Abstract
Free full text

Mechanism of substrate selection by a highly specific CRISPR endoribonuclease
Abstract
Bacteria and archaea possess adaptive immune systems that rely on small RNAs for defense against invasive genetic elements. CRISPR (clustered regularly interspaced short palindromic repeats) genomic loci are transcribed as long precursor RNAs, which must be enzymatically cleaved to generate mature CRISPR-derived RNAs (crRNAs) that serve as guides for foreign nucleic acid targeting and degradation. This processing occurs within the repetitive sequence and is catalyzed by a dedicated Cas6 family member in many CRISPR systems. In Pseudomonas aeruginosa, crRNA biogenesis requires the endoribonuclease Csy4 (Cas6f), which binds and cleaves at the 3′ side of a stable RNA stem–loop structure encoded by the CRISPR repeat. We show here that Csy4 recognizes its RNA substrate with an ~50 pM equilibrium dissociation constant, making it one of the highest-affinity protein:RNA interactions of this size reported to date. Tight binding is mediated exclusively by interactions upstream of the scissile phosphate that allow Csy4 to remain bound to its product and thereby sequester the crRNA for downstream targeting. Substrate specificity is achieved by RNA major groove contacts that are highly sensitive to helical geometry, as well as a strict preference for guanosine adjacent to the scissile phosphate in the active site. Collectively, our data highlight diverse modes of substrate recognition employed by Csy4 to enable accurate selection of CRISPR transcripts while avoiding spurious, off-target RNA binding and cleavage.
INTRODUCTION
Many bacteria and archaea employ small CRISPR (clustered regularly interspaced short palindromic repeats)-derived RNAs (crRNAs) as molecular sentries that base-pair with phage or plasmids and thereby trigger degradation of these foreign nucleic acids by CRISPR-associated (Cas) proteins (Horvath and Barrangou 2010; Marraffini and Sontheimer 2010; Al-Attar et al. 2011; Wiedenheft et al. 2012). CRISPR-derived precursor transcripts (pre-crRNAs) are processed enzymatically to generate the mature crRNAs that assemble into large ribonucleoprotein effector complexes (Brouns et al. 2008). In type I and type III CRISPR systems, as defined by Makarova et al. (2011), a single endoribonuclease from the Cas6 superfamily cleaves pre-crRNAs within each invariant repeat sequence to generate ~60-nucleotide (nt) products in which segments of the repeat sequence flank the target-binding spacer sequence (Brouns et al. 2008; Carte et al. 2008; Haurwitz et al. 2010; Gesner et al. 2011; Lintner et al. 2011; Sashital et al. 2011). crRNA biogenesis in type II systems requires RNase III, which cleaves double-stranded RNA (dsRNA) substrates formed by base-pairing between a small, noncoding RNA (tracrRNA) and the pre-crRNA (Deltcheva et al. 2011). Pre-crRNA processing is a hallmark of the CRISPR-Cas system, and the inactivation of these endoribonucleases results in a complete loss of immune system function (Brouns et al. 2008; Deltcheva et al. 2011; Sapranauskas et al. 2011).
We showed previously that Csy4, recently reclassified as Cas6f (Makarova et al. 2011), generates crRNAs in type I-F CRISPR systems (formerly the Yersinia pestis subtype) by cleaving pre-crRNAs at the bottom of stable stem–loops encoded by the CRISPR repeat (Fig. 1A; Haurwitz et al. 2010). The co-crystal structure of Csy4 from Pseudomonas aeruginosa UCBPP-PA14 bound to its pre-crRNA substrate (PDB ID: 2XLK) revealed a diverse set of molecular interactions that mediate RNA recognition (Fig. 1B). A highly basic α-helix docks into the major groove of the stem–loop and contains multiple arginine residues that form a network of hydrogen bonds with the RNA phosphate backbone along the 5′ strand of the stem. In a manner reminiscent of DNA-binding proteins, Csy4 interacts with the bottom two base pairs of the stem–loop through a direct readout mechanism involving formation of base-specific hydrogen bonds between the major groove faces of A19 and G20 and residues Gln104 and Arg102, respectively. The aromatic side chain of Phe155 stacks below the terminal base pair, thereby positioning the scissile phosphate within the active site. Together, these interactions enable Csy4 to recognize and cleave a single repetitive RNA sequence inside the cell, ensuring correct crRNA processing without off-target effects.
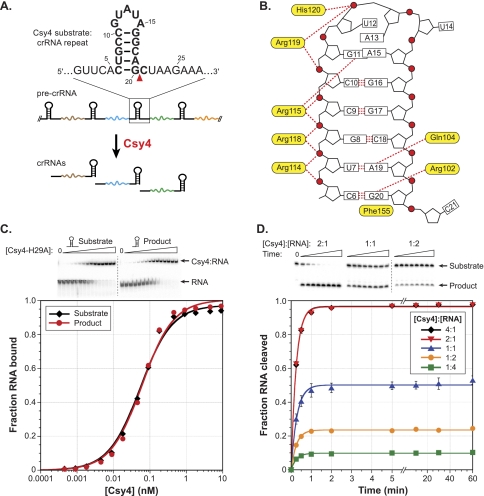
Csy4 binds its substrate and product with high affinity and functions as a single-turnover enzyme. (A) Csy4 cleaves within pre-crRNA repeat sequences (black) to generate mature crRNAs that contain a spacer sequence (colored line) flanked by fragments of the repeat. The substrate sequence and cleavage site (red triangle) are indicated above, with the crRNA construct previously used for crystallography shown in bold. (B) A schematic depicts protein:RNA contacts revealed by a co-crystal structure of Csy4 bound to a fragment of the crRNA repeat (PDB ID: 2XLK). Important amino acid residues are shown in yellow, and RNA nucleotides are numbered as in A. Red circles, pentagons, boxes, and red dotted lines denote phosphates, ribose groups, bases, and hydrogen-bonding interactions, respectively. (C) EMSAs (top) were performed with Csy4-H29A and the substrate and product of the cleavage reaction. The resulting data for these and all subsequent binding assays were fit with a standard binding isotherm to yield equilibrium dissociation constants (solid lines; see Materials and Methods), and average Kd and standard error of the mean (SEM) values from at least three independent experiments are reported in Supplemental Table 1. (D) RNA cleavage assays were conducted at five different enzyme:substrate molar ratios, and the extent of the reaction at various time points was assessed by denaturing PAGE (top). The resulting data for these and all subsequent cleavage assays were fit with a single exponential to yield first-order rate constants (solid lines; see Materials and Methods), and average kobs and SEM values from three independent experiments are reported in Supplemental Table 1. Error bars for each time point represent the standard deviation and are not always visible.
Bioinformatic analyses of Csy4-related Cas proteins together with existing CRISPR databases (Grissa et al. 2007) have revealed a potentially large number of enzyme variants whose substrate specificities have co-evolved with the RNAs encoded by CRISPR repeats. Gaining a thorough understanding of the selection mechanism by which Csy4 faithfully binds and cleaves its substrate should inform future work aimed at expanding the toolbox of these sequence-specific endoribonucleases. Furthermore, the propensity of many pre-crRNA repeat sequences to form small, stable stem–loops (Kunin et al. 2007) suggests that general principles of substrate recognition employed by Csy4 will be broadly applicable to other Cas6 family members that associate with structured repeats.
To determine the importance of sequence- and shape-specific RNA recognition during pre-crRNA processing, we investigated the relative contributions of substrate base-pair composition and geometry to binding and cleavage by Csy4. Here we show that Csy4 binds its substrate RNA with extremely high affinity (Kd ≈ 50 pM) and functions as a single-turnover enzyme. Single-stranded RNA (ssRNA) nucleotides that flank the stem–loop contribute negligibly to binding energy, but base-pair changes throughout the double-stranded stem and mutations to the loop sequence result in substantially weaker binding. We find that substrate recognition also involves the precise length of the stem, such that small base-pair insertions cause severe binding and/or cleavage defects due to their effects on helical geometry and substrate positioning. These findings reveal how Csy4 employs a unique set of molecular interactions to achieve highly specific selection of its pre-crRNA substrate while discriminating against similar, noncognate stem–loop structures.
RESULTS
Csy4 binds the crRNA repeat stem–loop with high affinity and functions as a single-turnover catalyst
Csy4 is a specialized ribonuclease that selects CRISPR transcripts from the cellular milieu for binding and cleavage. To determine the basis for this selectivity, we first examined the thermodynamic stability of the Csy4:RNA complex and the energetic contributions of protein:RNA interactions observed crystallographically (Fig. 1B). Using modified RNA substrates and/or Csy4 mutants, equilibrium dissociation constants (Kd) were measured using electrophoretic mobility shift assays (EMSA). The RNA substrates we tested derive from the invariant 28-nt repeat sequence found within pre-crRNAs generated from P. aeruginosa strain UCBPP-PA14 CRISPR locus 2 (Grissa et al. 2007), herein referred to as the crRNA repeat (Fig. 1A). We used the catalytically inactive Csy4-H29A mutant (Haurwitz et al. 2010) for experiments focused on analyzing the effects of changes to the RNA substrate, enabling investigation of RNA binding independent of cleavage. Wild-type (WT) Csy4 and Csy4-H29A bind a noncleavable RNA substrate with affinities that are within threefold of each other (Supplemental Fig. 1A).
Strikingly, Csy4 binds the full-length, WT-crRNA repeat substrate with extremely high affinity, characterized by an equilibrium dissociation constant of ~50 pM (Fig. 1C; Supplemental Table 1). Because Csy4 and the mature crRNA form part of the large Csy ribonucleoprotein complex responsible for target recognition (Wiedenheft et al. 2011a), we wondered whether Csy4 also retains high-affinity binding to the cleaved crRNA. Using a synthetic RNA corresponding to the 5′ product stem–loop structure, we found that Csy4 binds this RNA indistinguishably from the substrate (Fig. 1C). Thus, all protein:RNA interactions contributing favorably to binding energy occur upstream of the scissile phosphate. Analysis of substrates truncated in the 5′ ssRNA region allowed us to further demonstrate that nucleotides 1–4 of the crRNA repeat are completely dispensable for binding (Supplemental Fig. 2A), indicating that the high-affinity interaction we observe requires only the 15-nt stem–loop and one upstream nucleotide. We observed binding defects when A5 was mutated, suggesting that it might be specifically recognized. Indeed, a crystal structure of a Csy4:product RNA complex containing nucleotides 2–20 of the crRNA repeat sequence revealed base-specific hydrogen bonds between the Watson-Crick face of A5 and the peptide backbone of Leu139 (Supplemental Fig. 2B; RE Haurwitz, SH Sternberg, and JA Doudna, in prep.).
Considering the retention of Csy4 and crRNA in the Csy complex (Wiedenheft et al. 2011a), we speculated that tight association of Csy4 with its product may be an intrinsic mechanistic feature of Csy4 during crRNA biogenesis in type I-F CRISPR systems. To test this hypothesis, we carried out cleavage assays at a range of enzyme:substrate molar ratios and monitored both the rate and yield of product formation. As seen in Figure 1D, Csy4 completely lacks the ability to engage in multiple-turnover catalysis. The overall yield of the cleavage reaction remained directly proportional to the Csy4 concentration when present in sub-stoichiometric amounts relative to substrate, even with incubation times >200-fold longer than the reaction time constant. All time courses fit well to a single exponential decay and yielded uniform, first-order observed rate constants (kobs; Supplemental Table 1), which would only be the case in the absence of multiple-turnover behavior under conditions where the on-rate is not rate-limiting. These observations indicate that Csy4 remains product-bound after the reaction and is thereby strongly inhibited from performing additional rounds of RNA cleavage. Interestingly, crRNA repeat cleavage reached only 50% completion at an enzyme:substrate molar ratio of 1:1. A recent study used a two-hybrid system to demonstrate that Csy4 can interact with itself, but this result could not be repeated for all fusion constructs (Przybilski et al. 2011). While we cannot formally exclude the possibility that Csy4 might function as a dimer with one inactive subunit, our gel filtration experiments are consistent with purified Csy4 existing as a monomer (data not shown). Therefore, we speculate that the incomplete cleavage we observe reflects partial specific activity of purified WT-Csy4.
Protein determinants of high-affinity crRNA repeat binding and cleavage
The high-affinity interaction between Csy4 and the crRNA repeat substrate is tighter than many protein:RNA complexes studied to date. We were therefore interested in gaining a detailed understanding of the primary sources of binding energy, as informed by interactions identified from our crystal structure. We began by focusing on the bottom of the RNA stem, where the side-chains of Arg102 and Gln104 are each involved in two sequence-specific hydrogen bonds with the major groove faces of G20 and A19, respectively. Using a synthetic, noncleavable substrate that is bound indistinguishably from the WT-crRNA repeat (Supplemental Fig. 1B), EMSAs with Csy4-R102A and Csy4-Q104A mutants revealed that the binding energies contributed by these amino acids are quite distinct. The crRNA repeat binds >2000-fold more weakly to Csy4-R102A, representing a ΔΔG of 4.6 kcal/mol, whereas RNA binding by Csy4-Q104A is destabilized by only 1.4 kcal/mol relative to WT (Fig. 2A; Supplemental Table 2). This difference may be explained in part by the expected +1 charge on the arginine's guanidinium group at physiological pH. Whereas deletion of an uncharged hydrogen bond typically weakens binding between enzyme and substrate by 0.5–1.8 kcal/mol, charged hydrogen bonds generally contribute some 3–6 kcal/mol binding energy (Fersht 1987), in good agreement with our data.
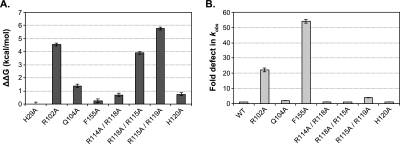
Amino acid contributions to binding energy and cleavage kinetics. (A) Csy4 residues involved in base-pair recognition and phosphate backbone contacts were mutated to alanine in order to assess their energetic contributions to binding. EMSAs were performed with a noncleavable crRNA repeat substrate containing a deoxyribonucleotide substitution at G20, and binding defects relative to Csy4-H29A were determined and converted to ΔΔG values (T = 298 K). Plotted are the average and SEM from at least three independent experiments. (B) First-order rate constants (kobs) for WT-crRNA repeat cleavage by each Csy4 mutant were determined. Cleavage data for R118A/R115A and R115A/R119A mutants showed biphasic kinetics and were fit with a double exponential decay to yield two rate constants (Supplemental Table 2), the faster of which is shown. Plotted are the average fold defects (relative to WT-Csy4) and SEM from three independent experiments. Average Kd, kobs, and SEM values are reported in Supplemental Table 2.
In addition to its interaction with Arg102, G20 of the crRNA repeat stacks onto the aromatic side-chain of Phe155. Stacking interactions between aromatic amino acids and nucleotides can contribute up to 5.5 kcal/mol of binding energy (Nolan et al. 1999; Auweter et al. 2006), but we were surprised to observe a negligible 1.5-fold binding defect (ΔΔG = 0.2 kcal/mol) with a Csy4-F155A mutant (Fig. 2A). Given the pre-crRNA processing defects we observed previously with Csy4-F155A (Haurwitz et al. 2010), these data suggest that Phe155 instead plays a role in achieving rapid cleavage kinetics. Indeed, under single-turnover conditions with saturating enzyme concentrations (see Materials and Methods), the F155A mutant led to an ~50-fold reduction in the observed cleavage rate constant (Fig. 2B). Csy4-R102A also exhibited an ~20-fold defect in cleavage kinetics, whereas the rate of cleavage by Csy4-Q104A was within 2.5-fold of WT (Fig. 2B). Collectively, these data suggest that, independent of their effects on binding energy, Phe155 and Arg102 are important for anchoring the G20 guanine in the active site and may thereby assist in positioning the ribose for subsequent activation of its 2′-OH nucleophile.
Moving up the crRNA repeat stem, we next focused on interactions observed in the crystal structure between the RNA and residues found in the α-helix that inserts into the major groove of the double-stranded stem (Fig. 1B). The guanidinium groups of Arg114, Arg115, Arg118, and Arg119 each present ≥2 hydrogen-bond donors within 3 Å of acceptors in the RNA phosphate backbone, yet their contributions to overall binding energy differ widely, as assessed through double R→A mutations. In particular, Arg114 and Arg118, which contact adjacent phosphates, contribute only 0.7 kcal/mol of binding energy, whereas alanine mutations at Arg115 and Arg119 led to a >15,000-fold binding defect (ΔΔG = 5.8 kcal/mol) (Fig. 2A). While all four residues are positioned to act as arginine forks, in that each side-chain contacts adjacent phosphates (Calnan et al. 1991), only Arg115 and Arg119 may simultaneously utilize all three nitrogen atoms of the guanidinium group as hydrogen bond donors. Arg115 hydrogen bonds to two phosphates in addition to the major groove face of G11, which forms part of the G·A sheared base pair at the bottom of the GUAUA pentaloop, and Arg119 is situated in a unique pocket of the loop where it interacts with phosphates separated by two nucleotides. His120 also interacts with a phosphate at the apex of the loop and contributes 0.8 kcal/mol of binding energy (Fig. 2A). The specific network of multi-dentate contacts between the arginine-rich helix and the RNA stem–loop suggests that high-affinity binding to the crRNA repeat is highly shape-specific, especially with regard to the tertiary structure of the loop. The large magnitude of the binding energy contributed by this protein helix enables Csy4 to maintain a tight grip on the substrate and product, but this interaction is not required for catalytic activity. Cleavage rates for the H120A and R→A mutants under saturating conditions were within fivefold of WT-Csy4 (Fig. 2B).
High-affinity crRNA repeat binding is sensitive to the loop structure
The direction of CRISPR loci transcription in P. aeruginosa has not been directly analyzed, and a recent report that detected mature crRNAs by Northern blot analysis used dsDNA probes that were not strand-specific (Cady and O'Toole 2011). Transcription in a direction opposite to that of our own predictions would generate pre-crRNAs containing the reverse complement of the crRNA repeat sequence. To determine whether Csy4 also recognizes and cleaves this potential substrate, we generated the reverse complement crRNA (rc-crRNA) repeat by in vitro transcription and tested its affinity for Csy4-H29A. We found that the rc-crRNA repeat binds Csy4 >105-fold more weakly than the WT-crRNA repeat (Fig. 3A) and is cleaved >750-fold more slowly (Supplemental Fig. 3A), strongly suggesting that the genuine Csy4 substrate in vivo is pre-crRNA transcribed in an orientation consistent with our previous work (Haurwitz et al. 2010). Northern blot analysis using single-stranded probes indeed confirmed the presence of crRNAs in P. aeruginosa UCBPP-PA14 with the repeat sequence we define in Figure 1A, but failed to detect transcripts from the opposite strand (Supplemental Fig. 4).
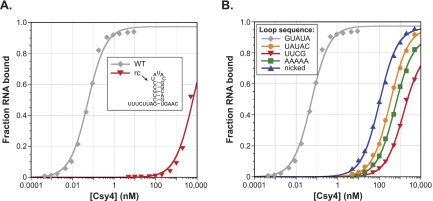
Importance of loop sequence for high-affinity RNA binding. (A) EMSAs demonstrate that Csy4 binds the reverse complement of the crRNA repeat (rc) >105-fold more weakly than the WT-crRNA repeat. (B) Mutant RNA substrates were generated by changing the WT loop sequence (GUAUA) to a quintuple mutant (UAUAC), the highly stable UUCG tetraloop, or a poly(A) pentaloop, or by removing the loop through use of a substrate nicked between U12 and A13. EMSAs reveal substantial defects associated with binding these mutant RNAs.
When comparing the two RNA sequences, the rc-crRNA repeat contains the identical five-base-pair stem sequence as the WT-crRNA repeat but with an additional predicted G–U wobble base pair below and different loop and flanking ssRNA sequences, indicating that one or more of these regions are specifically recognized by Csy4. Having already demonstrated the negligible binding defects resulting from deletion of flanking ssRNA nucleotides, we suspected that destabilized binding of the rc-crRNA repeat resulted primarily from the inability of Csy4 to interact productively with the UAUAC loop sequence and/or the unique tertiary structure it would impose on the RNA substrate. The GUAUA loop encoded by CRISPR locus 2 in P. aeruginosa UCBPP-PA14 forms a GNR(N)A pentaloop structure (Legault et al. 1998), in which U14 flips out of the loop to enable a GNRA tetraloop fold that involves sequential stacking of U12, A13, and A15 on the 3′ strand of the stem (Haurwitz et al. 2010). The CRISPR 3 locus encodes a GUGUA loop in the repeat sequence that is predicted to form the same pentaloop structure, and this crRNA structure is bound and cleaved indistinguishably from the substrate with a GUAUA loop (Supplemental Fig. 5). We hypothesized that Csy4 specifically recognizes this loop motif, and that other loop sequences unable to conform to a GNRA tetraloop fold would bind much more weakly.
To test this, we generated a panel of RNA substrates containing mutated loop sequences and tested their affinity for Csy4-H29A. In agreement with our hypothesis, Csy4 bound to each RNA at least 7000-fold more weakly than WT (Fig. 3B). Even a nicked RNA substrate formed from two oligonucleotides annealed in trans interacted more favorably with Csy4 than those containing a non-GNRA-like loop (Fig. 3B; Supplemental Fig. 6). These experiments confirm that high-affinity Csy4 binding relies in part on a precise substrate tertiary structure in the loop region, independently of base-specific contacts, and that the absence of a loop altogether is less detrimental to binding than the presence of a nonnative loop. It is interesting to note that, despite their weakened binding, RNAs with mutated loops were cleaved at rates within 2.5-fold of the WT-crRNA repeat at saturating Csy4 concentrations (Supplemental Fig. 3B). This was true even for a substrate containing the same loop (UAUAC) as the rc-crRNA repeat, which had a >750-fold defect in kobs. Since the stacking interaction between the terminal C–G base pair and the aromatic side chain of Phe155 is important for cleavage (Fig. 2B), we suspected that the additional base pair below the WT stem in the rc-crRNA repeat might impede Csy4 activity (see below).
Specificity within the crRNA repeat stem sequence during binding and cleavage
We were particularly interested in investigating the ability of Csy4 to discriminate between substrates containing the cognate five base pairs in the stem and those with similar but noncognate sequences. We therefore made all individual Watson-Crick base-pair substitutions at each position in the double-stranded stem and determined the energetic costs associated with binding each mutant RNA substrate relative to the WT-crRNA repeat using EMSAs (Fig. 4A). The data reveal that base-pair changes throughout the stem result in varying degrees of Csy4:RNA complex destabilization, ranging from 0.4 to 4.2 kcal/mol. The largest defects result from G–C and C–G substitutions at the ultimate and penultimate base pair, respectively, where Arg102 and Gln104 provide a direct readout mechanism of recognition and confer similar degrees of discrimination in spite of their unequal contributions to binding energy. To confirm this, we repeated binding experiments with RNA substrates containing substitutions at the bottom two base pairs using either Csy4-R102A or Csy4-Q104A (Fig. 4A). As expected, the overall specificity for particular base pairs at either position is lost when the amino acid specificity determinant is absent. The Csy4:RNA co-crystal structure did not reveal sequence-specific contacts with Watson-Crick base pairs in the upper part of the double-stranded stem (Haurwitz et al. 2010), but we observed substantial energetic penalties for binding substrates with base-pair substitutions in this region (Fig. 4A). Furthermore, the magnitude of these binding defects was highly sequence-dependent; when multiple base-pair substitutions were made in the top three base pairs simultaneously, binding defects ranged from seven- to almost 5000-fold (Supplemental Fig. 7), with the largest destabilization occurring when each C–G pair was mutated to its complement. These results reveal that substrate sequence specificity is mediated by Csy4 via a mechanism that does not rely exclusively on base-specific interactions.
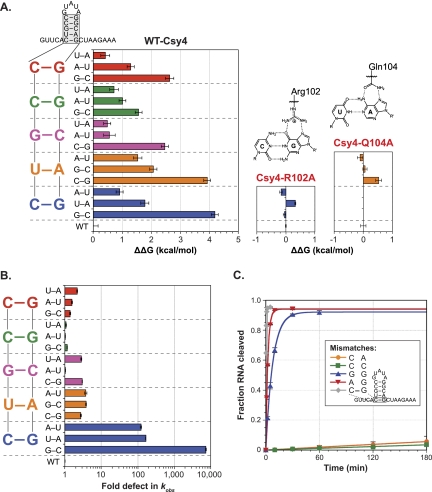
Substrate specificity within the crRNA repeat stem. (A) A library of mutated crRNA repeat substrates was generated containing all possible Watson-Crick base-pair substitutions at each position in the double-stranded stem. EMSAs were performed with Csy4-H29A and these RNA substrates, and the resulting binding defects relative to WT-crRNA repeat were determined and converted to ΔΔG values (T = 298 K). The WT stem sequence is shown at the left, with data for base-pair substitutions at each position color-coded similarly. Binding experiments with RNAs mutated at the bottom C–G or U–A base pair were repeated with Csy4-R102A (middle) or Csy4-Q104A (right), respectively; ΔΔG values were calculated relative to WT-crRNA repeat binding by each Csy4 mutant. Shown above are chemical structures of the interactions made by Arg102 and Gln104 with the WT base pairs. (B) Single-turnover cleavage assays were performed with the same library of RNA mutants as in A, and the resulting defects in kobs relative to WT-crRNA repeat were determined. The data are plotted as in A. (C) To investigate the importance of the terminal C–G base pair during cleavage, mismatched substrates were generated by mutating C6 or G20 individually and single-turnover cleavage assays were performed.
We next investigated whether these specificity determinants also influence the chemical cleavage reaction. To test this, we conducted single-turnover cleavage experiments with WT-Csy4 at saturating concentrations using the same library of RNAs as in Figure 4A and determined the first-order rate constants for RNA cleavage (kobs) relative to WT. In stark contrast to the observed binding specificity, rate constants governing the cleavage of RNA substrates with base-pair substitutions at any position other than the terminal position were within fourfold of WT (Fig. 4B). However, any mutation of the terminal C–G base pair in the stem–loop was detrimental for cleavage of the crRNA repeat, with kinetic defects ranging from ~100- to 7500-fold. To further dissect the importance of the terminal C–G base pair, we generated a series of RNA substrates containing mismatches at this position by mutating either C6 or G20 independently. Cleavage time courses with these substrates (Fig. 4C) clearly demonstrate the importance of G20, regardless of whether or not a base pair can form at the terminal position. RNA substrates containing C6A or C6G mutations were cleaved at rates within 40-fold of WT, whereas mutation of G20 to either adenosine or cytosine led to >10,000-fold defects.
Csy4 is highly selective for stem–loops of defined length
Having interrogated Csy4 for sequence specificity throughout the crRNA repeat, we also wondered whether Csy4 is sensitive to the length of the crRNA repeat stem. To test this, we inserted one or two base pairs at the top of the duplex region and tested these substrates for binding. Strikingly, just one or two additional G–C base pairs led to 1600- and 49,000-fold weaker binding affinities, respectively (Fig. 5A). This was particularly surprising because the crystal structure did not immediately suggest any obvious steric clashes that would result from insertions at the top of the stem. However, given the large energetic contribution of the arginine-rich helix to binding (Fig. 2C), we suspected that additional base pairs would disrupt protein–loop interactions and prevent stable docking of this helix into the major groove of the double-stranded stem. A-form dsRNA helices have deep and narrow major grooves that are generally inaccessible to proteins (Draper 1995), but exceptions occur in proximity to helix termini or asymmetric bulges, where the major groove can widen considerably (Weeks and Crothers 1993). We hypothesized that base-pair insertions cause narrowing of the major groove and thereby disrupt high-affinity interactions between the arginine-rich helix and crRNA repeat.
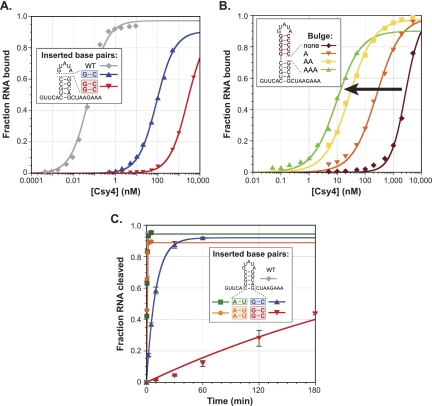
Stem length dependence during substrate binding and cleavage. (A) One or two G–C base pairs were inserted at the top of the stem between the closing C–G base pair and the GUAUA pentaloop, and EMSAs were performed. (B) To test the hypothesis that longer stems prevent stable binding of the arginine-rich helix via their effect on major groove accessibility, a substrate was generated that contains five G–C base pairs inserted above the WT stem. Subsequently, asymmetric adenosine bulges were inserted on the 3′ side of the duplex between the five-base-pair WT stem and the five-base-pair insertion. EMSAs reveal that binding affinities increase monotonically (black arrow) with bulges of increasing size. (C) One or two G–C or A–U base pairs were inserted below the terminal C–G base pair, and cleavage time courses were performed. Additional A–U base pairs have negligible effects on kobs, whereas two additional G–C base pairs result in ~1500-fold slower kinetics.
To test this idea, we generated an RNA construct that contains five G–C base pairs inserted atop the WT stem sequence while retaining the GUAUA pentaloop. This RNA was bound with an equilibrium dissociation constant of 4 μM (Fig. 5B), representing nearly a 105-fold defect relative to WT. We then introduced adenosine bulges of varying size on the 3′ side of the stem, at the junction between the WT five-base-pair stem sequence and the five G–C base-pair insertion. These types of asymmetric bulges within perfectly base-paired dsRNA helices have been shown previously to increase major groove accessibility progressively as a function of bulge size, as probed using diethylpyrocarbonate (DEPC) reactivity (Weeks and Crothers 1993). In excellent agreement with our hypothesis, we found that the binding affinity of Csy4 for these bulged substrates increased in concert with bulges of increasing size (Fig. 5B), suggesting that major groove widening enables stable docking of the arginine-rich helix. The inability to form favorable protein–loop interactions likely explains why bulged substrates are still bound >200-fold more weakly than the WT-crRNA repeat.
We also investigated the effects of inserting one or two base pairs at the bottom of the stem–loop below the terminal C–G base pair. We observed a range of binding defects, although these were milder than those resulting from insertions at the top of the stem (Supplemental Fig. 8A). Cleavage defects at saturating enzyme concentrations were highly dependent on sequence: Whereas substrates containing one or two A–U base-pair insertions were cleaved at rates within twofold of the WT substrate, one or two G–C base-pair insertions resulted in ~50- and ~1500-fold lower kobs values, respectively (Fig. 5C). Partial RNase T1 digestions and RNA hydrolysis ladders revealed that these RNA constructs were cleaved above the inserted base pair(s) and just below the WT C–G base pair (Supplemental Fig. 8B). Thus, Csy4-catalyzed cleavage likely requires prior melting of any additional secondary structures below the five-base-pair stem, such that the WT stem is correctly positioned in the binding pocket and the guanosine containing the 2′-OH nucleophile can productively interact with Arg102 and Phe155. In support of this interpretation, A–U and U–A base pairs are thermodynamically less stable than G–C and C–G base pairs at the termini of RNA duplexes (Xia et al. 1998) and are likely to be more susceptible to transient fraying (Snoussi and Leroy 2001), explaining the large magnitude of kobs differences for these distinct insertions.
Collectively, these data indicate that beyond sequence-specific recognition of its crRNA repeat substrate, Csy4 is finely tuned to bind and cleave stem–loop substrates containing just five base pairs within the dsRNA region, through at least two distinct mechanisms. First, binding energy contributed by the arginine-rich helix requires an accessible major groove, which depends on the double-stranded stem being properly spaced between interaction sites at its base (e.g., with Arg102) and the loop sequence. Second, rapid cleavage requires the positioning of a terminal C–G base pair within the active site and prior disruption of any additional secondary structures below.
DISCUSSION
The CRISPR-Cas adaptive immune system has evolved a sophisticated strategy for generating large libraries of short effector RNAs that target invasive genetic elements for destruction. Rather than requiring each crRNA to be individually transcribed, the repetitive CRISPR architecture allows large precursor transcripts to be successively processed by Cas endoribonucleases (in type I and III CRISPR systems) that are precisely tailored for specific recognition and cleavage of the invariant repeat sequence. Here we have defined the various molecular strategies employed by one such Cas enzyme—Csy4 (Cas6f) from P. aeruginosa UCBPP-PA14—to enable an impressive degree of affinity and specificity for its crRNA repeat substrate.
The Csy4:RNA complex is characterized by an ~50 pM equilibrium dissociation constant (Kd) and requires only a 16-nt stem–loop motif for tight binding. For comparison, U1A protein, MS2 coat protein, and the Nλ protein bind their RNA substrates with Kd values of 50 pM, 2.6 nM, and 5 nM, respectively (van Gelder et al. 1993; LeCuyer et al. 1995; Cilley and Williamson 1997). High-energy interactions are mediated almost exclusively within the major groove of a double-stranded RNA stem–loop, a region of A-form helices that is generally refractory to protein contacts because of its inaccessibility. Prior work used chemical probing to demonstrate that the termini of dsRNA contain uncharacteristically wide major grooves (Weeks and Crothers 1993), which explains how direct readout of A19 and G20 at their major groove edge is possible. Our data reveal that stable binding of the arginine-rich helix further up the stem is also highly sensitive to major groove accessibility, and that this requirement enables up to ~50,000-fold discrimination against hairpin substrates containing slightly longer stems. Four arginines within this α-helix are precisely positioned to contact multiple phosphates within the RNA backbone and adopt conformations reminiscent of the arginine fork first described for HIV-1 Tat protein by Frankel and colleagues (Calnan et al. 1991). This mode of multi-dentate interaction requires precise interatomic P–P distances, indicating that the network of hydrogen bonds formed by the arginine-rich helix depends on a very specific substrate conformation. Indeed, changes to the loop sequence or to the identity of base pairs in the upper part of the stem result in substantial binding defects, despite the general lack of base-specific contacts in this region. Substrate selection thus proceeds in large part via an indirect readout mechanism, whereby a particular RNA tertiary structure is recognized that is contingent on both primary sequence and the distinct helical geometry it imposes. Similar modes of substrate recognition have been described for a number of dsDNA-binding proteins (Otwinowski et al. 1988; Rohs et al. 2009).
Csy4 retains the same tight binding for both its substrate and product, and functions as a single-turnover catalyst due to potent product inhibition. These data strongly suggest that crRNA biogenesis in P. aeruginosa UCBPP-PA14 requires stoichiometric amounts of the processing endoribonuclease. Cleavage of the crRNA repeat substrate depends critically on the presence of a guanosine upstream of the scissile phosphate, independently of whether or not this nucleotide is base-paired, and is inhibited when additional secondary structure forms below the five-base-pair stem. The kobs defects we observed with Csy4-R102A and Csy4-F155A mutants indicate that the G20 base must be tightly locked in place within the enzyme active site in order to rapidly achieve chemical activation of the ribosyl 2′-OH. Other critical active site residues (Tyr176 and Ser148) have also been implicated in properly positioning the G20 ribose in an orientation that is compatible with nucleophilic attack on the downstream phosphodiester bond (RE Haurwitz, SH Sternberg, and JA Doudna, in prep.).
We recently reported that, together with six copies of Csy3 and single copies of both Csy1 and Csy2, Csy4 and the mature crRNA assemble into a large ribonucleoprotein complex (Csy complex) that is responsible for target recognition during the interference stage of the CRISPR pathway (Wiedenheft et al. 2011a). Our data are consistent with a model where the Csy4-bound crRNA serves as a nucleation point for assembling the remainder of the complex, which does not form independently of RNA (Wiedenheft et al. 2011a). Interestingly, Cse3 (Cas6e), the CRISPR-specific endoribonuclease from type I-E CRISPR systems, also acts as a single-turnover enzyme (Sashital et al. 2011) and forms part of the downstream target recognition effector complex (Cascade) (Brouns et al. 2008; Jore et al. 2011; Wiedenheft et al. 2011b). It is tempting to speculate that these related enzymes evolved to react stoichiometrically during pre-crRNA cleavage in order to ensure that the mature crRNA is not prematurely released into the cytoplasm but instead remains tightly sequestered by the Cas machinery. While this mechanistic feature may be intrinsic to certain Cas6 family members, it is not generalizable. Cas6 in type III-B CRISPR systems is not a component of the downstream effector complex (Cmr complex) (Hale et al. 2009), and Cas6 from type I-A CRISPR systems remains only loosely associated with the downstream effector complex (archaeal Cascade) (Lintner et al. 2011). Intriguingly, these differences correlate with the thermodynamic stability of hairpin structures encoded by CRISPR repeats typical of each subtype; repeats clustered based on sequence similarity that associate with type I-E and type I-F CRISPR systems encode highly stable RNA secondary structures, whereas those that associate with type I-A and type III-B systems encode RNAs predicted to be unstructured (Kunin et al. 2007).
CRISPR-specific endoribonucleases are unusual in that their biological function involves cleavage of a single, invariant substrate. As such, these enzymes have likely co-evolved with their target crRNA repeats to retain a high degree of substrate specificity, which serves to avoid spurious binding and/or cleavage of noncognate RNAs inside the cell. The work presented here highlights the diverse molecular strategies exploited by P. aeruginosa Csy4 (Cas6f) to generate this selectivity while maintaining an extremely high-affinity interaction with its ligand. The potential benefits of these attributes for molecular biology applications will be exciting to explore further. Finally, future work will be needed to determine whether the underlying principles of RNA stem–loop recognition exhibited by Csy4 are conserved among other Cas6 family members.
MATERIALS AND METHODS
Protein expression and purification
R102A, Q104A, F155A, and H29A Csy4 mutants were purified as described (Haurwitz et al. 2010). R114A/R118A, R118A/R115A, R115A/R119A, and H120A Csy4 mutants were generated using site-directed mutagenesis and purified essentially as described previously (Haurwitz et al. 2010), with the following exceptions. Protein genes encoded by the pHGWA vector (Busso et al. 2005) were overexpressed in BL21(DE3) cells. Following the second Ni-NTA affinity purification step, Csy4 mutants were purified by size exclusion chromatography using a single Superdex 75 (16/60) column (GE Healthcare) in 100 mM HEPES (pHRT 7.5), 500 mM KCl, 5% glycerol, 1 mM TCEP. Proteins were then concentrated and buffer-exchanged into 100 mM HEPES (pHRT 7.5), 150 mM KCl, 5% glycerol, 1 mM TCEP; snap-frozen in liquid nitrogen; and stored at −80°C.
Northern blot analysis
Total RNA was extracted from cultures of P. aeruginosa PAO1, P. aeruginosa UCBPP-PA14, and a csy4 deletion strain of P. aeruginosa UCBPP-PA14 (SMC3894) (Zegans et al. 2009) grown to exponential phase using the mirVana kit (Ambion). Duplicate samples of each RNA preparation (6 μg) were separated on adjacent lanes of a 15% denaturing polyacrylamide gel and subsequently transferred to a nylon membrane (Hybond-N+, GE Healthcare) using a semi-dry transfer cell (Bio-Rad). The single membrane was then cut in half to yield two membranes with identical samples. The membranes were pretreated with ULTRAHyb-Oligo Hybridization Buffer (Ambion) and probed with 5′-[32P]-radiolabeled DNA oligonucleotides corresponding to either the crRNA repeat sequence (5′-GTTCACTGCCGTATAGGCAGCTAAGAAA-3′) or the reverse complement of the crRNA repeat (5′-TTTCTTAGCTGCCTATACGGCAGTGAAC-3′). Membranes were washed twice with 2× saline-sodium citrate (SSC) buffer containing 0.5% SDS and visualized by phosphorimaging.
RNA transcription, purification, and 5′ radiolabeling
The following RNAs were synthesized by Integrated DNA Technologies: the noncleavable substrate, product RNA (Δ21–28), 5′ truncation constructs (Δ1–5, Δ1–4), the 5′-strand (nucleotides 1–12) and 3′-strand (nucleotides 13–28) used to generate the nicked substrate, the G20A mismatched substrate, and three substrates containing base-pair substitutions at the bottom of the stem (C6U/G20A, C6G/G20C, U7A/A19U). All other RNAs were transcribed in vitro using T7 polymerase and purified using denaturating polyacrylamide gel electrophoresis, according to the following protocol. Synthetic single-stranded DNA templates (Integrated DNA Technologies) containing the reverse complement of the desired crRNA repeat construct were annealed to a 1.5-fold molar excess of an oligonucleotide corresponding to the T7 promoter sequence (5′-TAATACGACTCACTATA-3′). Templates encoded an extra guanosine at the 5′ end of all constructs in order to ensure optimal transcription by T7 polymerase. This had no effect on binding affinities but did lead to a slight (~20%) increase in kobs for cleavage of the WT-crRNA repeat substrate. Transcription reactions (100 μL) were incubated at 37°C for 3–5 h and contained 1 μM template DNA, 100 μg/mL T7 polymerase, 1 μg/mL pyrophosphatase (Roche), 5 mM NTPs, 30 mM Tris-Cl (pHRT 8.1), 25 mM MgCl2, 10 mM dithiothreitol (DTT), 2 mM spermidine, and 0.01% Triton X-100. Reactions were then treated with 5 units of DNase (Promega) and incubated for an additional 30 min at 37°C before being loaded on a 15% urea-polyacrylamide gel. RNAs were excised from the gel and eluted into DEPC H2O overnight at 4°C. 5′ triphosphates were removed by incubating RNAs at 37°C for 1 h with 10 units of calf intestinal phosphate (New England Biolabs) in 1× NEBuffer 3, followed by phenol-chloroform extraction and ethanol precipitation. RNAs were resuspended in DEPC H2O and stored at −20°C.
For biochemical experiments, 10 pmol RNA were 5′-radiolabeled by incubating with 5 units T4 polynucleotide kinase (New England Biolabs) and ~3–6 pmol (~20–40 μCi) [γ-32P]-ATP (Promega) in 1× T4 polynucleotide kinase reaction buffer at 37°C for 30 min, in a 25 μL reaction. After heat inactivation (65°C for 20 min), reactions were spun through an illustra MicroSpin G-25 column (GE Healthcare) to remove ATP. Radiolabeled RNAs were diluted to ~100 nM stock concentrations with DEPC H2O and stored at −20°C.
Electrophoretic mobility shift assays
Protein concentrations were determined by taking multiple absorbance spectra using a NanoDrop spectrophotometer (Thermo Scientific), averaging A280nm values and converting to molar concentrations using the calculated Csy4 extinction coefficient (15,470 M−1 cm−1). Spectra were also recorded under denaturing conditions (6 M guanidine hydrochloride, 20 mM potassium phosphate buffer, pH 6.5), and absorbance values were within error of those taken under native conditions. Binding experiments were conducted in the following buffer: 20 mM HEPES (pHRT 7.5), 100 mM KCl, 5% glycerol, 0.01% Igepal-630, 1 mM DTT, and 0.1 mg/mL yeast tRNA (Sigma-Aldrich) to prevent nonspecific binding. After diluting concentrated 5′-[32P]-labeled RNA and Csy4 stock solutions into 1× binding buffer, trace amounts of RNA (≤0.05–0.2 nM, depending on construct and specific activity) were incubated with increasing concentrations of Csy4 in a 15 μL reaction at room temperature (~24°C) for one hour. Twelve microliters of each reaction were then loaded on a 10% native polyacrylamide gel containing 0.5× TBE buffer and resolved by running at 12 W for 90–120 min at 4°C in 0.5× TBE running buffer. Phosphor screens were exposed to dried gels and scanned with a Storm imager (GE Healthcare), and the intensities of unbound and Csy4-bound RNA were quantified using ImageQuant (GE Healthcare). The fraction of RNA bound at each Csy4 concentration was plotted as a function of Csy4 concentration, and binding data were fit with a standard binding isotherm using Kaleidagraph (Synergy Software), according to the equation:
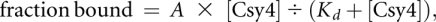
where A is the amplitude of the binding curve.
Binding experiments with the substrate nicked between U12 and A13 contained ~1 nM radiolabeled 3′-strand (nucleotides 13–28) and a 1000-fold excess (1 μM) of cold 5′-strand (nucleotides 1–12). For experiments with Kd values in the low pM range, binding data were also fit with the solution of a quadratic equation describing a bimolecular dissociation reaction, as described previously (Maag and Lorsch 2003), out of concern that [RNA] in these experiments was not sufficiently below the Kd to approximate [Csy4]total = [Csy4]free. This analysis returned values that agreed well with equilibrium dissociation constants determined from the standard binding isotherm equation, so these original values are reported. When fitting binding data with the rc-crRNA repeat, the amplitude was set equal to one because saturation could not be reached. Binding data with the RNA substrate containing a five G–C base-pair insertion showed apparent cooperativity and were fit with a modified binding equation using a variable Hill coefficient (n ≈ 1.5) and an amplitude fixed at one.
At least one binding experiment for each RNA or Csy4 mutant titrated Csy4 across a concentration range of three orders of magnitude centered around the Kd. Additional replicates typically tested five concentration points centered around the Kd and returned values in excellent agreement with those derived from a more complete titration. Kd values presented in the text and in Supplemental Tables 1 and 2 represent the average and standard error of the mean from at least three independent experiments. The average percent error for all reported Kd values is 10%. ΔΔG values for Csy4 or RNA mutants were calculated according to the equation:
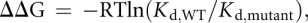
where R is the gas constant, T is temperature (set to 298 K), and Kd,WT/Kd,mutant is the ratio of Kd values for the WT and mutant construct.
RNA cleavage assays
Cleavage assays were conducted at room temperature (~24°C) in the following buffer: 20 mM HEPES, 100 mM KCl, 1 mM DTT at pHRT 7.5. Single-turnover cleavage experiments were 55 μL in volume and contained 0.5 nM 5′-[32P]-labeled RNA and a saturating concentration of Csy4 (typically 500 nM). At each desired time point, a 10 μL aliquot was removed and quenched by mixing it with 50 μL phenol:chloroform:isoamyl alcohol 25:24:1 at pH 8.0 (Sigma-Aldrich). The aqueous layer was mixed with an equal volume of formamide loading dye, heated to ~80°C for ~2 min, and separated on a 15% urea-polyacrylamide gel in 0.5× TBE running buffer. RNA was visualized by phosphorimaging, and the intensities of uncleaved and cleaved RNA were quantified using ImageQuant (GE Healthcare). The fraction of RNA cleaved at each time point was plotted as a function of time, and these data were fit with a single exponential decay curve using Kaleidagraph (Synergy Software), according to the equation:

where A is the amplitude of the curve, k is the first-order rate constant, and t is time. In order to avoid overestimating k in cases where the RNA was not quantitatively cleaved, the amplitude was fixed at one when fitting cleavage data for the substrate containing a G–C substitution at the bottom base pair, G20A and G20C mismatch mutants, and for the substrate with two G–C base pairs inserted below the stem–loop. Cleavage of the WT-crRNA repeat by Csy4-R118A/R115A and Csy4-R115A/R119A revealed biphasic kinetics, and the data were fit to a double exponential decay. The slower kinetic process may reflect a rate-limiting conformational change. Both rate constants are reported in Supplemental Table 2.
To ensure that Csy4 concentrations were saturating and that the on-rate for Csy4:RNA binding was not rate-limiting, cleavage experiments were repeated at fivefold higher enzyme concentrations and analyzed similarly. This analysis frequently returned slightly larger rate constants for RNAs with fast cleavage kinetics, which we attribute to slower quenching rates in the presence of more enzyme. Overall, rate constants for these experiments were generally within ~30% of those measured at the lower enzyme concentration. The precise nature of the rate-limiting step in our single-turnover cleavage assays is not known, and so first-order rate constants are reported as kobs. kobs values presented in the text and in Supplemental Tables 1 and 2 represent the average and standard error of the mean from three independent experiments. The average percent error for all reported kobs values is 4%.
Cleavage experiments with WT-Csy4 and WT-crRNA repeat at variable molar ratios (Fig. 1D) were conducted at a constant RNA concentration of 10 nM (0.25 nM 5′-radiolabeled RNA, 9.75 nM unlabeled RNA) and varying Csy4 concentrations (40, 20, 10, 5, 2.5 nM) in a final volume of 88 μL. Ten-microliter aliquots were removed and quenched at 0.25, 0.5, 1, 2, 5, 10, 30, and 60 min, and analyzed as described above. In determining the concentration of unlabeled RNA, hypochromicity of the stem–loop was corrected for by first hydrolyzing the RNA to nucleotides by incubating in 3 M NaOH at 50°C for one hour. Then, absorbance spectra were recorded using a NanoDrop spectrophotometer (Thermo Scientific), and A260nm values were averaged and converted to molar concentrations using the calculated extinction coefficient (295,900 M−1 cm−1). The 50% yield observed at an enzyme:substrate molar ratio of 1:1 may reflect Csy4 dimerization (Przybilski et al. 2011) or partial specific activity of purified WT-Csy4.
ACKNOWLEDGMENTS
The P. aeruginosa UCBPP-PA14 Δcsy4 strain was kindly provided by the G. O'Toole laboratory (Dartmouth Medical School). We thank K. Berry (Harvard Medical School), D. Sashital (The Scripps Research Institute), and other members of the Doudna laboratory for helpful discussions and critical reading of the manuscript. S.H.S. acknowledges support from the National Science Foundation and National Defense Science & Engineering Graduate Research Fellowship programs. J.A.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030882.111.
REFERENCES
- Al-Attar S, Westra ER, van der Oost J, Brouns SJJ 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): The hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 [Abstract] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH-T 2006. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res 34: 4943–4959 [Abstract] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [Abstract] [Google Scholar]
- Busso D, Delagoutte-Busso B, Moras D 2005. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 343: 313–321 [Abstract] [Google Scholar]
- Cady KC, O'Toole GA 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193: 3433–3445 [Europe PMC free article] [Abstract] [Google Scholar]
- Calnan B, Tidor B, Biancalana S, Hudson D, Frankel A 1991. Arginine-mediated RNA recognition: The arginine fork. Science 252: 1167–1171 [Abstract] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [Europe PMC free article] [Abstract] [Google Scholar]
- Cilley CD, Williamson JR 1997. Analysis of bacteriophage N protein and peptide binding to boxB RNA using polyacrylamide gel coelectrophoresis (PACE). RNA 3: 57–67 [Europe PMC free article] [Abstract] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [Europe PMC free article] [Abstract] [Google Scholar]
- Draper DE 1995. Protein-RNA recognition. Annu Rev Biochem 64: 593–620 [Abstract] [Google Scholar]
- Fersht AR 1987. The hydrogen bond in molecular recognition. Trends Biochem Sci 12: 301–304 [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, MacMillan AM 2011. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 18: 688–692 [Abstract] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172 10.1186/1471-2105-8-172 [Europe PMC free article] [Abstract] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [Europe PMC free article] [Abstract] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329: 1355–1358 [Europe PMC free article] [Abstract] [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [Abstract] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [Abstract] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61 10.1186/gb-2007-8-4-r61 [Europe PMC free article] [Abstract] [Google Scholar]
- LeCuyer KA, Behlen LS, Uhlenbeck OC 1995. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34: 10600–10606 [Abstract] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J 1998. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell 93: 289–299 [Abstract] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copié V, et al. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 286: 21643–21656 [Europe PMC free article] [Abstract] [Google Scholar]
- Maag D, Lorsch JR 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol 330: 917–924 [Abstract] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [Europe PMC free article] [Abstract] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [Europe PMC free article] [Abstract] [Google Scholar]
- Nolan S, Shiels J, Tuite J, Cecere K, Baranger A 1999. Recognition of an essential adenine at a protein-RNA interface: Comparison of the contributions of hydrogen bonds and a stacking interaction. J Am Chem Soc 121: 8951–8952 [Google Scholar]
- Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB 1988. Crystal structure of trp repressor/operator complex at atomic resolution. Nature 335: 321–329 [Abstract] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC 2011. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol 8: 517–528 [Abstract] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B 2009. The role of DNA shape in protein-DNA recognition. Nature 461: 1248–1253 [Europe PMC free article] [Abstract] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39: 9275–9282 [Europe PMC free article] [Abstract] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 18: 680–687 [Abstract] [Google Scholar]
- Snoussi K, Leroy JL 2001. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry 40: 8898–8904 [Abstract] [Google Scholar]
- van Gelder CW, Gunderson SI, Jansen EJ, Boelens WC, Polycarpou-Schwarz M, Mattaj IW, van Venrooij WJ 1993. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J 12: 5191–5200 [Europe PMC free article] [Abstract] [Google Scholar]
- Weeks KM, Crothers DM 1993. Major groove accessibility of RNA. Science 261: 1574–1577 [Abstract] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJR, Boekema EJ, Dickman MJ, et al. 2011a. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108: 10092–10097 [Europe PMC free article] [Abstract] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E 2011b. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477: 486–489 [Abstract] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA 2012. CRISPR RNA-guided gene silencing systems in bacteria and archaea. Nature (in press). [Abstract] [Google Scholar]
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37: 14719–14735 [Abstract] [Google Scholar]
- Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191: 210–219 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from RNA are provided here courtesy of The RNA Society
Full text links
Read article at publisher's site: https://doi.org/10.1261/rna.030882.111
Read article for free, from open access legal sources, via Unpaywall:
http://rnajournal.cshlp.org/content/18/4/661.full.pdf
Citations & impact
Impact metrics
Article citations
Engineering probiotic <i>Escherichia coli</i> Nissle 1917 to block transfer of multiple antibiotic resistance genes by exploiting a type I CRISPR-Cas system.
Appl Environ Microbiol, 90(10):e0081124, 10 Sep 2024
Cited by: 0 articles | PMID: 39254327
Symbolic recording of signalling and cis-regulatory element activity to DNA.
Nature, 632(8027):1073-1081, 17 Jul 2024
Cited by: 4 articles | PMID: 39020177 | PMCID: PMC11357993
The CRISPR-Cas System and Clinical Applications of CRISPR-Based Gene Editing in Hematology with a Focus on Inherited Germline Predisposition to Hematologic Malignancies.
Genes (Basel), 15(7):863, 01 Jul 2024
Cited by: 0 articles | PMID: 39062641 | PMCID: PMC11276294
Review Free full text in Europe PMC
CRISPR-Cas and CRISPR-based screening system for precise gene editing and targeted cancer therapy.
J Transl Med, 22(1):516, 30 May 2024
Cited by: 1 article | PMID: 38816739 | PMCID: PMC11138051
Review Free full text in Europe PMC
New design strategies for ultra-specific CRISPR-Cas13a-based RNA detection with single-nucleotide mismatch sensitivity.
Nucleic Acids Res, 52(2):921-939, 01 Jan 2024
Cited by: 7 articles | PMID: 38033324 | PMCID: PMC10810210
Go to all (105) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(2 citations)
PDBe - 2XLKView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence- and structure-specific RNA processing by a CRISPR endonuclease.
Science, 329(5997):1355-1358, 01 Sep 2010
Cited by: 433 articles | PMID: 20829488 | PMCID: PMC3133607
Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA.
EMBO J, 31(12):2824-2832, 20 Apr 2012
Cited by: 63 articles | PMID: 22522703 | PMCID: PMC3380207
Cutting it close: CRISPR-associated endoribonuclease structure and function.
Trends Biochem Sci, 40(1):58-66, 18 Nov 2014
Cited by: 78 articles | PMID: 25468820
Review
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.
FEMS Microbiol Rev, 39(3):428-441, 19 May 2015
Cited by: 137 articles | PMID: 25994611 | PMCID: PMC5965381
Review Free full text in Europe PMC
Funding
Funders who supported this work.





