cycle is regulated by a series of protein complexes composed of cyclins and cyclin-dependent kinases (CDKs), the activity of which is suppressed by a group of CDK inhibitors (CKIs), such as p27Kip1 and p16Ink4A.20,21 p27, as an inhibitor of CDKs, acts as a central regulator of cellular proliferation, differentiation and malignant transformation. p27 is a tumor suppressor, and low levels of expression or mislocalization to the cytoplasm correlate with tumor recurrence and poor prognosis in some forms of cancer.22–25 p27 is a short-lived molecule, and its cellular levels are largely mediated by the ubiquitin-protesome system, and several E3 ligases responsible for p27 degradation have been identified.26 SCFSkp2 is perhaps best-characterized for the degradation of p27 in the nucleus during the S-G2 phase of the cell cycle.27–29 In addition to SCFSkp2, the Kip1 ubiquitination-promoting complex (KPC) mediates ubiquitylation of p27 in the cytoplasm during the G1 phase, thus allowing its cytoplasmic destruction following nuclear export.30
Several studies have also implicated the Cul4A-based E3 ligase complex in the ubiquitylation and degradation of p27, although the substrate specificity factors for this pathway have yet to be identified.31–33 Cul4A associates with the multi-propeller adaptor protein DDB1, which binds specific substrate receptors also known as DCAFs (DDB1-Cul4-associated factors).34–38 DDB (damaged DNA binding protein) was purified as a heterodimer, which contains the subunits DDB1 and DDB2 and has high affinity for UV-damaged DNA.39 The function of this complex has been primarily associated with the global genome repair (GGR) pathway of nucleotide excision repair.40,41 However, more recent studies have shown that DDB can associate with Cul4A and target proteins such as XPC, H3, H4 and DDB2 for ubiquitylation.42–44 In addition, by interacting with different substrate receptors or DCAFS, Cul4A-DDB1 can induce proteolysis of proteins involved in various cellular pathways, particularly those involved in chromatin biology.36
Here, we show that Artemis is a component of the Cul4A-DDB1 E3 ligase complex via a direct interaction with DDB2. Additionally, Artemis interacts with p27, and both DDB2 and Artemis are required for the ubiquitylation and degradation of p27 mediated by the Cul4A-DDB1 complex. Furthermore, we show that the regulation of p27 by Artemis and DDB2 is critical for cell cycle progression in normally proliferating cells and in response to serum withdrawal. These findings define a novel pathway for the regulation of p27 in the nucleus during the G1 cell cycle phase and define a new function for Artemis as an effector of Cullin-based E3-ligase mediated ubiquitylation and, therefore, a cell cycle regulator in unperturbed, proliferating cells.
Results
Artemis interacts with the DDB heterodimer via DDB2.
To investigate the function of Artemis, we initiated a search for cellular interacting partners using GST pull-down and mass spectrometry. DDB1 was specifically detected in the transfected lysates, suggesting that it might be an Artemis-interacting protein (Fig. 1A). To confirm this interaction, we first immunoprecipitated (IPed) endogenous Artemis from the normal human fetal lung cell line MRC5 and showed that DDB1 co-IPed with it (Fig. 1B). In addition, HEK293 cells were treated with UV, IR or mock treated and reciprocal co-immunoprecipitations (co-IPs) were performed. DDB1 and Artemis co-IPed with each other indicating that Artemis interacts with DDB1 both before and after DNA damage (Fig. 1C and D). These experiments demonstrate that Artemis and DDB1 form a complex, and that this interaction is not affected by DNA damage.
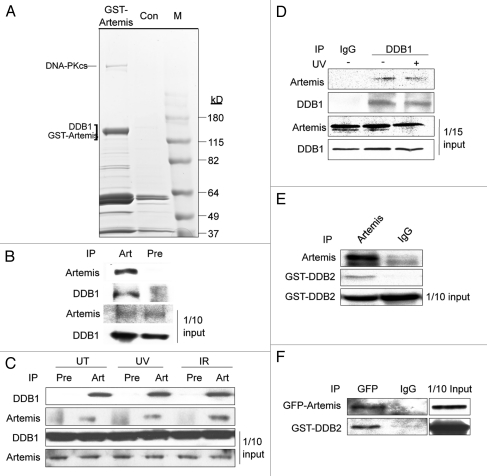
Artemis interacts with DDB1 and DDB2 in vivo. (A) Gel stained with Coomassie blue showing proteins from a GST-Artemis pull-down experiment. GST-Artemis expressed in HEK293 cells was pulled-down by glutathione sepharose beads (left lane). A pull-down from untransfected cells was used as a control (middle lane). Labeled band indicate proteins identified by mass spectrometric analysis. “M” indicates marker proteins. (B) Immunoblot showing that Artemis co-IPs with DDB1 in the primary fetal lung cell line MRC5. (C) Immunoblot showing that endogenous DDB1 interacts with Artemis in vivo before and after UV or IR irradiation. HEK293 cells were treated with UV (25 J/m2), IR (10 Gy) or mock treated (UT). Cells were harvested for co-IP analysis two hours post-irradiation. “Pre” indicates pre-immune serum. (D) Reciprocal co-IP experiment showing that endogenous Artemis interacts with DDB1 in vivo before and after UV damage. HEK293 cells were treated with UV (25 J/m2) or left untreated. Cells were incubated for two hours after treatment and subsequently harvested for analysis. (E) Immunoblot showing that endogenous Artemis interacts with ectopically expressed DDB2. HEK293 cells were transfected with GST-DDB2, and 48 h later, cells were harvested for co-IP experiments. (F) Co-IP experiment showing the interaction between Artemis and DDB2. GsT-DDB2 and GFP-Artemis were co-transfected into HEK293 cells, and 48 h later, cells were harvested for analysis.
DDB1 was originally purified as a heterodimer with DDB2 that binds to chromatin after UV-induced DNA damage. Since Artemis interacts with DDB1 in vivo, we examined whether Artemis also interacts with DDB2. The result of a co-IP experiment indicated that Artemis interacts with DDB2 (Fig. 1E). Furthermore, an interaction between ectopically expressed GFP-Artemis and GST-DDB2 was observed, indicating that Artemis interacts with both DDB1 and DDB2 in vivo (Fig. 1F).
To determine which subunit of the DDB complex directly interacts with Artemis, we conducted pull-down assays with 35S-methionine-labeled proteins translated in vitro. Although Artemis and DDB1 interacted with each other in vivo, these proteins did not co-IP in vitro (Fig. 2A and B), although, as expected, DDB1 co-IPed with Cul4A (Fig. 2A and lower part). However, GST-DDB2 was co-IPed by Artemis antibody (Fig. 2C), indicating a direct interaction between Artemis and DDB2. This direct interaction was confirmed by a GST-pull down assay using recombinant GST-Artemis and MBP-DDB2 expressed and purified from E. coli (Fig. 2D). These results demonstrate a direct association between Artemis and DDB2, but not with DDB1, indicating that DDB2 subunit mediates the interaction with Artemis.
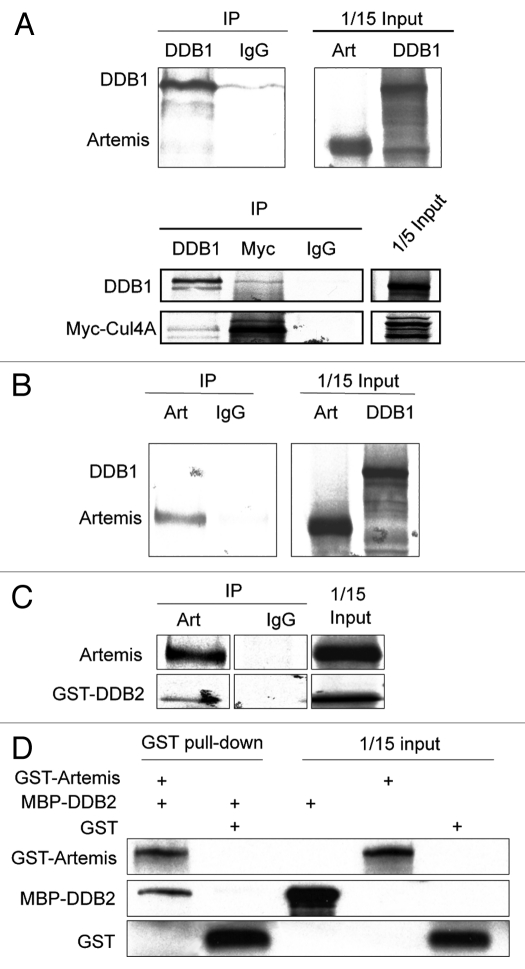
Artemis interacts directly with DDB2. (A and B) Artemis does not interact with DDB1 in vitro. Artemis and DDB1 labeled with 35S-methionine were expressed in vitro by transcription-coupled translation. Artemis and DDB1 were mixed and IPed with DDB1 antibody (A, upper part) or Artemis antiserum (B) as indicated. A control experiment shows the interaction between DDB1 and Cul4A (A, lower part). Bound proteins were eluted, separated by SDS-PAGE and detected by autoradiography. (C) Artemis directly interacts with GST-DDB2 in vitro. Artemis and GST-DDB2 were in vitro translated and labeled with 35S-methionine. Synthesized proteins were mixed and incubated as indicated for immunoprecipitation. Parts show the autoradiography of labeled proteins. (D) Purified recombinant Artemis and DDB2 directly interact. GST-Artemis, GST and MBP-DDB2 proteins were expressed and purified from E. coli. Purified proteins were mixed as indicated, and pull-down assays were performed and analyzed by immunoblotting.
Artemis and DDB2 mediate the degradation of p27.
DDB1 and DDB2 are known components of the Cul4A-based E3 ligase.34 Because Artemis interacts with DDB, we next investigated whether Artemis plays a role in regulating the stability of Cul4A-DDB1 substrates. Cul4A-DDB1 has been shown to promote the degradation of the CDK inhibitor, p27.31 However, the substrate receptor(s) remains unknown. We depleted expression of Artemis or DDB2 in HeLa cells and examined p27 levels (Fig. 3A and lanes 1–4 and 8–11). Interestingly, p27 was significantly increased in cells with decreased Artemis or DDB2 expression compared with control cells treated with a non-specific siRNA. This increase in p27 was reduced when an Artemis construct resistant to the Artemis specific siRNA was expressed in the cells depleted of endogenous Artemis (Fig. 3A and lanes 5–7). Additionally, p27 levels were also increased in Artemis−/− mouse embryonic fibroblasts (MEFs) compared with wild-type MEFs (Fig. 3B). Moreover, when Artemis or DDB2 was overexpressed in HeLa cells, p27 was significantly decreased (Fig. 3C).
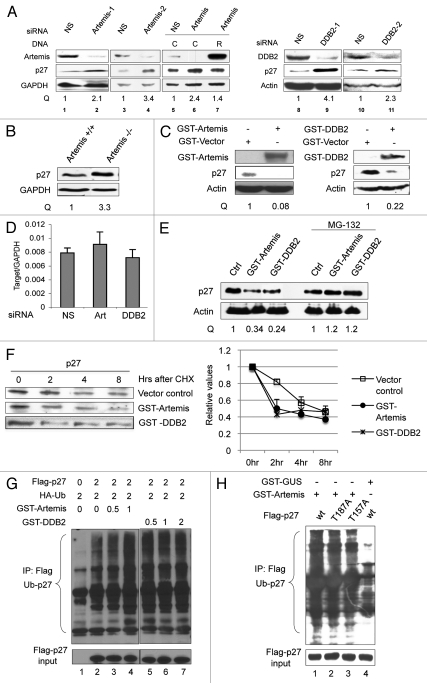
Artemis and DDB2 regulate p27 protein levels via a ubiquitin-mediated pathway. (A) P27 accumulates in Artemis or DDB2 depleted cells. HeLa cells were transfected with control (NS), Artemis or DDB2 siRNas. Forty-eight hours after transfection, cells were harvested and cell lysates were subjected to immunoblot analysis. Artemis-1 and Artemis-2, DDB2-1 and DDB2-2 indicate distinct siRNAs. As a control, 24 h after siRNA transfection cells were transfected with control plasmid DNA (C) or an Artemis construct refractory to Artemis siRNA (R). Cells were then incubated for an additional 24 h before harvesting. GAPDH and Actin indicate loading controls. “Q” indicates the relative band intensities of p27 normalized to GAPDH or actin levels. (B) p27 accumulates in Artemis null MEF cells. Lysates prepared from Artemis+/+ and Artemis−/− MEF cells were subjected to immunoblot analysis. (C) Overexpression of Artemis or DDB2 reduces p27 levels. HeLa cells were transfected with GST-Artemis (left part) or GST-DDB2 (right part) plasmid DNAs or treated with mock transfections. Forty-eight hours after transfection, cells were harvested and cell lysates were subjected to immunoblot analysis. (D) P27 mRNA is stable in Artemis or DDB2 depleted cells. mRNA was isolated from HeLa cells 48 h after transfection with control (NS), Artemis or DDB2 siRNAs. P27 mRNA levels were determined by real-time PCR. Results were normalized using GAPDH as an internal control. (E) Artemis and DDB2 regulate p27 protein levels through proteosome mediated degradation. HeLa cells were transfected with GST-Artemis or GST-DDB2 DNAs. After 48 h cells were treated with 20 mM MG-132, and 5 h later, cells were harvested and lysates subjected to immunoblot analysis. (F) Overexpression of either Artemis or DDB2 decreases the half-life of p27 compared with control. HeLa cells were transefected with GST-Artemis or GST control vectors. Thirty-six hours after transefection, cycloheximide (100 µg/ml) was added and the decrease in p27 protein level analyzed as a function of time by immunoblot analysis (left part). Quantitation of immunoblots are shown (right part). (G) Artemis and DDB2 promote ubiquitylation of p27 in vivo. HeLa cells were transfected with indicated amounts of Flag-p27, HA-Ub and GST-Artemis or GST-DDB2 DNAs. Cells were harvested 48 h after transfection, and an in vivo ubiquitination assay was performed using anti-Flag M2 agarose. Ubiquitylated proteins were detected using an HA antibody. Flag-p27 indicates a loading control at 10% of input. (H) Artemis promotes ubiquitylation of p27 independent of its phosphorylation on T187 or T157. HeLa cells were transfected with Flag-tagged wild-type (lane 1 and 4), T187A (lane 2) or T157A (lane3) p27, HA-Ub, GST-Artemis and GST-GUS. Forty-eight hours after transfection, an in vivo ubiquitylation assay was performed as indicated in Figure 4F.
p27 is regulated at both transcriptional and post-translational levels.45–48 Thus, to rule out the possibility that p27 accumulation in the absence of Artemis or DDB2 was a result of increased transcription, real-time PCR was performed in cells depleted of Artemis or DDB2. As shown (Fig. 3D), p27 mRNA levels remained stable in transfected cells, indicating that the observed accumulation of p27 is regulated at a post-transcriptional level. In addition, upon treatment with the protease inhibitor MG-132, the differential expression of p27 between control and Artemis or DDB2 overexpression was abolished (Fig. 3E). Finally, we performed experiments to determine the half-life of p27 upon overexpression of either Artemis or DDB2 and showed that it was reduced compared with transfection of a vector control (Fig. 3F). We conclude that alterations in expression of Artemis or DDB2 affect the stability of p27, and this is due to a post-translational pathway.
Next, we investigated the direct involvement of Artemis and DDB2 in p27's ubiquitylation in vivo. Overexpression of either Artemis or DDB2 promoted the ubiquitylation of p27 in the presence of MG132 (Fig. 3G and compare lanes 3 and 4 with lane 2, and lanes 5–7 with lane 2). These findings are consistent with our observations that overexpression of Artemis or DDB2 reduces p27 levels in vivo (Fig. 3C).
It has been shown that p27 is phosphorylated at numerous sites, and these modifications are important for regulation of its stability and cellular localization. For example, phosphorylation of p27 at Thr187 by CDK2 is required for SCFSkp2-mediated ubiquitylation.27,49,50 Also, AKT-mediated phosphorylation of p27 at Thr157 is responsible for its cytoplasmic retention.51 To test whether phosphorylation of these sites is required for Artemis-mediated p27 ubiquitylation, wild-type, Thr157A or Thr187A mutants of p27 were overexpressed. The presence of GST-Artemis promoted the ubiquitylation of both wild-type and the mutant p27s equally, whereas, in the absence of GST-Artemis the ubiquitylation was greatly reduced (Fig. 3H). This result suggests that Artemis can promote the degradation of p27 in the nucleus, and that phosphorylation of p27 at Thr187 is not required for this activity.
Artemis and DDB2 regulate p27 via the Cul4A-based E3 ligase complex.
Several studies have implicated Cul4A and DDB1 in the degradation of p27.31–33 To determine if Artemis and DDB2 act with Cul4A-DDB1, we examined the effect of Cul4A or DDB1 overexpression on p27 in cells depleted of Artemis or DDB2. When cells were treated with control siRNA, co-transfection of Cul4A or DDB1 destabilized p27, as did co-transfection of GST-Artemis or GST-DDB2 (Fig. 4A). However, p27 remained largely unchanged when either Artemis or DDB2 siRNAs were used to treat the co-transfected cells (Fig. 4A). Moreover, GST-DDB2 did not promote p27's degradation when it was overexpressed in Artemis-depleted cells, nor did GST-Artemis overexpression in the DDB2 depleted cells (Fig. 4A), indicating that they are involved in a common pathway. Furthermore, in a reciprocal experiment we depleted Cul4A and transfected GST-DDB2 or GST-Artemis DNA. The overexpression of either Artemis or DDB2 did not cause reduction of p27 in Cul4A depleted cells compared with a control siRNA (Fig. 4B). These results indicate that Artemis and DDB2 are both required for Cul4A-DDB1-mediated degradation of p27.

Artemis and DDB2 regulate p27 protein levels through the Cul4A-based E3 ligase complex. (A) Artemis and DDB2 are required for p27 degradation by the Cul4A-based E3 ligase complex. HeLa cells were transfected with control, Artemis or DDB2 siRNAs. After 24 h, cells were transfected with the indicated DNa, and 24 h later, cells were harvested for immunoblot analysis. (B) Cul4A is required for Artemis or DDB2-mediated p27 degradation. HeLa cells were transfected with control or Cul4A siRNAs, and 24 h later, cells were transfected with the indicated DNA. Cells were harvested after an additional 24 h for immunoblot analysis.
Artemis interacts directly with p27.
Our results demonstrate an interaction between Artemis and the DDB complex, and that Artemis is involved in the Cul4A-DDB1-mediated degradation of p27. We, therefore, examined whether Artemis interacts with the substrate p27. A co-IP experiment showed that p27 and Artemis interact in vivo (Fig. 5A). To determine if Artemis and p27 directly interact, we purified GST-Artemis and MBP-p27 from E. coli cells and performed a pull-down assay. This experiment indicated that Artemis and p27 directly interact (Fig. 5B). These results and those shown above suggest that Artemis may function in substrate recruitment in the Cul4A-DDB1DDB2 ligase complex (Fig. 5C).
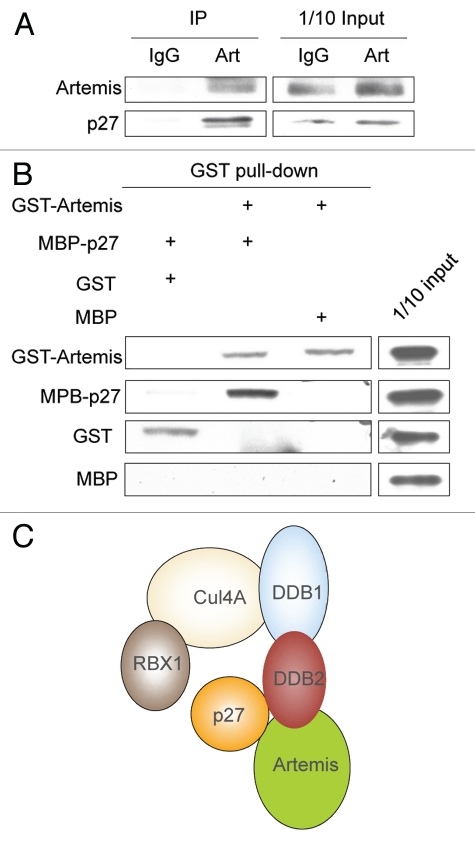
Artemis interacts with p27. (A) Artemis interacts with p27 in vivo. Immunoblot analysis showing co-IP experiment performed in lysates from HEK293 cells. (B) Artemis directly interacts with p27 in vitro. GST, GST-Artemis, MBP and MBP-p27 were expressed in E. coli. Purified proteins were mixed and subjected to GST pull-down assays. (C) Putative structural model for the roles of DDB2 and Artemis in the ubiquitylation of p27 by the Cul4A-DDB1 complex.
Numerous substrates have been identified for the Cul4A-DDB1 ligase complex.36,52 Here, we have demonstrated the involvement of Artemis in this complex to promote the ubiquitylation of p27. To determine if this role of Artemis is substrate specific, we examined CDT1, a known substrate of the Cul4A-DDB1Cdt2 complex53 in Artemis-depleted cells. CDT1 is known to be degraded by Cul4A-DDB1Cdt2 both during S phase and after UV irradiation. However, CDT1 levels in cells with or without UV treatment were not affected by Artemis depletion (Fig. S1A) Moreover, interaction between Artemis and CDT2, the receptor protein for CDT1 in the Cul4A-DDB1 complex, was not detected (Fig. S1B). These results suggest that Artemis is specifically involved in the degradation of particular substrates, such as p27, as opposed to playing a constitutive role in Cul4A-DDB1-mediated substrate ubiquitylation.
Artemis and DDB2 regulate G1 cell cycle progression.
p27 has a crucial role in the G1-S transition by interacting with and inhibiting cyclin E-CDK2.54 Since we have shown above that Artemis and DDB2 regulate the stability of p27, we next examined the effects of these proteins on transition through G1. hTERT-RPE1 cells, a telomerase-immortalized primary human cell line, were employed for cell cycle analysis.55,56 First, we depleted the expression of Artemis and/or p27 and determined the population of S-phase cells by BrdU incorporation (Fig. 6A). Depletion of p27 led to the expected increase in S-phase cells compared with a control, while depletion of Artemis led to a decrease in the S-phase population. When both Artemis and p27 were depleted together, these differences in S-phase population were similar to the control cells, which suggests that Artemis can regulate the G1/S transition in normally proliferating cells by affecting p27 stability.
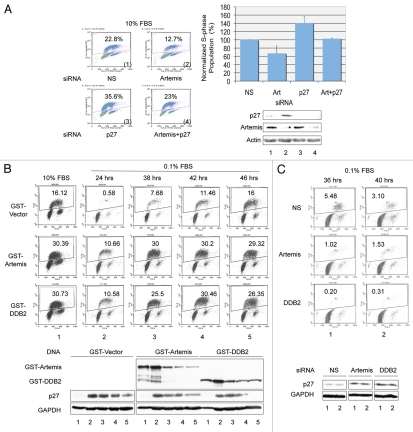
Artemis and DDB2 regulate cell cycle progression. (A) Depletion of Artemis causes a p27-dependent G1 arrest. hTERT-RPE1 cells were transfected with Artemis and/or p27 specific siRNA or non-specific (NS) siRNA. Forty-eight h after transfection, BrdU was added to label S-phase cells. Thirty minutes after incubation with BrdU, cells were harvested and fixed. Cell cycle analysis was performed by FACS (upper panel), and results from three independent experiments are shown graphically (lower left panel). Samples collected in the cell cycle analysis were subjected to immunoblot analysis for p27 and Artemis (lower right panel). Upper numbers in the panels indicate the percentage of S-phase cells. (B) Artemis and DDB2 affect the G1-G0 transition via regulation of p27. hTERT-RPE1 cells were transfected with GST-Artemis or GST-DDB2 constructs. Thirty h after transfection, cells were placed into DMEM F-12 media containing 0.1% FBs. Thirty minutes prior to harvest at the indicated times, BrdU was added to the media. Cell cycle profiles were determined by FACS analysis (upper panel). Lower panel: Samples collected in the cell cycle analysis were subjected to immunoblot analysis for p27 (lower panel). (C) Artemis and DDB2 affect the G1-G0 transition via regulation of p27. Upper panel: hTERT-RPE1 cells were transfected with Artemis, DDB2, or non-specific siRNAs (NS). Twenty-four h after transfection cells were placed into media containing 0.1% FBS for the indicated time. BrdU was added into the culture 30 min before harvest, and cell cycle analysis was performed by FACS (upper panel). Samples collected in the cell cycle analysis were subjected to immunoblot analysis for p27 (lower panel).
In response to the absence of mitogenic signals, p27 accumulates and cells arrest in a quiescent or G0 state. Downregulation of p27 is required for arrested cells to re-enter the cell cycle. To determine if Artemis and DDB2 can affect the G1 to G0 transition, we overexpressed these proteins in hTERT-RPE1 cells. Consistent with our previous observations, overexpression of Artemis or DDB2 destabilized p27 resulting in an approximate 2-fold increase in the S-phase population compared with control cells (Fig. 6B and column 1). Upon reduction of the serum concentration to 0.1% for 24 h, cells expressing either Artemis or DDB2 exhibited a 20-fold increase in the S-phase population compared with control cells (Fig. 6B and column 2). An interesting peculiarity of the hTERT-RPE cells is that after 24 h in 0.1% serum, they appeared to adapt and return to the cell cycle, as indicated by an increase S-phase population (Fig. 6B and vector). Overexpression of either Artemis or DDB2 accelerated this re-entry into the cell cycle (Fig. 6B and columns 3–5). Furthermore, in a reciprocal experiment, depletion of Artemis or DDB2 by siRNA impeded cell cycle reentry of hTERT-RPE cells kept in 0.1% serum, consistent with increased levels of p27 (Fig. 6C). These results support the conclusion that Artemis and DDB2 regulate p27 in response to the withdrawal of mitogenic signals.
Discussion
Artemis is a component of Cul4A-DDB1 E3 ligase complex.
Artemis has previously been shown to participate in multiple cellular pathways including V(D)J recombination, NHEJ, HR and cell cycle checkpoint responses.7,10,11,16–18,57,58 These prior studies have generally indicated a role for Artemis in response to DNA damage or an involvement in nucleic acid processing. However, in this study we have defined a novel function for Artemis as a component of the Cul4A-DDB1 E3 ubiquitin ligase complex and as a mediator, together with DDB2, of p27 regulation during the unperturbed cell cycle.
Using a proteomics screen we identified DDB1 as an interacting partner of Artemis and showed that this interaction was mediated directly via DDB2, which is one of many substrate specificity factors or DCAFs that interact with DDB1.36–38 The Cul4A-DDB1DDB2 E3 ligase has been shown to have a role in the repair of UV-induced DNA damage, however, neither UV nor IR affected the interaction between Artemis and DDB1. This observation is of interest for several reasons. First, because Artemis has been shown to be extensively phosphorylated after UV, this result indicates that these phosphorylations do not affect this interaction. Previously, we have shown that phosphorylation of Artemis by UV enhances its interaction with the F-box protein Fbw7 and the subsequent ubiquitylation of cyclin E.17 Additionally, the DDB2 protein has been shown to be rapidly degraded after UV,44 nevertheless, the interaction between Artemis and DDB1 remains unchanged. Moreover, this interaction was also unaffected when DDB2 expression was depleted using a specific siRNA (data not shown). This poses the likelihood that in addition to DDB2, other substrate specificity factors mediate the interaction between Artemis and DDB1. One likely candidate is the DCAF CSA, which is a repair protein involved in the transcription-coupled pathway of NER.34,59,60 Similar to DDB2, it is a WD40 protein that acts as a substrate receptor for the Cul4A-DDB1 ligase. In contrast to DDB2, CSA does not undergo degradation after UV, and we have observed a direct interaction between Artemis and CSA in vitro (data not shown). This observation indicates that in addition to DDB2, CSA and possibly other DCAFs can bridge the interaction between Artemis and DDB1 and, therefore, provide an explanation for the unchanged interaction between Artemis and DDB1 after UV. These findings suggest that Artemis is involved in the ubiquitylation of additional substrates of Cul4A-DDB1 other than p27. However, Artemis is not a constitutive member of the Cul4A-DDB1 complex as it appears not to be involved in the ubiquitylation of substrates such as CDT1 or p21, which is mediated by the CDT2 substrate specificity factor.
Artemis and DDB2 regulate the degradation of p27 and G1-S cell cycle progression via the Cul4A-DDB1 complex.
As a CDK inhibitor, p27 levels, localization and activity are largely regulated through post-translational mechanisms.26,61 SCFSkp2 and KPC1/2 are two well-established E3 ligases that promote the degradation of p27.27,30 These two complexes act distinctly; SCFSkp2 functions in the nucleus and targets p27 phosphorylated by CDK2 at Thr187 during S and G2 phases, while cytoplasmic KPC1/2 ubiquitylates p27 that is exported from the nucleus upon Ser10 phosphorylation during early G1 phase. Recent studies have also indicated a possible involvement of Cul4A-DDB1 in p27 degradation.31–33 Our results presented here not only confirm these previous findings, but also reveal that both Artemis and DDB2 are required for p27 degradation via the Cul4A-DDB1 complex. These observations provide direct evidence to support the hypothesis that in complex with Artemis, Cul4A-DDB1DDB2 (Cul4A-DDB1DDB2-Artemis) can function as a third E3 ligase responsible for p27 degradation.
Why do cells require a third E3 ligase to mediate the degradation of p27? As a Cip/Kip family member, p27 can bind and inhibit CDK2.54 P27 levels are high in G0 phase, which is concomitant with low CDK activity, whereas, p27 levels decrease via the ubiquitin-proteasome pathway when cells re-enter the cell cycle and progress to S phase, thereby allowing the activation of the CDK2 kinase. CDK2 phosphorylates p27 at Thr187, which is then recognized and ubiquitylated by SCFSkp2.50 Thus, p27 inhibits the activity of the kinase that potentiates its degradation by SCFSkp2. Moreover, Skp2 is not expressed until late G1 phase, and Skp2−/− cells exhibit normal G0-G1 progression and p27 degradation.30,62 This evidence indicates that a SCFSkp2-independent p27 degradation pathway(s) is employed by cells, and this speculation led to the discovery of the KPC1/2 complex.30 Upon cell cycle re-entry, p27 is phosphorylated on serine 10 by KIS kinase.63 This phosphorylation signals the nuclear export of p27, and cytoplasmic p27 is ubiquitylated and degraded by KPC1/2. The subcellular redistribution and subsequent degradation decreases p27 levels in the nucleus, thereby relieving the inhibitory effect on CDK2 to permit its activation, which in turn facilitates SCFSkp2-mediated p27 degradation. However, only a fraction of p27 is phosphorylated on serine 10 and trans-located into the cytoplasm in early G1 phase. In addition, nuclear retained p27 can still be degraded independent of serine 10 phosphorylation.62 Therefore, the existence of another E3 ligase to downregulate p27 in the nucleus during G1 phase appears necessary. Our findings here indicate that Cul4A-DDB1DDB2-Artemis fulfills this role, and that it is likely involved in regulating both G1-S and G1-G0-S transitions.
As further support for this model, Thr to Ala mutations of p27 at Thr187 and Thr157 did not affect the level of Artemismediated ubiquitylation compared with wild-type p27. These results indicate that Artemis-mediated targeting of p27 can occur during early G1 phase in the absence of activated CDK2-cyclin E. Furthermore, phosphorylation of Thr157 by AKT results in cytoplasmic sequestration of p27.51 However, Artemis can effectively promote the ubiquitylation of nuclear localized T157A mutant of p27, indicating that Cul4A-DDB1DDB2-Artemis has activity toward nuclear p27.
We have shown that Artemis acts with the Cul4A-DDB1DDB2 ligase to mediate degradation of p27, however, it is unlikely that Artemis has a function parallel to a DCAF. First, Artemis does not directly interact with DDB1, which has proved to be the case for all the known DCAFs. In addition, a motif search of Artemis did not reveal any WD40 repeats or WDXR motifs that are typically found in DCAFs. Moreover, in our study Artemis interacts with the DCAF, DDB2, as well as the substrate p27, and both Artemis and DDB2 are jointly required for p27 degradation. Taken together, these results suggest that Artemis is likely acting as an additional substrate specificity factor for p27 degradation by Cul4A-DDB1DDB2. Several published results support the concept that an additional factor is required for the recruitment of substrates by CRLs. For example, SCFSkp2 requires CKS1 to target the ubiquitylation of p27;64,65 CKS1 binds to p27 phosphorylated at Thr187, which is an essential element of the SCFSkp2-mediated degradation of p27. Also, the Cul4A-DDB1CDT2 ligase complex requires PCNA in order to interact with the substrates CDT1 and p21 and mediate their ubiquitylation.37,66 Similarly, Artemis interacts with both DDB2 and p27, which leads us to propose that Artemis functions in the Cul4A-DDB1DDB2 complex to help recruit p27 in the nucleus during G1 without the necessity of phosphorylation at the Thr187 residue (Fig. 5C).
Dual nature of Artemis.
Artemis was first identified as the causative gene for a subset of SCID patients with defective V(D) J recombination.7 Subsequent studies have shown that one of its significant functions is to act as a nuclease in V(D)J recombination and in the repair of DSBs by NHEJ and HR. Findings from our laboratory, however, have demonstrated that Artemis has additional roles in the regulation of the mammalian cell cycle including promoting normal recovery from DNA damage-induced checkpoint responses and as a negative regulator of p53 in response to oxidative or oncogenic stress.16,17,19,57 These prior studies all demonstrated a role for Artemis in response to cellular stress of one form or another. The studies reported here add to Artemis' repertoire of functions by showing that it is also a negative regulator of 27 via the Cul4A-DDB1DDDB2 E3 ligase. A distinct difference with this latter discovery is that Artemis acts to regulate p27 in normally proliferating cells and to promote the G1-to-S or G0-to S-phase transitions. In summary, we have described a novel E3 ubiquitin ligase complex requiring Artemis and DDB2 in association with Cul4A-DDB1 that negatively regulates the tumor suppressor p27. These results together with our previous findings demonstrating that Artemis acts with SCFFbw7 to regulate cyclin E degradation clearly define a role for Artemis in Cullin-based protein ubiquitylation.17
Materials and Methods
Cell culture.
HeLa and HEK293 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. hTERT-RPE1 cells were cultured in DMEM-F12 containing 10% FBS. Preparation of MEF cells, genotyping and cell culture were performed as described in reference 67.
Inhibition of gene expression by siRNA.
siRNAs were synthesized by Dharmacon. The sequence of Artemis siRNAs was previously reported in reference 19. The sequence of the DDB2-1 siRNA was 5′-CCA GAA GAC CUC CGA GA, and that of DDB1 siRNA was 5′-GCA GAA UCG ACU CAA UAA A. Additional siRNAs from Sigma were used, DDB2-2 (00101645) and p27 (00113637). SiRNAs for Cul4A (sc-44355) were purchased from Santa Cruz Biotechnology. SiRNAs were transfected into proliferating cells using oligofectamine RNA transfection reagent (Invitrogen).
DNA constructs and DNA transfection.
Constructs expressing myc-tagged mouse Cul4A and hDDB2 were obtained from Pengbo Zhou68 and Yoshihiro Nakatani,34 respectively. DDB2 was cloned into the pENTR3C vector for use in the Gateway system (Invitrogen). Human DDB1 full-length cDNA in pCMVSPORT6 vector was purchased from Open Biosystems and subcloned into the pENTR11 vector. Artemis cDNAs were prepared as previously described in reference 16 and 18. Flag-p27 T157A, T157D, T187A and T187D were generated by PCR-based mutagenesis (Stratagene). DNA transfections were performed using FuGENE6 transfection reagent (Roche Molecular Biochemicals).
Antibodies.
Artemis polyclonal antibodies and Artemis phospho-specific antibodies were previously described in reference 16 and 18. Other antibodies include goat anti-DDB1 (abCAM), rabbit anti-DDB2, mouse and rabbit anti-GFP, rabbit anti-p27 (Santa Cruz Biotechnology), monoclonal antibody for BrdU (Becton Dickinson), mouse anti-HA (Roche), mouse anti-Flag (Sigma), mouse anti-MBP (New England Biolab) and mouse anti-GST (Cell Signaling). Rabbit anti-Cul4A, anti-DDB1 and anti-Cdt1 antibody were kindly provided by Yue Xiong (University of North Carolina). Secondary antibodies conjugated with horse-radish peroxidase were obtained from Amersham-Pharmacia.
Protein expression and in vitro translation.
For coupled in vitro transcription-translation experiments, Artemis, DDB1, DDB2 and Cul4A cDNAs were cloned into pDEST vectors under the T7 promoter. Proteins labeled with 35S-methionine were produced in vitro using the T7 TNT Coupled Reticulocyte Lysate System (Promega).
Bacterially expressed GST-Artemis and MBP-DDB2 were purified using GST beads (Amersham) or amylose resin (New England Biolabs), respectively.
Immunoprecipitation, GST pull-down assay and mass spectrometric analyses.
Co-IP experiments were performed as previously described in reference 18. For in vitro IP experiments, 35S-methionine-labeled proteins were mixed and were incubated on ice for 1 h. Two hundred ml 1x PBS together with the indicated antibody was added, and the reactions were incubated at 4°C for 1 h. Ten µl of protein A-Sepharose CL-4B beads were added, and the reactions were rotated for another hour at 4°C. Bound proteins were eluted and separated on SDS-PAGE for autoradiography.
For in vitro GST pull-down assays, purified proteins were mixed and incubated at 4°C overnight in the presence of BSA, and then GST beads were added to the mixture. After a 1 h incubation, beads were washed, and bound proteins were eluted and separated by SDS-PAGE.
For mass spectrometric analysis HEK293 cells were transfected with a GST-tagged Artemis. Transfected or control cells were lysed with Net-N buffer and lysates were incubated with GST beads. Immunocomplexes were resolved by SDS-PAGE and subjected to mass spectrometric analysis for identification.
In vivo ubiquitylation assay.
HeLa cells were transfected with HA-ubiquitin, GST-Artemis and Flag-p27. Forty-eight h after transfection, 20 µM MG-132 was added to the media. Cells were harvested 5 h later in lysis buffer (50 mM Tris PH 7.5, 0.5 mM EDTA, 1% SDS and 1 mM DTT). Lysates were boiled for 10 min and diluted 10 times in Net-N buffer containing protease inhibitors (Roche). Immunoprecipitations were performed using anti-Flag M2-conjugated agarose (Sigma).
Serum starvation and cell cycle analyses.
For serum starvation experiments, hTERT-RPE1 cells were grown in DMEM-F12 media containing 10% FBS. Twenty-four h after transfection, media was removed and cells were washed with 1x PBS twice. The media was changed to DMEM-F12 with 0.1% FBS 24 h later, cells were harvested and fixed in 70% ethanol, and DNA was labeled by propidium iodide for FACS analysis.
To determine cell cycle distribution, cells were labeled with BrdU at a concentration of 10 µM for 30 min prior to harvesting and fixation. Cells were then incubated in PBS containing 4% BSA and 0.2% Triton X-100 for 1 h and processed for BrdU staining and FACS analysis as described in reference 16.
RNA purification and RT-PCR.
Total cellular RNA was extracted using the RNAqueous-4PCR kit (Ambion). One µg of RNA was reverse-transcribed into cDNA, and the cDNA was subjected to real-time quantitative PCR using the SYBR green supermix and iCycler iQ Real-time PCR detection system (Bio-Rad). All results were normalized to GAPDH.
Acknowledgments
This work was supported by National Cancer Institute grants CA096574 and CA097175.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
Articles from Cell Cycle are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.4161/cc.10.23.18227
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.4161/cc.10.23.18227?needAccess=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4161/cc.10.23.18227
Article citations
Artemis inhibition as a therapeutic strategy for acute lymphoblastic leukemia.
Front Cell Dev Biol, 11:1134121, 31 Mar 2023
Cited by: 1 article | PMID: 37082620 | PMCID: PMC10111164
A protein with broad functions: damage-specific DNA-binding protein 2.
Mol Biol Rep, 49(12):12181-12192, 03 Oct 2022
Cited by: 4 articles | PMID: 36190612 | PMCID: PMC9712371
Review Free full text in Europe PMC
Hypoxia-inducible CircPFKFB4 Promotes Breast Cancer Progression by Facilitating the CRL4DDB2 E3 Ubiquitin Ligase-mediated p27 Degradation.
Int J Biol Sci, 18(9):3888-3907, 06 Jun 2022
Cited by: 5 articles | PMID: 35813480 | PMCID: PMC9254479
Gentiopicroside targets PAQR3 to activate the PI3K/AKT signaling pathway and ameliorate disordered glucose and lipid metabolism.
Acta Pharm Sin B, 12(6):2887-2904, 06 Jan 2022
Cited by: 28 articles | PMID: 35755276 | PMCID: PMC9214054
DDB2 regulates DNA replication through PCNA-independent degradation of CDT2.
Cell Biosci, 11(1):34, 08 Feb 2021
Cited by: 8 articles | PMID: 33557942 | PMCID: PMC7869461
Go to all (20) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A.
Cancer Res, 68(13):5014-5022, 01 Jul 2008
Cited by: 83 articles | PMID: 18593899
Cullin-4A·DNA damage-binding protein 1 E3 ligase complex targets tumor suppressor RASSF1A for degradation during mitosis.
J Biol Chem, 286(9):6971-6978, 04 Jan 2011
Cited by: 29 articles | PMID: 21205828 | PMCID: PMC3044953
Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome.
Mol Cell Biol, 26(7):2531-2539, 01 Apr 2006
Cited by: 82 articles | PMID: 16537899 | PMCID: PMC1430311
E3 Ubiquitin Ligases as Molecular Targets in Human Oral Cancers.
Curr Cancer Drug Targets, 16(2):130-135, 01 Jan 2016
Cited by: 9 articles | PMID: 26560119
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: P01 CA097175
Grant ID: CA096574
Grant ID: CA097175
Grant ID: R01 CA096574






