Abstract
Free full text

miRNA-34c regulates Notch signaling during bone development
Abstract
During bone homeostasis, osteoblast and osteoclast differentiation is coupled and regulated by multiple signaling pathways and their downstream transcription factors. Here, we show that microRNA 34 (miR-34) is significantly induced by BMP2 during osteoblast differentiation. In vivo, osteoblast-specific gain of miR-34c in mice leads to an age-dependent osteoporosis due to the defective mineralization and proliferation of osteoblasts and increased osteoclastogenesis. In osteoblasts, miR-34c targets multiple components of the Notch signaling pathway, including Notch1, Notch2 and Jag1 in a direct manner, and influences osteoclast differentiation in a non-cell-autonomous fashion. Taken together, our results demonstrate that miR-34c is critical during osteoblastogenesis in part by regulating Notch signaling in bone homeostasis. Furthermore, miR-34c-mediated post-transcriptional regulation of Notch signaling in osteoblasts is one possible mechanism to modulate the proliferative effect of Notch in the committed osteoblast progenitors which may be important in the pathogenesis of osteosarcomas. Therefore, understanding the functional interaction of miR-34 and Notch signaling in normal bone development and in bone cancer could potentially lead to therapies modulating miR-34 signaling.
INTRODUCTION
Bone development and homeostasis requires tight regulation of gene activation and repression in response to various signaling pathways. Along with Wnt signaling, bone morphogenic proteins (BMPs) are potent morphogens that activate transcriptional programs of mesenchymal stem cells (MSC) to commit to the osteoblast lineage (1,2). BMP2 can activate key transcription factors, including Runt related transcription factor 2 (Runx2) and Osterix (Osx), during osteoblastogenesis (3,4). In addition, Notch signaling has been implicated in osteoblastic and osteoclastic differentiation in several in vitro studies (5–7). In vivo, our and others’ loss and gain of Notch1 function studies show that Notch signaling acts in a temporal and context-dependent manner in osteoblast differentiation. Gain of function in the early osteoprogenitor compartment leads to inhibition of osteoblastic commitment and low bone mass. In contrast, gain of function in committed osteoblastic cells stimulates proliferation of immature osteoblasts while inhibiting terminal osteoblastic differentiation to result in an osteosclerotic phenotype (8–10). Meanwhile, a deficiency of Notch signaling in committed osteoblasts down-regulates osteoprotegerin (Opg), thereby stimulating osteoclastogenesis in a non-cell-autonomous manner to cause an age-related osteoporosis (8).
Notch signaling is involved in maintaining the balance between cell proliferation and differentiation, and altered Notch signaling has been associated with various disorders, including cancer (11–14). Hajdu–Cheney syndrome, a rare skeletal disorder characterized by severe osteoporosis, is due to the gain-of-function mutations in NOTCH2 (15,16). The loss of function in JAG1 or DLL3, Notch ligands, leads to a spectrum of vertebral patterning defects, such as those seen in Alagille syndrome or spondylocostal dysostosis (17–19). Furthermore, Notch can act as an oncogene and a tumor suppressor depending on its expression levels and timing in a context-dependent manner (20,21). Aberrant Notch signaling is involved in solid and hematological tumor formation, and activating mutations of Notch1 in humans can cause T-cell acute lymphoblastic leukemia (22). Interestingly, human biopsy specimens of osteosarcomas show transcriptional up-regulation of NOTCH1, JAG1 and NOTCH downstream targets (HES1 and HEY2) (23,24). Furthermore, inhibition of the Notch pathway via γ-secretase inhibitors decreases osteosarcoma cell proliferation in vitro and in vivo in xenografts models. Also, loss of p53 in a mouse model showed increased Notch signaling, suggesting a negative interaction between Notch and p53 (23).
MicroRNAs (miRNAs) are single-stranded small RNAs that down-regulate the expression of target genes by either mRNA degradation or translational inhibition (25). The global effects of miRNAs on bone development have been studied by excision of Dicer in mice using Col1a1-Cre and Osteocalcin (Oc)-Cre. Conditional deletion of the Dicer in osteoprogenitors by Col1a1-Cre prevented their differentiation and compromised fetal survival at E15.5. In contrast, Oc-Cre excision of Dicer in mature osteoblasts delayed perinatal bone formation without compromising survival (26). In addition, several other microRNAs were reported to be involved in bone homeostasis. For example, osteoblast-specific gain of miR-206 inhibits osteogenesis in mice by targeting Cx43 (27); and miR-29b inhibits anti-osteogenic factors, such as HDAC4 and Tgfβ-3, to promote osteogensis by modulating the expression of bone extracellular matrix protein in vivo (28). Here, we focused on how Notch signaling is regulated by miRNAs during osteoblast differentiation.
In this study, we show that the expression of microRNA 34 (miR-34c) is distinctively induced in premyoblast C2C12 cells during osteogenic differentiation stimulated by BMP2. Also, osteoblast-specific gain of miR-34c mice show an age-related osteoporotic phenotype that is due to both a defect in osteoblast mineralization and an increase in osteoclastogenesis. Importantly, we identified that miR-34c can target multiple members of the Notch signaling pathway. Consistent with this, the osteoclastic phenotype in miR-34 transgenic mice is reminiscent of the consequences of loss of Notch function in osteoblasts. Together, these data demonstrate that miR-34c plays a critical role in bone homeostasis in part through modulating Notch signaling.
RESULTS
miRNA-34b and miRNA-34c are up-regulated and directly target Notch1 during BMP2-induced C2C12 osteoblast differentiation
To identify novel miRNA candidates involved in osteoblast differentiation, we performed a miRNA microarray using total RNA from C2C12 premyoblast mesenchymal cells treated for 72 h with BMP2 or vehicle (control). A similar approach has been successfully used in the identification of Osx, a key osteogenic transcription factor (29). After a comprehensive analysis of the microarray data, 34 miRNAs were differentially expressed between control and BMP2-treated groups (Fig. 1A). Consistent with other studies (27,30), myogenic miRNAs including miR-206, miR-1, miR-133b and miR-133a were significantly down-regulated, suggesting a proper osteoblast lineage commitment occurred after BMP2 treatment. Interestingly, we found that miR-34c and miR-34b-5p (miR-34b) were increased upon BMP2 treatment (Fig. 1B). Consistent with our observation, miR-34c expression was reported to be induced and maintained during osteoblast differentiation of MC3T3-E1, a preosteoblast cell line (31). Furthermore, miR-34c and miR-34b were also found up-regulated in terminally differentiated osteoblasts from bone marrow stromal cell (BMSC) under mineralizing conditions by quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) (Fig. 1B). Both miR-34b and miR-34c are well conserved across species and expressed as a common primary transcript (32,33). Along with miR-34a, miR-34b and -34c have been well characterized as tumor suppressive miRNAs, and their transcription is regulated by the direct recruitment of p53 to their promoters. They display tumor suppressive effects in various cancers, including osteosarcoma (34–36).
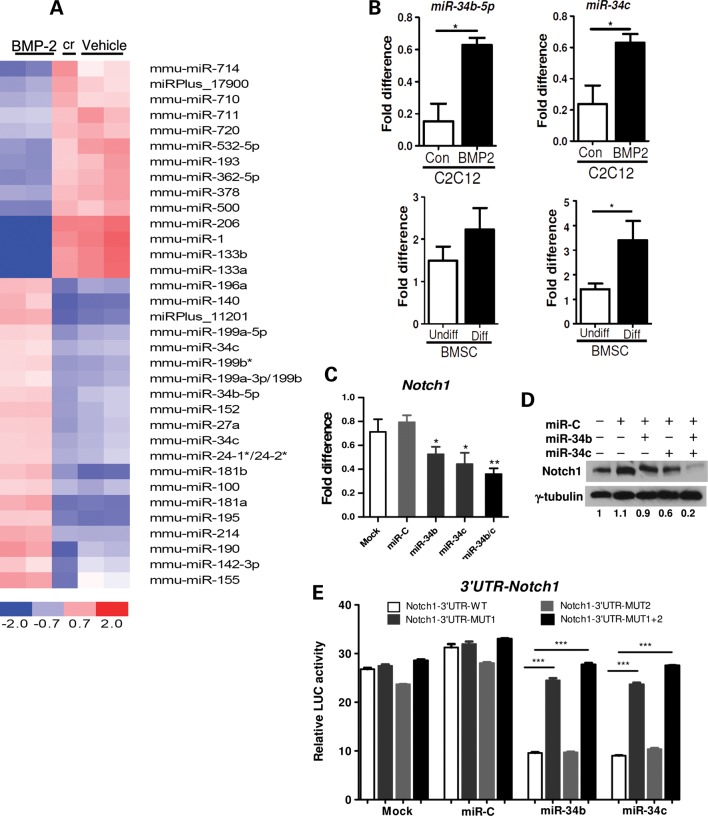
miR-34b and -34c are induced during BMP2-mediated C2C12 osteoblast differentiation and directly target 3′-UTR of Notch1. (A) miRNA microarray (Exiqon, miRCURY LNA array v.9.2) was performed with total RNA isolated from BMP2-treated and vehicle-treated C2C12 cells (n = 2 biological replicates). The heat map diagram shows the result of two-way hierarchial clustering of miRNAs and samples. The color scale shown at the bottom illustrates the relative expression level of miRNA across samples. Cr indicates a dyeswap. (B) Validation of the array data for miR-34b and -34c was performed with RNA used for the array using a Taqman miRNA assay, normalized to sno202 (n = 3). *P < 0.05 compared with C2C12 control (con). Also RNA from undifferentiated (Undiff) and differentiated (Diff) of BMSCs was analyzed by Taqman miRNA assay (n = 3). *P < 0.05 compared with Undiff-BMSC. (C) C2C12 cells were transfected with miR-34b, miR-34c, miR-C (control) or transfection reagent only (Mock). After 48 h, Notch1 transcription level was quantified by qRT–PCR (normalized to B2m) (n = 4).*P < 0.05, **P < 0.01 compared with miR-C. (D) Western blots for Notch1 and β-tubulin (control) were performed on total cell lysates. Numbers indicate the quantification of relative Notch1/β-tubulin band intensity ratios. (E) HeLa cells were co-transfected with the luciferase reporter containing either wild-type (WT) 3′-UTR or mutant (Mut1, Mut2 or Mut1 + 2) 3′-UTR Notch1 and miR-34b, -34c or miR-C (n = 3). ***P < 0.001. Three independent experiments were performed.
Using TargetScan and miRBase, we found that miR-34b and 34c target the 3′ untranslated region (UTR) of Notch1. Two putative target sites for miR-34b and 34c were found at position −187 to −193 and −800 to −806 in the 3′-UTR of Notch1 (Supplementary Material, Fig. S1A). Consistent with this putative targeting, we found that Notch1 expression decreased during osteogenesis of C2C12 cells, concomitantly with an increased miR-34b and 34c expression (data not shown). To test whether Notch1 is a target of miR-34b and 34c, the miR-34b, 34c or negative control mimics were transfected in C2C12 cells. The miR-34b or 34c mimic-transfected cells showed a significant decrease in Notch1 expression at both the transcript and protein levels (Fig. 1C and D, Supplementary Material, Fig. S1B and C). Moreover, the levels of Notch1 mRNA and protein were further reduced when miR-34b and 34c mimics were co-transfected (Fig.1C and D). Next, to examine whether miR-34b and -34c directly regulate Notch1 by binding to these two putative sites at the 3′-UTR of Notch1, the entire 3′-UTR region was cloned into the luciferase reporter vector psiCHECK2 and co-transfected with the miR-34b, -34c or negative control mimic in HeLa cells. The results showed that miR-34b and -34c could effectively suppress the luciferase reporter activity. Furthermore, the mutations in these two putative miR-34b/c-binding sites (referred to as Mut1 and Mut2) significantly impaired the binding of miR-34b/c, relieving the repression of luciferase activity. These data suggest that miR-34b and/or -34c directly bind to these two sites in the 3′-UTR of Notch1 and thereby regulate Notch1 (Fig. 1E). Interestingly, we also observed that a mutation at the proximal site (Mut1), which resides close to the AU-rich elements (AREs), was critical for the miR-34b- or miR-34c-mediated repression compared with the distal site (Mut2). The relief of repression was additive when the distal site was mutated along with the proximal site (Fig. 1E). This observation is consistent with a recent finding that neighboring cis-elements of miRNA-binding sites, such as AREs, can potentiate miRNA repression (37). Here, we demonstrated that Notch1 is a direct target of miR-34b or -34c during BMP2-mediated osteoblast differentiation of C2C12 cell.
Gain of miR-34c in osteoblasts manifests aged-related osteoporosis in vivo
To further understand how miR-34 regulates bone homeostasis by targeting Notch signaling, we generated transgenic (Tg) mice over-expressing miR-34c in osteoblasts using the 2.3 kb collagen type 1 (Col1a1) promoter (38). Two transgenic mice lines were established (Lines 1 and 2). The over-expression level of miR-34c in transgenic mice was determined by qRT–PCR (Supplementary Material, Fig. S2A). By microCT analysis, we found that the miR-34c Tg mice (Line 1) exhibited a low bone mass in both long bones (Fig. 2A and B) and vertebrae (Supplementary Material, Fig. S2C) at 6 months of age, but not at 8 weeks of age (Supplementary Material, Fig. S2B). Consistent with the reduced bone volume/total volume (BV/TV) of miR-34c Tg mice, other parameters such as trabecular thickness (Tb.Th) and trabecular number (Tb.N) were reduced, while trabecular separation (Tb.Sp) was increased in femurs and vertebrae (Fig. 2A and Supplementary Material, Fig. S2C). However, there was no difference in these parameters earlier at 8 weeks of age (data not shown). Unlike Line 1, Line 2 showed a significant reduction in BV/TV at 8 weeks of age likely due to a dosage effect of the miR-34c transgene (Supplementary Material, Fig. S2B). Overall, these gross microCT analyses suggest that the elevated expression of miR-34c in osteoblasts leads to an age-related osteoporotic phenotype in the transgenic mice. Furthermore, bone histomorphometric analysis in the vertebrae of miR-34c Tg mice showed a significant reduction in osteoblast numbers and surfaces (Fig. 2C). This observation was confirmed by a BrdU-labeling assay, in which primary osteoblasts from miR-34c Tg calvaria exhibited decreased BrdU incorporation rate, suggesting that osteoblast proliferation was also affected in miR-34c Tg mice (Supplementary Material, Fig. S3). According to calcein double-labeling analysis, miR-34c Tg mice exhibited significantly decreased bone formation rate (BFR/BS) and mineralizing surface (MS/BS), and trended towards a lower mineral apposition rate (MAR) (Fig. 2D). These results indicate that the overall decreased bone formation in miR-34c Tg mice is primarily due to decreased osteoblast surface, but not due to a defect in the new bone matrix formation from individual osteoblast. These data are consistent with the significantly decreased Ob.S/BS in miR-34c Tg mice (Fig. 2C). Therefore, the gain of miR-34c in osteoblasts decreases osteoblast numbers or proliferation and ultimately decreases new bone formation.
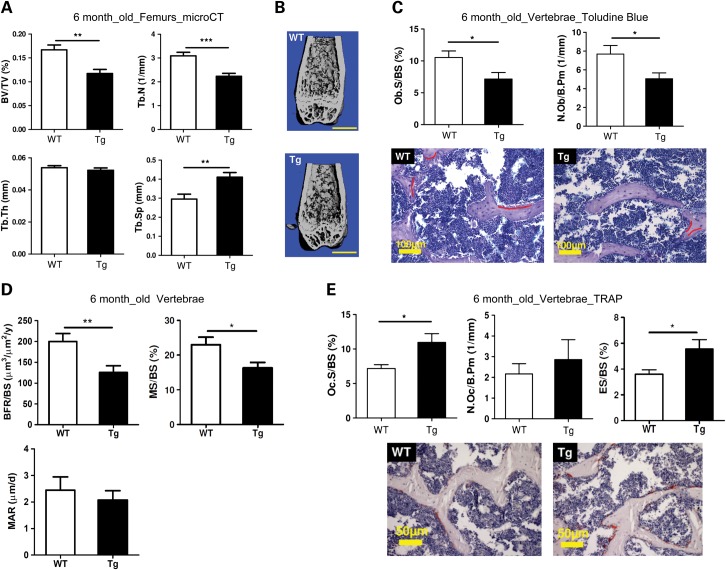
Osteoblast-specific miR-34c transgenic mice manifest osteoporosis. (A) Micro-CT analysis of 6-month-old miR-34c Tg mice show decreased BV/TV, Tb.N and Tb.Th and increased Tb.Sp compared with WT (n = 10–13 per group). **P < 0.01, ***P < 0.001. (B) Micro-CT reconstruction of distal femur from 6-month-old miR-34c Tg mice show decreased trabecular bone. Scale bar, 1 mm. (C) Histomorphometric analysis of the vertebrae of 6-month-old miR-34c Tg and WT mice (n = 8–12 per group) reveal decreased Ob.S/BS and N.Ob/B.Pm. *P < 0.05. Error bars are SD. Toludine blue staining of 6-month-old vertebrae section from miR-34c Tg and WT showed decreased osteoblast staining. Osteoblasts are underlined in red. Scale bar, 100 μm. (D) BFR/BS, MS/BS and MAR were decreased in miR-34c Tg. *P < 0.05, **P < 0.01. Error bars are SEM. (E) Oc.S/BS, N.Oc/B. Pm and ES/BS were increased in miR-34c Tg. *P < 0.05. Error bars are SD. TRAP staining (red) of 6-month-old vertebrae section from miR-34c Tg and WT, indicating increased osteoclast staining. Scale bar, 50 μm.
Along with decreased osteoblast numbers and surfaces, the osteoclast surfaces were significantly increased and osteoclast numbers were also increased, albeit not significantly, in the miR-34c Tg mice (Fig. 2E). Furthermore, a significantly increased eroded surface (ES/BS) was found in miR-34c Tg mice, suggesting an increased bone resorption (Fig. 2E). These data indicate that the gain of miR-34c in osteoblasts not only attenuates osteoblast differentiation but also promotes osteoclastogenesis in a non-cell-autonomous manner, thus leading to an imbalance of bone formation and resorption. Overall, these results show that miR-34c can regulate bone remodeling by affecting both osteoblasts and osteoclasts in vivo.
miR-34c inhibits osteoblast differentiation and increases osteoclastogenesis in vitro
To further dissect the mechanisms by which miR-34c regulates osteoblast differentiation as well as osteoblast–osteoclast (Ob–Oc) coupling, we performed in vitro mineralization assays using BMSCs, which were differentiated for 21 days in the mineralization media containing ascorbic acid and β-glycerophosphate. The expression of miR-34c was significantly higher in differentiated osteoblasts from BMSCs isolated from miR-34c Tg versus WT (Fig. 3A). Compared with WT, miR-34c Tg cells showed markedly reduced mineralization by both Alizarin red and Von Kossa staining (Fig. 3B and D). This finding is consistent with a recent study of ectopic expression of miR-34c in MC3T3-E1 cells, which also exhibited decreased mineralization (31).
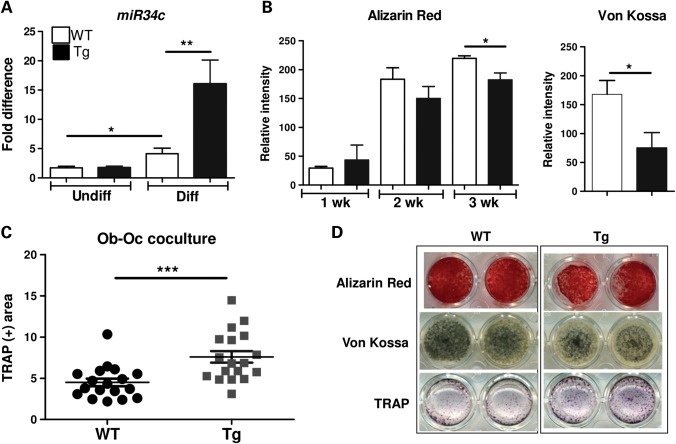
miR-34c inhibits osteoblast differentiation and increases osteoclastogenesis. (A) Induction of miR-34c was quantified by qRT–PCR after osteoblast differentiation from both WT and miR-34c Tg BMSC (n = 5 per group). *P < 0.05, **P < 0.01. (B) Alizarin Red and Von Kossa staining were performed weekly and quantified using ImageJ (n = 6 per group).*P < 0.05. (C) Co-cultures of WT or miR-34c Tg mesenchymal-derived osteoblasts and WT splenocytes were performed in the presence of 10−8 m of Vit.D3 and stained for TRAP-positive cells at day 10 (n = 18 per group). ***P < 0.001. (D) Representative stained image. Error bar is SEM.
Activated osteoblasts stimulate osteoclastogenesis by a coupling mechanism, which is critical for the maintenance of bone homeostasis. Osteoblasts induce osteoclast differentiation by expressing RANKL, a key osteoclastogenic factor. Conversely, the mature osteoblasts also produce osteoprotegrin (OPG), a RANKL antagonist, to inhibit osteoclastogenesis. To understand how gain of miR-34c in osteoblasts influences osteoclastogenesis, we performed osteoblast–osteoclast co-culture assays followed by tartrate-resistant acid phosphatase (TRAP) staining to quantify osteoclastogenesis. Compared with WT, miR-34c Tg osteoblasts induced greater numbers of osteoclasts from WT splenocytes (used as osteoclast precursor cells) (Fig. 3C and D). This result indicated that gain of miR-34c in osteoblasts induces osteoclast differentiation in a non-cell-autonomous fashion, i.e. through an osteoblast–osteoclast coupling mechanism. Interestingly, the dysregulation of bone homeostasis observed in miR-34c Tg mice partially phenocopies the loss of Notch function in osteoblasts, which resulted in an age-related osteoporosis due to increased osteoclast activity but not due to impaired osteoblast function (8). Hence, while the targeting of Notch explains the osteoclastic effect, it does not account for the osteoblastic effect as loss of Notch signaling in osteoblasts does not affect osteoblastic differentiation. The in vivo osteoblast phenotype in the transgenic mice suggests, perhaps not surprisingly, that miR-34c regulates osteoblast differentiation also in a cell-autonomous fashion via the targeting of components other than Notch signaling.
miR-34c targets the Notch signaling pathway during bone development in vivo
To identify the molecular mechanism of miR-34c-mediated regulation of osteoblast differentiation in an unbiased and global approach, we performed transcriptome profiling (Illumina) using total RNA isolated from P2 calvarial tissue of Tg or WT mice. After normalization of the microarray data, a list of down-regulated genes in miR-34c Tg calvaria were analyzed using DAVID Bioinformatics Resources (39). Consistent with our miR-34c targeting of Notch1, the Notch signaling pathway was significantly modulated (P-value = 4.1E−3) according to a KEGG pathway analysis. The Illumina microarray data were subsequently validated by qRT–PCR. Interestingly, Notch1 and Notch2 receptors were significantly reduced in the Tg calvarial tissue, while Notch3 and Notch4 levels were unchanged (Fig. 4A). Similar to Notch1, Notch2 also contains a well-conserved miR-34c-binding site in its 3′-UTR region (Supplementary Material, Fig. S4A) and has been shown to be targeted by miR-34a in glioblastoma (40). However, miR-34c-binding sites are not found in the 3′-UTR of Notch3 and Notch4. This again suggests that the 3′-UTR of Notch1 or Notch2 is critical and specific for the action of miR-34c in osteoblasts. In addition, we found that the Jag1, a ligand of the Notch1 and a target of miR-34a (41), was also significantly down-regulated (Fig. 4B). Interestingly, Jag1 also contains a miR-34c-binding site at its 3′-UTR (Supplementary Material, Fig. S4B). Consistent with a decreased Notch signaling, canonical Notch downstream targets, Hey1, Hes2 and Hes5, were also found to be decreased in our Illumina microarray data (Supplementary Material, Fig. S4C). The validation of targets based on microarray data suggests that, during bone homeostasis, multiple components of Notch signaling are regulated by miR-34c in a post-transcriptional manner.
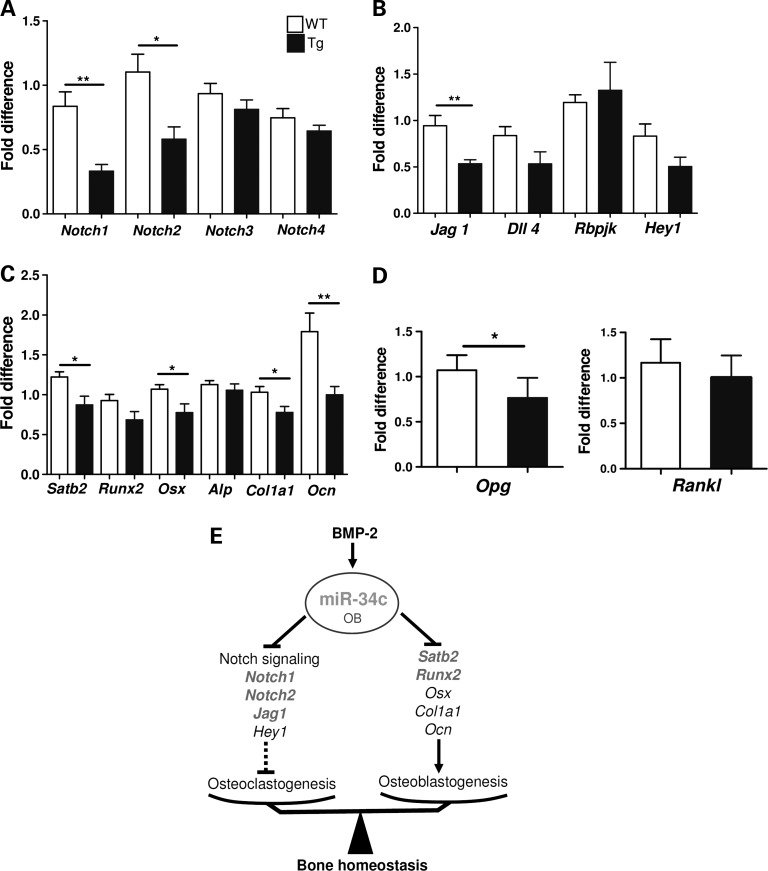
miR-34c targets the Notch signaling pathway during osteoblast development in vivo. Total RNA was extracted from P2 calvarial tissues of WT and miR-34c Tg mice. The expression level of Notch receptors (A), Notch ligands (Jag1 and Dll4), Rbpjk and Hey1 (B), osteoblast markers (C) and Opg and Rankl (D) were quantified by qRT–PCR and normalized to B2m (n = 6). *P < 0.05, **P < 0.01. Error bar is SEM. (E) Diagram of miR-34c-mediated regulation of bone development. MiR-34c regulates both osteoclasts and osteoblasts via targeting multiple factors in the Notch signaling pathway and Runx2 and Satb2, respectively, during skeletal development. The direct targets of miR-34c are in bold.
Consistent with the reduced osteoblast mineralization observed in miR-34c Tg mice, the qRT–PCR validation of microarray data also showed significant decreases in osteoblast differentiation markers, such as Osx, Col1a1 and Osteocalcin (Ocn) (Fig. 4C). Interestingly, Runx2, a master osteoblast transcription factor, which was recently reported as a miR-34c target in an in vitro study (31), was also reduced. Additionally, we found that the expression of Satb2, a nuclear matrix protein, was dramatically decreased. Furthermore, miR-34c can bind to the 3′-UTR of Satb2 according to TargetScan (Supplementary Material, Fig. S4D). SATB2 has been shown to facilitate osteoblast differentiation by scaffolding RUNX2 and ATF4 to the Ocn promoter and facilitating its transcription (42). Therefore, reduced expression of Satb2 and Runx2 in miR-34c Tg mice may contribute to dysregulation of Ocn transcription (Fig. 4C). These data suggest that miR-34c affects osteoblast differentiation through a Notch-independent pathway, likely through the direct targeting genes, including Satb2 and Runx2.
Both our histomorphometric analysis of osteoclasts and osteoblast–osteoclast co-culture assays suggest that gain of miR-34c in osteoblasts increased osteoclastogenesis. Because the balance of RANKL versus OPG production from osteoblasts is one of the most important determinants of osteoclastogensis, we analyzed their expression level in calvarial tissue of miR-34c Tg and WT mice. We found similar levels of Rankl mRNA between Tg and WT mice, but the expression of Opg mRNA was significantly reduced in the Tg mice (Fig. 4D). Hence, the increased Rankl/Opg ratio from the miR-34c transgenic osteoblasts is one potential mechanism responsible for the elevated osteoclastogenesis in the Tg mice. This non-cell-autonomous effect of osteoblasts on osteoclastogenesis is consistent with earlier studies of loss of Notch signaling in committed osteoblasts, which also decreased Opg levels to increase the Rankl/Opg ratio (8,43). Therefore, miR-34c suppresses Notch signaling in osteoblasts leading to increase osteoclastic differentiation via inhibition of osteoprotegerin expression.
In summary, we have identified miR-34c as a miRNA enriched during osteogenesis. Osteoblast-specific miR-34c Tg mice show an age-related osteoporotic phenotype, due to a reduced terminal differentiation of osteoblasts and increased bone resorption via elevated osteoclastogenesis. Therefore, miR-34c has a critical role in bone homeostasis by affecting both osteoblasts and osteoclasts in vivo. In the former, it may act by targeting Satb2 and Runx2, while in the latter, it acts by inhibiting multiple factors of the Notch pathway (Fig. 4E).
DISCUSSION
To date, the global roles of miRNAs in bone development have been investigated by excision of Dicer in mice using Col1a1-cre and Oc-cre (26) which inhibited osteoblast differentiation and delayed perinatal bone formation, respectively. These global deletion approaches suggest the importance of miRNAs in bone development in general; however, there are few mouse models that demonstrate either the physiological or pathological role of miRNAs in bone (27,28). Here, we have identified miR-34c as one of the enriched miRNAs during BMP2-mediated osteoblast differentiation of C2C12 cells. In our study, osteoblast-specific miR-34c Tg mice show an age-related osteoporotic phenotype, a result of reduced proliferation and terminal differentiation of osteoblasts and increased bone resorption due to increased osteoclastogenesis. The hyper-resorptive component of the miR-34c Tg mice phenotype phenocopies the loss of Notch in osteoblasts. In osteoblast-specific Notch mutants, age-related osteoporosis was found to be due to increased osteoclast activity (8). Our microarray data confirmed that miR-34c regulates Notch signaling in a post-transcriptional manner (Fig. 4E). Furthermore, we showed that the defect in the osteoblast differentiation of miR-34c Tg mice was due to the cell-autonomous effect by the predicted targeting of Satb2 and Runx2 rather than Notch signaling (Fig. 4E). Overall, this study revealed the role of miR-34c in bone homeostasis by affecting both osteoblasts and osteoclasts in vivo by regulating multiple targets in osteoblast. The temporal and spatial regulation of multiple targets by miRNAs is critical for maintaining homeostasis; however, the dysregulation of miRNAs expression can also contribute to cancer. Dysregulation of Notch signaling during bone development can lead to an increased proliferation of immature cells, thus leading to the increased formation of woven bone (8). Furthermore, we previously reported that Notch signaling components were significantly up-regulated in primary human osteosarcoma samples (23,44). Hence, it is not surprising that miR-34 expression has been reported to be decreased in primary osteosarcoma samples due to the genetic and epigenetic alteration of miR-34 genes (34). MiR-34c-mediated post-transcriptional regulation of Notch signaling in osteoblasts is one possible mechanism to modulate the proliferative effect of Notch in the committed osteoblast progenitors which may be important in the pathogenesis of osteosarcomas. Therefore, understanding the functional interaction of miR-34 and Notch signaling in normal bone development and in bone cancer could potentially lead to therapies modulating miR-34 signaling.
MATERIALS AND METHODS
Primary cell culture conditions, Luciferase reporter assay, western blot assay and other standard methods are described in detail in the Supplementary Material.
Microarray and data processing
C2C12 cells were maintained in the α-MEM (Hyclone) medium, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Osteogenic differentiation of C2C12 cells was induced by supplementing 300 ng/ml BMP2 (R&D system) in the growth medium. C2C12 cells were induced to osteoblast differentiation or maintained in the growth medium as described above for 72 h. Total RNA was extracted by TRIzol (Invitrogen), and further purified by mirVana miRNA isolation kit (Amibion). A total of 2 μg of RNA was used for microarrays. We used two replicates for both the control and the BMP2-treated C2C12 cells. The miRNA array (miRCURY LNA array version 9.2) was performed as a service provided by Exiqon service (Vedbaek, Denmark). The miRNA array results were normalized by Exiqon.
mRNA microarray assay was performed using Illumina whole-genome expression arrays (MouseWG-6 v2.0 Expression BeadChip). Three biological replicates were used for each WT and gain of miR-34c. Array data analysis was performed using the Lumi package within the R statistical package. A variance-stabilizing trans-formation was performed on the data set, and extracted expression values were quantile-normalized. Differentially expressed genes were determined using the t-test under the Limma package within R. The down-regulated gene list from miR-34c Tg mice was analyzed using DAVID Bioinformatics Resources. The raw data can be obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/). GEO accession number for miRNA microarray and mRNA microarray are GSE37036 and GSE36780, respectively.
qRT–PCR analysis
For qRT–PCR analysis, total RNA from calvaria of P2 mice was extracted with Trizol and cDNA was synthesized using Superscript III First Strand RT-PCR kit (Invitrogen), and the quantitative real-time PCR was performed on a LightCycler instrument (Roche). β2-microglobulin was used as the internal control to normalize gene expression. For miRNA qRT–PCR, TaqMan MicroRNA Assays (Applied Biosystems) were used to quantify the expression of mature miR-34b-5p (Assay ID: 002617) and miR-34c (Assay ID: 000428). Amplication and detection were performed using 7500HT Fast Real-Time PCR system (Applied Biosystems) and using the TaqMan® Universal PCR Master Mix, without AmpErase® UNG (Applied Biosystems). Gene expression was normalized with internal control sno202 RNA.
Transgenic mice
A 370 bp fragment of genomic DNA containing the miR-34c precursor was subcloned downstream of the 2.3 kb osteoblast-specific Col1a1 promoter in a vector containing the tyrosinase minigene and the woodchuck posttranscriptional regulatory element (WPRE) sequence. Transgenic founders were generated by pronuclear injections in FVB/N embryos. We identified the transgenic mice at birth by eye pigmentation and confirmed by PCR using primers specific for the WPRE sequence. These studies were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Skeletal analyses and bone histomorphometry
We performed toludine blue, Von Kossa and TRAP stains of 5–7 μm undecalcified lumber vertebrae plastic sections by using standard protocols. The static and dynamic histomorphometric analysis was performed by calcein injection (15 mg/kg) with an interval of 7 days after the first injection and data were analyzed on the OsteoMeasure histomorphometry system (Osteometrics, Decatur, GA, USA). We analyzed micro-computed tomography (μCT) scans of the trabecular bone of the lumber vertebrae and femurs by the μCT system (μCT-40, Scanco Medical, Wayne, PA, USA).
FUNDING
This work was supported by the National Institute of Health (AI053831 to Y.B., AR061565 to J.T., HD22657 to B.H.L., DE016990 to B.H.L.); and the Cancer Prevention Institute of Texas (RP101017 to B.H.L.).
ACKNOWLEDGMENTS
We thank members of the Brendan Lee laboratory for the critical review for this manuscript and helpful discussion; Micheal Starbuck (Bone Histomorphometry Core, MD Anderson Cancer Center) for technical supports and discussion for bone histomorphometry analysis.
REFERENCES
Articles from Human Molecular Genetics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/hmg/dds129
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/hmg/article-pdf/21/13/2991/17255093/dds129.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/hmg/dds129
Article citations
Molecular Signaling Pathways and MicroRNAs in Bone Remodeling: A Narrative Review.
Diseases, 12(10):252, 12 Oct 2024
Cited by: 0 articles | PMID: 39452495 | PMCID: PMC11507001
Review Free full text in Europe PMC
Integrative genetic and genomic networks identify microRNA associated with COPD and ILD.
Sci Rep, 13(1):13076, 11 Aug 2023
Cited by: 0 articles | PMID: 37567908 | PMCID: PMC10421936
Deletion of osteopontin in non-small cell lung cancer cells affects bone metabolism by regulating miR-34c/Notch1 axis: a clue to bone metastasis.
Eur J Histochem, 67(3), 26 Jul 2023
Cited by: 2 articles | PMID: 37491944 | PMCID: PMC10476534
Monocyte subpopulations display disease-specific miRNA signatures depending on the subform of Spondyloarthropathy.
Front Immunol, 14:1124894, 17 Apr 2023
Cited by: 1 article | PMID: 37138886 | PMCID: PMC10149963
Brain-derived extracellular vesicles promote bone-fat imbalance in Alzheimer's disease.
Int J Biol Sci, 19(8):2409-2427, 29 Apr 2023
Cited by: 10 articles | PMID: 37215980 | PMCID: PMC10197897
Go to all (143) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE37036
- (1 citation) GEO - GSE36780
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Overexpression of miR-34c inhibits high glucose-induced apoptosis in podocytes by targeting Notch signaling pathways.
Int J Clin Exp Pathol, 8(5):4525-4534, 01 May 2015
Cited by: 38 articles | PMID: 26191142 | PMCID: PMC4503014
MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells.
Stem Cells, 32(4):902-912, 01 Apr 2014
Cited by: 115 articles | PMID: 24307639
Maintenance of Bone Homeostasis by DLL1-Mediated Notch Signaling.
J Cell Physiol, 232(9):2569-2580, 31 Mar 2017
Cited by: 8 articles | PMID: 27735989 | PMCID: PMC5485010
Notch and the regulation of osteoclast differentiation and function.
Bone, 138:115474, 08 Jun 2020
Cited by: 34 articles | PMID: 32526405 | PMCID: PMC7423683
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: AI053831
NIAMS NIH HHS (2)
Grant ID: R03 AR061565
Grant ID: AR061565
NICHD NIH HHS (1)
Grant ID: HD22657
NIDCR NIH HHS (1)
Grant ID: DE016990





