Abstract
Free full text

Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges
Abstract
Bacterial flagellin is a dominant innate immune activator of the intestine. Therefore, we examined the role of the intracellular flagellin receptor, NLRC4, in protecting the gut and/or driving inflammation. In accord with NLRC4 acting via transcription-independent pathways, loss of NLRC4 did not reduce the rapid robust changes in intestinal gene expression induced by flagellin administration. Loss of NLRC4 did not alter basal intestinal homeostasis nor predispose mice to development of colitis upon administration of an anti-IL-10R monoclonal antibody. However, epithelial injury induced by dextran sulfate sodium (DSS) in mice lacking NLRC4 resulted in more severe disease indicating a role for NLRC4 in protecting the gut. Moreover, loss of NLRC4 resulted in increased mortality in response to flagellate, but not aflagellate Salmonella infection. Thus, despite not being involved in rapid intestinal gene remodeling upon detection of flagellin, NLRC4-mediated inflammasome activation results in production of IL-1β and IL-18, two cytokines that protect mice from mucosal and systemic challenges.
INTRODUCTION
Bacterial flagellin can be recognized by 2 distinct signaling pathways of the innate immune system 1. TLR5-mediated detection of extracellular flagellin results in activation of canononical MyD88-mediated MAPK and NF-KB signaling resulting in transcriptional activation of a panoply of genes related to inflammation and host defense. Detection of intracellular flagellin by NLRC4, also referred to as Interleukin converting enzyme Protease Activating Factor (IPAF), results in activation of caspase-1 resulting in post-translational processing/secretion of IL-1β and IL-182, 3. TLR5 is functionally expressed on the basolateral surface of intestinal epithelial cells where it serves to activate pro-inflammatory gene expression upon translocation of bacteria beyond the epithelium 4. While expression of TLR5 seems to be variable in hemopoietic cells, it is functionally present on intestinal CD11c+ (likelyDC or macrophages) thus being in accord with the notion that flagellin is a dominant innate immune activator, capable of acting on multiple cell types, of the gut5. In contrast, functionally significant expression of NLRC4 has yet to be observed in cells other than macrophages and was specifically not observed in cultured intestinal epithelial cells (IECs) 6, 7.
Studies of TLR5-deficient mice (T5KO mice) have revealed that this receptor plays a key role in protecting the gut from pathogens and, moreover, that loss of TLR5 renders mice unable to control their commensal microbiota thus resulting in increased pro-inflammatory gene expression that can manifest as spontaneous colitis or eventuate in insulin resistance 8, 9. In contrast, the extent to which NLRC4 plays a role in protecting the intestine is largely unknown. Intestinal NLRC4 is functionally expressed in the intestine as mice deficient in NLRC4 (N4KO mice) lacked intestinal production of IL-1β and IL-18 in response to purified flagellin 7. Whether loss of NLRC4 alters basal gut phenotype or the host response to a challenge, or both, remains undefined. Thus, the goal of this study was to define the role of NLRC4 in maintaining homeostasis of the gut and protecting the host from challenges to this organ. We observed that NLRC4 was not required to maintain basal homeostasis, nor did its absence predispose mice to immune-mediated colitis. However, NLRC4 plays a role in protecting the gut from chemical-induced acute colitis as well as the mortality that results from dissemination of Salmonella beyond the gut.
MATERIALS AND METHODS
Mice
Generation of WT, T5KO, N4KO and T5/N4-DKO was previously described 7. Mice over-expressing the natural antagonist of IL-18, IL-18 binding protein (IL-18 BP), were engineered by Charles Dinarello and colleagues (University of Colorado, Denver, CO) 10 and generously given to us. Mice were bred and maintained at Emory University and used between 6 and 8 weeks of age. All experiments involving animals were approved by the Emoryand Georgia State Universities animal use committees.
Bacteria
Wild-type Salmonella enterica serovar Typhimurium (SL3201) and its isogenic mutant (aflagellate, phenotype: nonmotile; genotype: fliC−/fljB−) were grown in Luria-Bertani (LB) medium under microaerophilic conditionsas previously described 11.
Flagellin administration and mRNA preparation
Flagellin (FliC) from WT Salmonella enterica serovar Typhimurium (SL3201, fljB−) was purified through sequential cation and anion-exchange chromatography and purity was verified as previously described 4. WT, T5KO, N4KO and T5/N4-DKO mice (n=6) were given either 0.2 mL PBS or flagellin (10μg/mouse in 0.2 mL PBS) intraperitoneally. After 1h, mice were euthanized and colon was taken and stored in RNAlater (Invitrogen) for 1 day. Total mRNA was isolated from colonic tissues using TRIzol (Invitrogen) and purified using the RNeasy® Plus Mini kit (Qiagen) according to the manufacturer’s instructions 6.
Microarray
Microarray analyses were performed at the Emory Biomarker Microarray Core. Briefly, mRNA samples were reverse-transcribed, amplified, labeled and used to probe MouseWG-6 v2 chips purchased from Affymetrix (Santa Clara, CA). Samples were assayed using a Molecular Devices Gene Pix (4100A) (Silicon Valley, CA) and raw fluorescence readings were processed by an algorithm designed to reduce spurious readouts of gene activation. Microarray data were quantile-normalized using freely available scripts written in R (http://R-project.org). Significantly altered genes were identified using Significance Analysis of Microarrays algorithm (SAM) and assessed by hierarchal clustering and principle component analysis using Spotfire Decision Site for Functional Genomics software (TIBCO, Somerville, MA) to determine relatedness of gene expression patterns resulting from loss of NLRC4. Upregulated genes, only observed in WT and N4KO mice, were then analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (http://david.abcc.ncifcrf.gov/) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to determine which pathways were significantly up-regulated after flagellin injection.
Quantitative RT-PCR
RNA was quantified as previously described 12. Briefly, previous purified total mRNA were measured in Realplex4 (Eppendorf) using QuantiFast™ SYBR®Green RT-PCR Kit (Qiagen) with specific mouse oligonucleotides. The sense and antisense oligonucleotides used were, respectively: 36B4, 5′-TCCAGGCTTTGGGCATCA-3′ and 5′-CTTTATCAGCTGCACATCACTCAGA-3′; pro-IL-1β, 5′-TTGACGGACCCCAAAAGATG-3′ and 5′-AGAAGGTGCTCATGTCCTCAT-3′; CXCL1, 5′-TCGCGAGGCTTGCCTTGACC-3′ and 5′-AGACGGTGCCATCAGAGCAG-3′; ccl2, 5,- GATCACCAGCAGCAGGTGTCC-3′ and 5′-CAAAGGTGCTGAAGACCTTAGG-3′; ccl7, 5′-AGAAACAAAAGATCCCCAAGAGG-3′ and 5′-GGGGTTTTCATGTCTAAGTATGC-3′. All results were normalized to the unaffected housekeeping 36B4 gene.
Administration of anti-Il-10R monoclonal antibody (IL-10R mAb)
IL-10R neutralizing mAb (1B1.3a) was purchased from BioXcell (West Lebanon, NH, USA). WT, N4KO and T5/N4-DKO mice (n=6) were treated with 1mg of IL-10R mAb (i.p.) weekly for 4 weeks as previously described 12, 13. As a control, mice that did not receive IL-10R mAb were injected with sterile PBS. Mouse body mass was measured weekly. After euthanasia, colitis severity was assessed by measuring spleen and colon weight as well as colonic myeloperoxidase (MPO) activity and cytokine production.
Dextran Sulfate Sodium (DSS) treatment
Six to eight week-old WT and N4KO male mice (≈22 g) received 2% (w/v) DSS (MW = 36,000–50,000 kDa, MP Biomedicals, Solon, OH) in drinking water for 6 days to induce colon injury. Rectal bleeding was assessed by Hemoccult II test (SKD SARL). Blood score ranges from 0 (no blood) to 4 (visible rectal bleeding). Colitis progression was also assessed by colonoscopy to look for ulcerations.
Streptomycin pretreated Salmonella induced gastroenteritis
Wild-type Salmonella enterica serovar Typhimurium (SL3201) were grown and streptomycin pretreated Salmonella induced gastroenteritis was induced in WT, N4KO, T5/N4-DKO and MyD88KO mice as previously described 14. Briefly, mice were fasted for 4h before streptomycin treatment (10mg per mice by gavage). One day after, mice were infected by gavage using 108 CFU of Salmonella per mice. Then, mice were euthanized 48h post infection and cecal inflammation was assessed.
Colon culture
Following euthanasia, colons (1 cm) were removed, cut open longitudinally, washed in HBSS and cultured in RPMI 1640 medium containing 1% penicillin and streptomycin 15. After 24h incubation at 37°C with 5% CO2, the supernatants were centrifuged at 4°C and used for assaying cytokines by ELISA.
Tissue myeloperoxidase (MPO) assay
Neutrophil influx in tissue was accessed by assaying the enzymatic activity of MPO, a widely used marker for neutrophils. Briefly, tissue (50 mg/mL) was thoroughly washed in PBS and homogenized in 0.5% hexadecyltrimethylammonium bromide (Sigma) in 50 mM PBS, (pH 6.0), freeze-thawed 3 times, sonicated and centrifuged. MPO was assayed in the clear supernatant by adding 1 mg/mL of dianisidine dihydrochloride (Sigma) and 5×10−4% H2O2 and the change in optical density measured at 450nm. Human neutrophil MPO (Sigma) was used as standard. One unit of MPO activity was defined as the amount that degraded 1.0 μmol of peroxide/min at 25°C 16.
ELISA
IL-1β and CXCL1 ELISAs were performed using kits purchased from R&D Systems (R&D Systems, Minneapolis, MN) according to the manufacturer’ instructions. The minimum detectable doses (pg/mL) were CXCL1 (15.6) and IL-1β (15.6). IL-18 was quantitated with R&D systems quantikine kit (minimum detection was 25 pg/mL).
Histology
Following euthanasia, cecum or colon were fixed for 24h in 10% buffered formalin at room temperature and then subjected to Hematoxylin & Eosin (H&E) staining on tissue sections of 5μm thickness. H&E stained slides were scored by a pathologist (IN) blinded to the study protocol as previously described. 17 Briefly, slides were scored for the presence/absence of active inflammation, the intensity of inflammation (average number of neutrophils and the number of fields that were involved), the extent of inflammation (mucosa, submucosa or serosa), the presence/absence of ulceration, architectural disarray and the pattern of involvement.
Low-dose oral Salmonella infection
Wild-type Salmonella enterica serovar Typhimurium (SL3201) and its aflagellate isogenic mutant, fliC−/fljB−, were used to infect WT and N4KO mice. Briefly, mice were fasted 4h and infected orally by gavage with either flagellate or aflagellate S. Typhimurium (1×106 or 1×108 CFU/mouse). Mice were euthanized after loss of more than 30% of their initial body weight. Salmonella Typhimurium translocation was also measured at days 3 and 7 post infection by counting the Salmonella CFU present in the spleen after plating on LB agar plate.
Statistical analysis
Significance was determined using the Student’s t test, the Mann Whitney t test or one-way ANOVA(GraphPad Prism software). Differences were noted as significant *p<0.05.
RESULTS
Role of NLRC4 in mediating intestinal gene expression
Bacterial flagellin is considered a dominant innate immune activator in the gut in that both intestinal epithelial cells and intestinal phagocytes are highly responsive to flagellin and unresponsive to the canonical TLR agonist LPS 6. Intestinal recognition of flagellin is mediated in part by TLR5, which is functionally expressed on the basolateral surface of intestinal epithelial cells 4, 18 and CD11c+ lamina propria cells, presumed to be DC 5. In response to systemic challenge with purified flagellin or Salmonella, flagellin also activates the NLRC4 inflammasome, whose expression has, so far, only been observed in macrophages 19, 20. Mice lacking TLR5 (T5KO) are prone to developing spontaneous colitis although whether colitis actually manifests in such mice is dependent upon environmental factors, particularly the composition of the gut microbiota 9, 12, 21. However, regardless of whether T5KO mice exhibit spontaneous colitis, loss of TLR5 results in altered basal gene expression in the colon that can be most readily appreciated via use of microarray-based analysis of colon gene expression 12. To begin to investigate the role of NLRC4 in the gut, we bred mice engineered to lack NLRC4 (N4KO), generated on a C57BL/6 background, into our colony of T5KO mice thus allowing eventual generation of mice lacking NLRC4 and/or TLR5 as well as WT littermates that began life with common ancestry and, therefore, similar microbiotas. In contrast to our colony of T5KO mice, N4KO mice lacked indicators of spontaneous colitis such as loose stools, colomegaly and splenomegaly, and, moreover, were indistinguishable from WT littermates based on myeloperoxidase (MPO) assay and histopathological analysis (Supplemental figures 1 A–C). Moreover, microarray analysis performed in triplicate indicated that loss of NLRC4 had only minor effects on basal colonic gene expression (Supplemental figure 1 D - full data set will be posted in GEO). Thus, in contrast to TLR5, loss of NLRC4-mediated recognition of flagellin may not have important consequences for intestinal phenotype in the absence of an overt challenge.
Next, to better understand the role of NLRC4 relative to TLR5 upon intestinal challenge, we first examined the roles of these receptors in regulating intestinal gene expression in vivo upon direct exposure to flagellin. Our approach was to administer flagellin in a manner known to be sufficient to activate both TLR5 and NLRC4 but to assay changes in gene expression at the mRNA level at a very early time (1h) that was highly likely to reflect changes in gene transcription in cells that had detected flagellin (we reasoned that gene expression at later times would include many changes that resulted from responses to cytokines produced by directly responding cell). Mice (WT, T5KO, N4KO and T5/N4-DKO), were systemically exposed to flagellin (10 μg/mouse) via intraperitoneal injection, their colons harvested 1 h later and mRNA immediately extracted. The 10 μg dose was utilized because we previously demonstrated it was sufficient to robustly and specifically activate both NLRC4 and TLR5. Microarrays were performed on biological triplicates (i.e. 3 mice per condition - analyzed individually). The complete set of results has been posted on National Library of Medicine’s Gene Expression Omnibus <http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34492> and a complete unbiased bioinformatics analysis can be found in supplementary data. The main message of this data set, which can be most easily appreciated when data is viewed as a heat map (Figure 1A) is that flagellin-induced changes in intestinal gene expression at this early time-point are fully dependent on TLR5 and largely independent of NLRC4. More specifically, none of the 226 genes that the Significance Analysis of Microarrays (SAM) algorithm indicated were induced by flagellin in WT mice, were significantly upregulated in mice lacking TLR5. Similar unresponsiveness to flagellin was observed in mice lacking both NLRC4 and TLR5 whereas, in contrast, mice lacking only NLRC4 exhibited robust flagellin-induced gene expression at this time point (195 genes significantly upregulated by SAM) thus confirming that TLR5, rather than NLRC4 mediates this rapid induction of gene expression. Such TLR5-activated genes were analysed using Kyoto Encyclopedia of Genes and Genomes (KEGG) program to determine which pathways were significantly upregulated in response to flagellin (Table 1). Both WT and N4KO mice exhibited the same pattern of significant upregulated pathways. Selected genes from the microarray were examined byquantitative RT-PCR to confirm the microarray results (Figures 1B–E). Thus, the immediate gene expression remodeling that occurs in the intestine in response to detection of soluble flagellin is mediated by TLR5 rather than NLRC4.
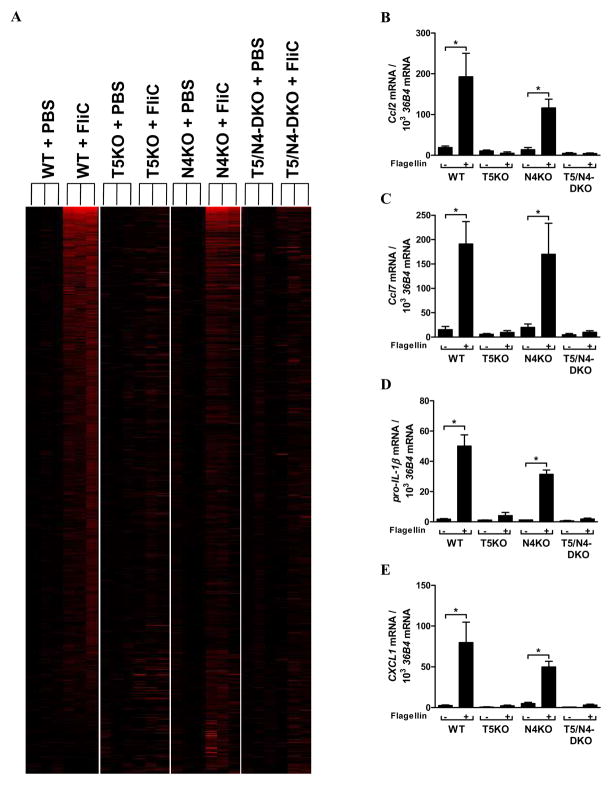
Indicated strains of mice (n= 3) were administered 10 μg of flagellin (FliC) in 0.2 mL PBS via intraperitoneal injection. 1h later, mice were euthanized, colons immediately removed and mRNA extracted and subjected to microarray analysis of global gene expression. (A) Transcription profiles in colons of WT, T5KO, N4KO and T5/N4-DKO mice have been compared to colons from vehicle-treated genotype-matching mice. Heat map shows the results of all 3 mice per condition assayed. Illustration of genes induced with >1.5-fold change (in red) was built using the TreeView software. (B–E) Quantitative reverse transcription-PCR (qRT-PCR) was used to confirm mRNA synthesis of selected genes. * p<0.05.
Table I
Gene expression level has been assessed by microarray using MouseWG-6 v2 chips (Affymetryx) in the colon of WT, T5KO, N4KO and T5/N4-DKO mice 1h after intraperitoneally flagellin injection in comparison to the matching genotype mice receiving only PBS. Upregulated genes, only observed in WT and N4KO mice, were then analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (http://david.abcc.ncifcrf.gov/) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to determine which pathways were significantly up-regulated after flagellin injection.
| Pathway | WT (Fold enrichment) | N4KO (Fold enrichment) |
|---|---|---|
| NOD-like receptor signaling pathway | 16.7 | 18.0 |
| Toll-like receptor signaling pathway | 10.5 | 9.5 |
| Cytosolic DNA-sensing pathway | 8.7 | 9.3 |
| Cytokine-cytokine receptor interaction | 7.8 | 5.6 |
| Chemokine signaling pathway | 7.0 | 6.1 |
| T cell receptor signaling pathway | 5.4 | 5.1 |
| MAPK signaling pathway | 4.5 | 5.2 |
Role of NLRC4 in colitis
Regardless of whether they are maintained in a manner that results in observable incidence of spontaneous colitis, loss of TLR5 predisposes mice to developing overt colitis upon ablation of IL-10 signaling 9, 12 and increases the severity of DSS-induced colitis 22. Thus, we investigated the extent to which loss of NLRC4 signaling might affect development of colitis in such conditions. First, we neutralized IL-10 signaling for 4 weeks via weekly injections of an IL-10R mAb. Such treatment was recently observed to uniformly result in robust colitis (100% penetrance) in 2 distinctly-generated strains of T5KO mice12. In contrast to T5KO mice, and like WT mice, N4KO mice subjected to IL-10 blockade lacked evidence of colitis in that they continued to gain weight normally and showed neither gross nor histopathologic evidence of colitis although, like WT mice, they showed increases in MPO activity in response to IL-10R neutralization (Figures 2A–E). Such IL-10R mAb-induced T5KO colitis was also associated with elevated levels of intestinal or systemic production of the inflammasome cytokines IL-1β or IL-18, respectively (Figures 2F and G), which was not observed in the N4KO mice after IL-10R mAb treatment. A previous study has already shown that IL-10R mAb-induced colitis in T5KO mice correlates with colonic IL-1β production and requires the IL-1 receptor11. We hypothesized that such IL-1β production might reflect elevated expression/activity of NLRC4 that might result from loss of TLR5. However, while loss of NLRC4 upon a T5KO background modestly reduced the intestinal expression of IL-1β and IL-18 upon IL-10R mAb (Figures 2F and G), mice lacking TLR5 and NLRC4 still developed robust colitis in response to IL-10R neutralization arguing against this possibility and suggesting that NLRP3 or another inflammasome pathway compensates for lack of NLRC4.
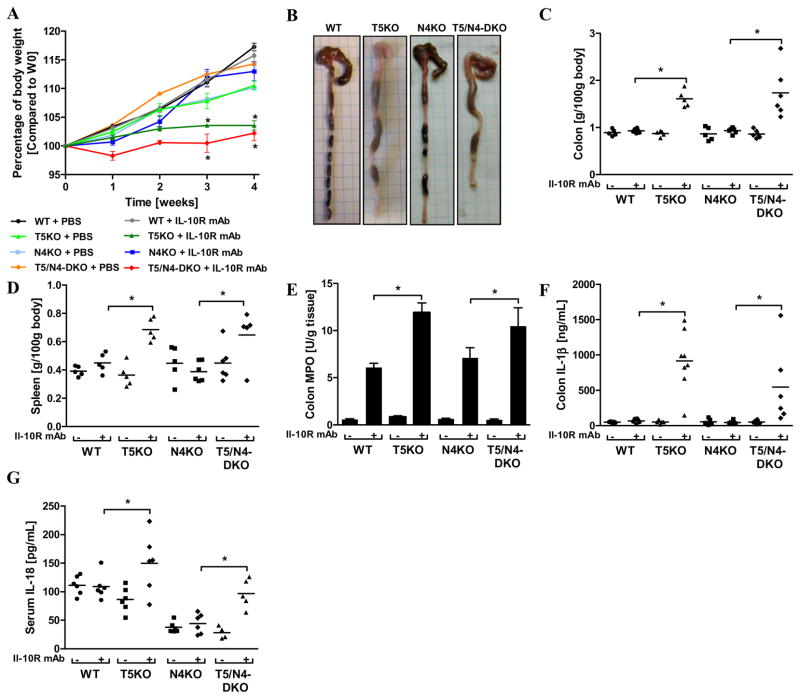
Wild-type, T5KO, N4KO and T5/N4-DKO mice (n=5–6 mice per group) were intraperitoneally injected weekly with 1mg of IL-10R mAb or PBS (vehicle) for 4 weeks. (A) Body mass was monitored weekly during the treatment. (B) Gross picture of colon. (C) Following euthanasia, colon was isolated and mass measured. (D) Spleen mass. (E) Colonic MPO activity. (F) Colon was cultured for 24h, at which time supernatant was assayed for IL-1β by ELISA. The data is representative of 2 independent experiments. (G) Levels of IL-18 were measured in the sera by ELISA. Data show mean ± SEM of values obtained from individual mice.* p<0.05.
We next examined the relative roles of TLR5 and NLRC4 in DSS-induced colitis, which results in focal destruction of the gut epithelium and subsequent colitis characterized by weight loss, bloody stools, splenomegaly, colomegaly and increased colonic MPO levels. In accordance with work of Steiner and colleagues 22, T5KO mice exhibited more severe disease than WT mice based on clinical-type indicators and histopathological analysis (Figure 3). N4KO were observed to be more even sensitive to this chemical colitogen exhibiting severe colitis in response to the relatively modest dose of 2% DSS (Figures 3A–E). Specifically, N4KO mice exhibited colonoscopic evidence of colitis and histopathologic scoring confirmed greater severity of colitis in N4KO mice (Figures 3F–G). The higher histological score of the N4KO mice largely reflected that the colitis in N4KO mice was transmural whereas it was more focal in WT mice. Exposure of WT and N4KO mice to a higher concentration of DSS (3.5%) resulted in robust colitis in both WT and N4KO mice that did not significantly differ in its severity between these genotypes (Data not shown). Exposure of WT, T5KO, N4KO and T5/N4-DKO mice to a lower concentration of DSS (1.25%) did not result in discernible colitis within 7 days. However, prolonged DSS treatment eventuated in uniform death in mice lacking TLR5 and/or NLRC4, but not WT mice (Figure 3H), providing additional evidence that, like T5KO mice, N4KO mice exhibit more severe disease upon exposure to this chemical colitogen. To better understand how NLRC4 protected against DSS-induced disease, we measured cytokine production in this model (Figures 3I–K). Whereas loss of TLR5, by itself, resulted in reduced production of the neutrophil chemoattractant CXCL1 (perhaps explaining why histology/MPO showed reduced neutrophils but greater ulcerations), loss of NLRC4 resulted in reduced production of IL-18 suggesting this inflammasome cytokine may protect against this chemical colitogen.
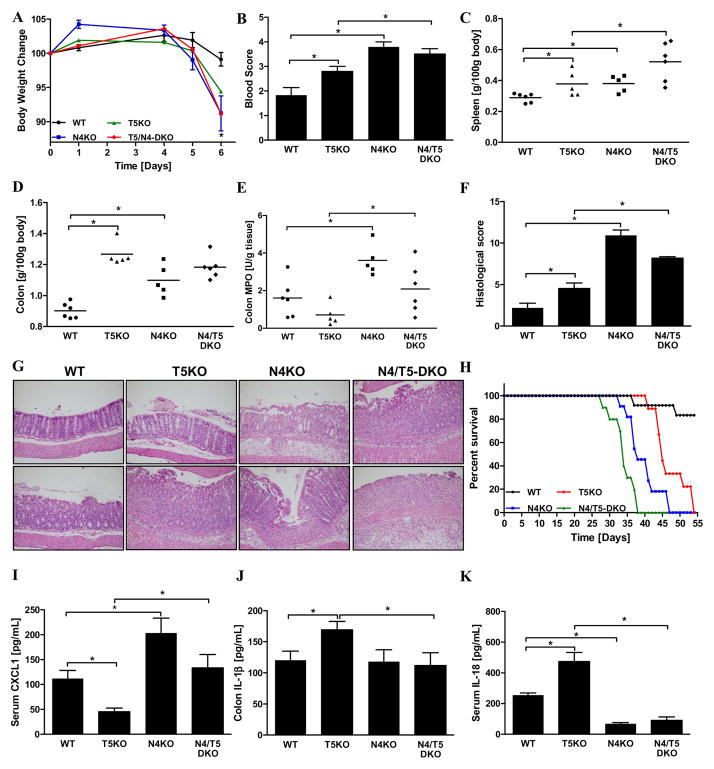
Wild-type, T5KO, N4KO and T5/N4-DKO mice (n=5–6 mice per group) received water containing 2% DSS ad libitum throughout the experimental period (6 days). (A) Percentage of body weight change over the experimental period. (B) Rectal bleeding by assaying blood in stool. (C) Following euthanasia, spleen was isolated and mass measured. (D) Colon mass. (E) Colonic MPO activity. (F) Inflammation severity has been monitored in the colon by calculating a histological score as described in Methods. (G) Representative H&E-stained histological observations of mouse colon following 2% DSS treatment. (H) DSS-induced mortality in mice given a low dose of 1.25% DSS. (I) Levels of serum CXCL1 were measured by ELISA. (J) Colon was cultured for 24h, at which time supernatant was assayed for IL-1β by ELISA. The data is representative of 3 independent experiments. (K) Levels of IL-18 were measured in the sera by ELISA. Data show mean ± SEM of values obtained from individual mice. * p<0.05.
Role of NLRC4 in Salmonella infection
Intestinal recognition of flagellin protects against enteric and systemic disease in that infection by aflagellate Salmonella results in more severe intestinal and systemic disease in multiple mouse strains 11. Greater Salmonella-induced intestinal disease was also observed in mice lacking TLR5 although the extent to which the aflagellate phenotype can be phenocopied by deletion of TLR5 varies considerably in different studies likely reflecting that the extent to which loss of TLR5 alters basal gene expression/phenotype is environment dependent 23. The above-described observation that loss of NLRC4 does not markedly affect basal intestinal gene expression, at least in our vivarium, minimizes this sort of caveat thus allowing us to use these KO mice to examine the role of NLRC4 in Salmonella infection. Mice (WT, T5KO, N4KO and T5/N4-DKO) were subjected to the streptomycin/Salmonella model, which results in robust cecal inflammation characterized by edema and neutrophil recruitment within 2 days of infection. Whether assessed via cecum weight, MPO activity or histological scoring, loss of neither TLR5 nor NLRC4 did not have a significant effect on the robust inflammation in this model (Figures 4A–E). However, interestingly, loss of both NLRC4 and TLR5 or loss of MyD88, which will ablate signaling by TLR5 and inflammasome cytokines (as well as other TLRs) dramatically reduced Salmonella-induced inflammation as assessed by MPO and histopathologic scoring. Such protection against Salmonella-induced acute inflammation correlated with lack of production of the inflammasome cytokine IL-1β whereas increased CXCL1 was clearly not required for inflammation in this model (Figures 4F and G). That ablation of the 2 known pathways of flagellin recognition phenocopies MyD88 deficiency in this model supports the notion that flagellin is a major pro-inflammatory actor during infection by flagellated pathogens and that inflammation can be driven by either innate immune detector capable of recognizing this protein.
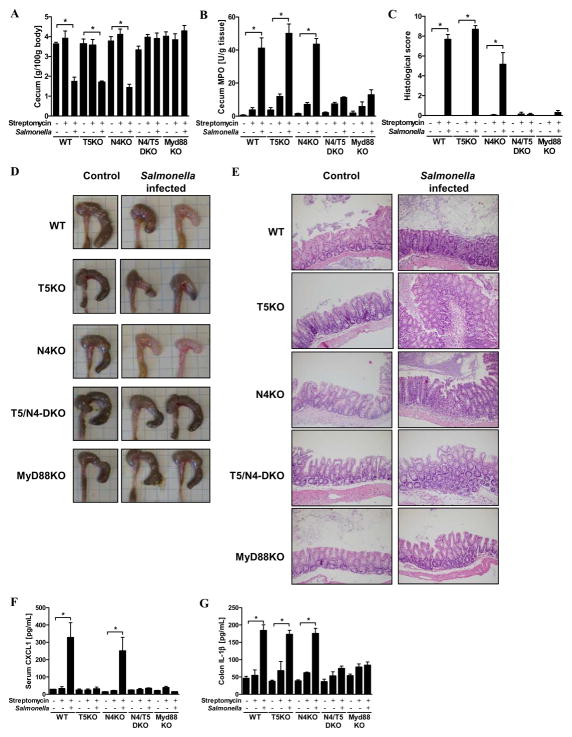
Wild-type, T5KO, N4KO, T5/N4-DKO and MyD88KO mice (n=5–6 mice per group) were pretreated with streptomycin (10 mg) and, 24h later, infected orally with flagellate S. Typhimurium (108 CFU/mice). (A) Following euthanasia (48h post infection), cecum was isolated and mass measured. (B) Cecum MPO activity. (C) Inflammation severity has been monitored in the cecum by calculating a histological score as described in Methods. (D) Gross picture of cecum. (E) Representative H&E stained histological observations of cecum following treatment. (F) Levels of serum CXCL1 were measured by ELISA. (G) Colon was cultured for 24h, at which time supernatant was assayed for IL-1β by ELISA. The data is representative of 2 independent experiments.* p<0.05.
Next, we compared WT and N4KO mice in response to low-dose oral challenge with Salmonella. As in the above-described experiment, we utilized the SL3201 strain that has been used in many papers to study the role of flagella expression in Salmonella virulence 24, 25. Use of a relatively low dose of 106 CFU resulted in death in about 50% of WT mice and 100% of N4KO mice (Figure 5A). Loss of NLRC4 on a TLR5-deficient background resulted in similarly increased lethality (100%) within 3 weeks of infection (Figure 5B). Increasing the infectious dose by 100-fold resulted in almost uniform lethality in both WT and N4KO mice although the rate of death was significantly increased in N4KO mice (Figure 5C). To determine the extent to which the greater mortality exhibited by N4KO mice might reflect increased Salmonella dissemination beyond the intestine, we examined bacterial loads in the spleen at days 3 and 7. We found that bacterial loads in the spleen were indistinguishable on day 3 and significantly elevated on day 7 in N4KO mice suggesting loss of NLRC4 may result in more severe mortality via failure to control extra-intestinal Salmonella replication (Figures 5D and E). Noting that loss of NLRC4 was not observed to significantly affect mortality in response to Salmonella strain SL134420, we infected our mice with this strain and also observed no difference between WT and N4KO mice (Data not shown) suggesting that the role played by NLRC4 in response to this pathogen is dependent upon bacterial strain. In addition to flagellin, NLRC4 is involved in detection of the Salmonella protein PrgJ 26. Thus, to determine the extent to which loss of NLRC4 reflected lost detection of flagellin, we infected WT and N4KO mice with an aflagellate mutant (SL3201 fliC−/fljB−). No difference in mortalitywas observed between WT and N4KO mice (Figure 5F) indicating the role of NLRC4 in protecting against Salmonella observed herein reflects recognition of flagellin during the course of the infection. While the broadly appreciated role of IL-1β in host defense suggests this cytokine likely contributes to NLRC4-mediated defense against Salmonella infection 27, 28, the role of IL-18 is less well studied. To begin to address whether the reduced ability of N4KO mice to withstand Salmonella infection reflected lost production of IL-18, we examined the extent to which loss of IL-18 signaling phenocopied that of N4KO mice. Specifically, we examined Salmonella-induced mortality in mice engineered by Charles Dinarello and colleagues 10 to over-express the natural antagonist of IL-18, IL-18 binding protein (IL-18 BP). These mice are maintained as heterozygous thus producing IL-18 BP and WT control offspring and eliminating the potential of a non-specific difference resulting from paternally-altered microbiota skewing our results. As shown in figure 5G, IL-18 BP mice exhibited increased severity to Salmonella infection reminiscent of that of N4KO mice suggesting that loss of IL-18 signaling mediates, in part, N4KO protection against Salmonella infection.
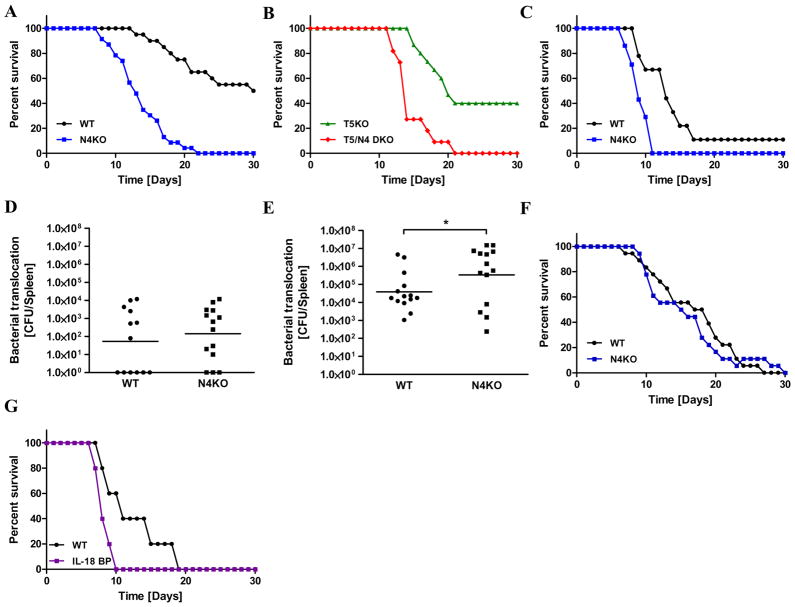
Wild-type (in black), N4KO (in blue), T5KO (in green) and T5/N4-DKO (in red) mice (n=6–20 mice per group) were orally infected with flagellate S. Typhimurium (106 or 108 CFU/mice). (A) Salmonella-induced mortality using an infectious low-dose (106 CFU/mice) by gavage in WT and N4KO mice. (B) Salmonella-induced mortality using an infectious low-dose (106 CFU/mice) by gavage in T5KO and T5/N4-DKO mice. (C) Salmonella-induced mortality using an infectious dose of 108 CFU/mice by gavage in WT and N4KO mice. (D) Bacterial translocation by numbering Salmonella CFU present in the spleen at day 3 post infection. (E) Salmonella CFU present in the spleen at day 7 post infection. (F) WT and N4KO mice were infected by gavage with aflagellate Salmonella low-dose (106 CFU/mice) and monitored for 30 days. (G) Salmonella-induced mortality using an infectious dose of 108 CFU/mice by gavage in WT and IL-18BP mice (over-express the natural antagonist of IL-18, IL-18 binding protein). * p<0.05.
DISCUSSION
The notion that bacterial flagellin may be a major pro-inflammatory determinant in the gut suggested the possibility that blockade of innate immune recognition of flagellin might be a reasonable strategy to treat inappropriate intestinal inflammation. This concept is supported by the observation that, in some genetic backgrounds, carriage of a SNP that results in 75% loss of TLR5 function correlated with reduced incidence of Crohn’s disease 29. However, complete loss of TLR5 in mice resulted in a tendency to develop spontaneous colitis 9. While the incidence of such colitis was dependent upon the composition of the microbiota in which the colony is maintained, T5KO were uniformly predisposed to developing colitis upon blockade of the anti-inflammatory cytokine IL-10 and developed more severe colitis upon exposure to the chemical colitogen DSS 9, 12, 22. Lastly, T5KO mice harboring a microbiota that did not result in colitis still exhibited elevated pro-inflammatory gene expression that may promote metabolic syndrome 8. Together, these observations in mice suggest that blockade of TLR5 in humans might result in unacceptable risks of disease development including inducing the diseases that we and others originally speculated it might prevent. Consequently, we have begun to explore the possibility that inhibition of the inflammasome pathway of flagellin recognition might have therapeutic value in attenuating gut inflammation. Overall, our results don’t broadly support the concept that specific inhibition of NLRC4 would be useful in treating/preventing inflammation. Rather, our observations shed light on the function of NLRC4 in the gut and serve as another example that blocking pro-inflammatory pathways can worsen outcomes of inflammatory diseases and increase the risk of severe infectious disease.
In contrast to the case for T5KO mice, breeding of N4KO mice into our colony resulted in neither evidence of spontaneous colitis nor markedly altered basal colonic gene expression. The lack of a significant role for NLRC4 in gut homeostasis seems unlikely to reflect an absolute inability of intestinal NLRC4 to recognize soluble flagellin as systemic exposure to flagellin induces NLRC4-dependent intestinal section of IL-1β and IL-187. However, it may simply reflect that NLRC4-mediated detection of soluble flagellin requires 100–1000 fold higher concentrations of flagellin than does TLR5 and that the transient local breaches that occasionally occur in the gut epithelium are not sufficient to achieve concentrations of soluble flagellin sufficient to activate NLRC4. A related possibility would be that occasional breaches of the mucosa by bacteria themselves are not sufficient, or that perturbing bacteria do not persist long enough within intestinal phagocytes, to result in NLRC4 activation. In either case, our observation that TLR5 plays a much greater role in basal gut gene expression than does NLRC4 is in accord with the paradigm posited by Aderem and colleagues that TLR5 deals with many challenges by flagellated bacteria whereas NLRC4 serves as a “red alert” that a serious infection has occurred 1.
The association of inflammasome cytokines IL-1β and IL-18 with the extent of inflammation in clinical human gut inflammation and numerous murine colitis models, including T5KO mice, led us to speculate that deletion of NLRC4 might reduce inflammation in general and protect T5KO mice from inflammation in particular. However, such protection was not observed. Rather, we observed increased severity of colitis in N4KO mice in response to DSS that eventuated in enhanced mortality. Such enhanced severity of colitis did not speak to the role of IL-1β in driving inflammation per se. Rather, that intestinal levels of IL-1β continued to correlate well with severity of colitis, and T5/IL-1R-DKO mice are protected from colitis, suggests ablation of NLRC4 simply eventuates in activation of the inflammasome via an alternate pathway that still results in production of IL-1β. In contrast, multiple studies have observed that loss of the NLRP3 inflammasome actually reduced levels of IL-1β in DSS colitis 30, 31 suggesting NLRC4 likely plays a modulatory rather than dominant role in producing this cytokine. In contrast, the severe DSS-induced colitis in mice lacking NLRC4 occurred despite a dramatic absence of IL-18 suggesting that IL-18 may be protective in the gut and NLRC4 may have a non-redundant role in producing this inflammasome cytokine. Yet, the role of inflammasome cytokines in DSS colitis is apparently complex and environment-dependent as NLRP3KO mice developed aggravated disease in one study and were protected in another 30, 31. Thus, further development of pharmacologic strategies to treat/prevent inflammation via inflammasome blockade require more basic understanding of the mechanisms that mediate inflammation/restitution in the gut.
That loss of NLRC4 by itself did not reduce inflammatory pathology in any of the models studied parallels observations that loss of TLR5 by itself also provided no protection against inflammation in either IL-10 deficiency-induced colitis 9, 12, DSS colitis 22 or streptomycin/salmonella-induced intestinal inflammation 23. However, herein we observed that loss of both TLR5 and NLRC4 dramatically reduced severity of inflammation in the latter model. Such lack of inflammation in T5/N4-DKO was reminiscent of the large reduction in inflammation seen in MyD88KO mice in this model. In contrast, MyD88KO mice exhibit markedly greater disease in response to DSS 32. The lack of an obvious role for TLR5/NLRC4 and MyD88 in protecting the gut in this model of acute inflammation may reflect that it lacks the severe epithelial damage that characterizes the DSS model. In any case, considering that T5/N4-DKO mice should signal normally via other TLRs, the phenocopying of MyD88KO 23, 33 by T5/N4-DKO in this model is in accord with the notion that flagellin is indeed a dominant pro-inflammatory determinant in the gut and that inability to signal in response to flagellin results in failure to recruit immune cells in response to this pathogen. While such “ignorance” of a pathogen seems likely to have dire consequences, it suggests the possibility that blocking all innate immune recognition of flagellin may have value in select scenarios.
The role of TLR5 in host defense against oral administration of Salmonella Typhimurium, widely used as a model of typhoid-like illness, has been difficult to ascertain with T5KO mice as considerably different results have been reported by different investigators likely reflecting differences in basal phenotype and/or vivaria-specific differences in microbiota composition 21, 23, 34. We speculate that lack of basal phenotype in N4KO mice will prevent such variability. Herein, we observed that N4KO mice exhibited a clear increase in mortality in response to oral administration of Salmonella Typhimurium. Such increased mortality was seen over a range of infectious doses, was specific for flagellated Salmonella Typhimurium and correlated with greater splenic CFU at d7, but not d3, post-infection. One possible reason for why we saw increased splenic CFU at only the latter time is that there is delayed loss of gut defense/barrier integrity in N4KO mice, perhaps reflecting a role for IL-18 in gut integrity, resulting in increased Salmonella translocation. However, given the reduced ability of N4KO macrophages to kill Salmonella, perhaps a simpler explanation is that NLRC4 may be important in bacterial killing in the spleen but not the gut. In accordance, enhanced mortality of N4KO mice was also observed in response to intraperitoneal infection (Data not shown). Elegant pioneering studies by Miao et. al demonstrated that a key means by which NLRC4 protects against systemic Salmonella infection is by driving macrophage pyroptosis that eventuates in bacterial killing by neutrophils 20. However, interestingly, Miao et. al only observed a protective role for NLRC4 against Salmonella engineered to maintain flagellin expression throughout infection. Given that we also did not see a difference between WT and N4KO mice when using their WT (SL1344) Salmonella strain, we speculate that the clear difference we see with our preferred WT (SL3201) strain may reflect reduced ability to rapidly turn off flagella expression in mice. Regardless, our observation indicates that any attempts to pharmacologically block NLRC4 signaling may increase the risk of severe infection.
Acknowledgments
This work was supported by NIH grants DK061417 and DK083890 to A.T. Gewirtz. F.A. Carvalho and M. Vijay-Kumar are recipients of, respectively, Research Fellowship and Career Development awards from the Crohn’s and Colitis Foundation of America. We also acknowledge NIH Digestive Disease Research and Development Center (DDRDC) grants to Emory University (DK064399). We thank Daniel Moore, Catherine Paul and Sindhu Srinivasan for excellent technical support.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/mi.2012.8
Read article for free, from open access legal sources, via Unpaywall:
http://www.mucosalimmunology.org/article/S1933021922013071/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/mi.2012.8
Article citations
Enhanced Effect of <i>β</i>-Lactoglobulin Immunization in Mice with Mild Intestinal Deterioration Caused by Low-Dose Dextran Sulphate Sodium: A New Experimental Approach to Allergy Studies.
Nutrients, 16(20):3430, 10 Oct 2024
Cited by: 0 articles | PMID: 39458426 | PMCID: PMC11510979
NLRC4, inflammation and colorectal cancer (Review).
Int J Oncol, 65(4):99, 06 Sep 2024
Cited by: 0 articles | PMID: 39239759 | PMCID: PMC11387119
Review Free full text in Europe PMC
The NLR family of innate immune and cell death sensors.
Immunity, 57(4):674-699, 01 Apr 2024
Cited by: 11 articles | PMID: 38599165
Review
The Angiotensin II Receptor Neprilysin Inhibitor LCZ696 Inhibits the NLRP3 Inflammasome By Reducing Mitochondrial Dysfunction in Macrophages and Alleviates Dextran Sulfate Sodium-induced Colitis in a Mouse Model.
Inflammation, 47(2):696-717, 06 Feb 2024
Cited by: 1 article | PMID: 38319541
Epithelial regulation of microbiota-immune cell dynamics.
Mucosal Immunol, 17(2):303-313, 28 Feb 2024
Cited by: 0 articles | PMID: 38428738
Review
Go to all (91) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE34492
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Bacterial Outer Membrane Vesicle-Mediated Cytosolic Delivery of Flagellin Triggers Host NLRC4 Canonical Inflammasome Signaling.
Front Immunol, 11:581165, 18 Nov 2020
Cited by: 25 articles | PMID: 33312172 | PMCID: PMC7708323
Type 1 interferon-dependent repression of NLRC4 and iPLA2 licenses down-regulation of Salmonella flagellin inside macrophages.
Proc Natl Acad Sci U S A, 117(47):29811-29822, 11 Nov 2020
Cited by: 7 articles | PMID: 33177235 | PMCID: PMC7703570
The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus.
Nature, 477(7366):596-600, 14 Sep 2011
Cited by: 781 articles | PMID: 21918512
Nlrc4/Ipaf/CLAN/CARD12: more than a flagellin sensor.
Int J Biochem Cell Biol, 42(6):789-791, 11 Jan 2010
Cited by: 29 articles | PMID: 20067841 | PMCID: PMC2862870
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIDDK NIH HHS (10)
Grant ID: R01 DK083890
Grant ID: DK083890
Grant ID: R01 DK061417-09
Grant ID: DK061417
Grant ID: R24 DK064399
Grant ID: R01 DK061417
Grant ID: R01 DK061417-10
Grant ID: R01 DK083890-02
Grant ID: DK064399
Grant ID: R01 DK083890-01A2





