Abstract
Free full text

Mechanism of foreign DNA selection in a bacterial adaptive immune system
Associated Data
Summary
In bacterial and archaeal CRISPR immune pathways, DNA sequences from invading bacteriophage or plasmids are integrated into CRISPR loci within the host genome, conferring immunity against subsequent infections. The ribonucleoprotein complex Cascade utilizes RNAs generated from these loci to target complementary “non-self” DNA sequences for destruction, while avoiding binding to “self” sequences within the CRISPR locus. Here we show that CasA, the largest protein subunit of Cascade, is required for non-self target recognition and binding. Combining a 2.3 Å crystal structure of CasA with cryo-EM structures of Cascade, we have identified a loop that is required for viral defense. This loop contacts a conserved 3-base pair motif that is required for non-self target selection. Our data suggest a model in which the CasA loop scans DNA for this short motif prior to target destabilization and binding, maximizing the efficiency of DNA surveillance by Cascade.
Introduction
Effective immunity against infection requires detection and neutralization of foreign transgressors while avoiding reactions to self. In CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) immune systems, RNA-guided complexes bind complementary regions in invasive nucleic acids, targeting them for destruction ((Barrangou et al., 2007; Brouns et al., 2008; Garneau et al., 2010; Marraffini and Sontheimer, 2008) and reviewed in (Deveau et al., 2010; Jore et al., 2011a; Karginov and Hannon, 2010; Terns and Terns, 2011; Wiedenheft et al., 2012)). CRISPR loci comprise repetitive elements separated by short “spacer” segments of foreign DNA acquired during previous infections (Fig. 1a) (Barrangou et al., 2007; Bolotin et al., 2005; Jansen et al., 2002; Mojica et al., 2005). Following transcription of the CRISPR locus, a CRISPR-specific endoribonuclease cleaves the transcript within the repeat sequence (Brouns et al., 2008; Carte et al., 2010; Carte et al., 2008; Deltcheva et al., 2011; Gesner et al., 2011; Haurwitz et al., 2010; Przybilski et al., 2011; Sashital et al., 2011). The resulting RNA (crRNA) combines with a number of CRISPR-associated (Cas) proteins to form a targeting complex (Brouns et al., 2008; Deltcheva et al., 2011; Hale et al., 2012; Hale et al., 2009; Lintner et al., 2011; Wiedenheft et al., 2011b; Zhang et al., 2012). In Type I CRISPR systems (Makarova et al., 2011), this targeting complex binds double-stranded (ds)DNA containing complementary “proto-spacer” sequences, preparing the target for its subsequent destruction by the effector nuclease Cas3 (Beloglazova et al., 2011; Brouns et al., 2008; Mulepati and Bailey, 2011; Sinkunas et al., 2011).
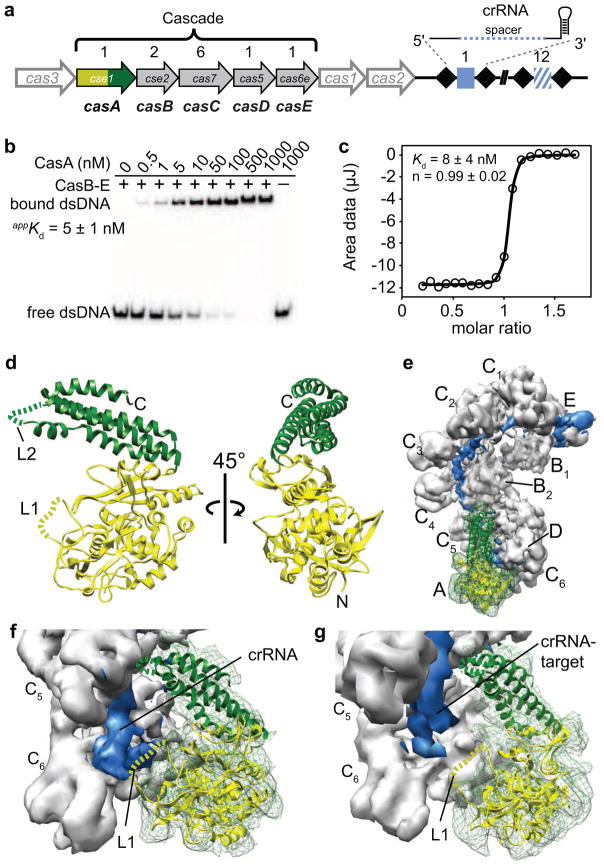
CasA is required for dsDNA binding. (a) Schematic of E. coli Cas/CRISPR locus. Cas genes A–E (gene family indicated within each arrow) encode the Cascade subunits, which are present in the stoichiometry indicated above each gene. The downstream CRISPR, comprising 12 unique spacers (blue squares) separated by repetitive sequences (black diamonds), is transcribed and processed into 61-nt crRNAs. (b) Electrophoretic mobility shift assay for dsDNA binding as CasA associates with Cascade. CasA was titrated from 0.1–1000 nM and the CasBCDE-crRNA complex (CasB-E) and O1—O2 dsDNA substrate (Fig. S1a) were held constant at 1000 nM and 20 pM, respectively. (c) Isothermal titration calorimetry data for WT CasA binding to the CasBCDE-crRNA subcomplex. Integrated data were fit to a single exponential function to determine the Kd. (d) Crystal structure of CasA from T. thermophilus at 2.3 Å resolution. N-terminal domain (green); C-terminal domain (yellow); disordered loops (dashed lines), including residues 130-143 (L1, yellow) and 401-410 (L2, green). (e) Crystal structure of CasA docked into 8 Å cryo-EM structure of E. coli Cascade. Cryo-EM density for the crRNA is shown in blue, and density for CasA is shown in green mesh throughout. (f) Close-up of CasA crystal structure docked into Cascade structure with no target bound. (g) Closeup of CasA crystal structure docked into Cascade structure with target bound.
In Escherichia coli, the protein subunits of the crRNA-guided targeting complex Cascade (CasA, CasB1-B2, CasC1-C6, CasD and CasE) assemble with a 61-nucleotide (nt) crRNA that comprises a 32-nt spacer sequence flanked by portions of the adjacent repeat sequences (Fig. 1a) (Brouns et al., 2008; Jore et al., 2011b). Base pairing between the crRNA spacer sequence and the complementary target strand of a virus or plasmid results in displacement of the non-target DNA strand (Jore et al., 2011b). Recognition and binding require target-crRNA complementarity at positions 1–5 and 7–8 of the 5′-end of the crRNA spacer sequence (Semenova et al., 2011). This short, high-affinity “seed” segment is thought to facilitate rapid genome scanning and detection of bona fide targets (Semenova et al., 2011; Wiedenheft et al., 2011b). Target binding must rely on crRNA—DNA base pairing and not specific protein-DNA interactions, as target sequences are highly variable. However, the mechanism by which a crRNA gains access to the complementary sequence in a dsDNA target remains unknown.
An important consideration in CRISPR immunity is how Cascade avoids binding to complementary sequences within the host CRISPR locus. A conserved proto-spacer adjacent motif (PAM), which is present in the target, but not the CRISPR locus, allows for distinction between foreign and host DNA through an unknown mechanism (Deveau et al., 2008; Gudbergsdottir et al., 2011; Marraffini and Sontheimer, 2010; Mojica et al., 2009; Semenova et al., 2011). In E. coli, the PAM is located upstream of the target sequence (positions -3 to -1), and exhibits stringent requirements for Cascade target binding and viral resistance (Semenova et al., 2011). The mechanism of PAM recognition likely occurs through protein-DNA interactions, as the PAM is not fully complementary to the crRNA. Structures of Cascade determined by single particle cryo-electron microscopy revealed that CasA is located at one end of the complex near the 5′ end of the crRNA, suggesting that it could contact target DNA near the PAM (Wiedenheft et al., 2011a).
Here we present the 2.3 Å crystal structure of CasA from Thermus thermophilus together with biochemical and in vivo experiments demonstrating its central role in non-self DNA target binding. When docked into cryo-EM reconstructions of Cascade, the CasA structure reveals a flexible loop that is adjacent to the crRNA and target. Mutations in this loop reveal a three amino acid motif that interacts with the PAM sequence and is required for viral resistance at the levels of both Cascade assembly and DNA target recognition. In addition, we present evidence that PAM recognition may be linked to dsDNA destabilization. Together, our data suggest a concerted mechanism for genome surveillance and detection of non-self DNA targets by Cascade.
Results and Discussion
CasA is required for non-self target binding
In addition to binding a DNA target complementary to the spacer sequence of the crRNA, Cascade also displays a non-specific DNA binding activity that has been proposed to promote genome surveillance by localizing the complex onto DNA (Jore et al., 2011b). Previously, it was shown that CasA is required for this non-specific binding activity (Jore et al., 2011b). However, because the importance of the PAM was unknown at the time, the effect of CasA on specific binding of non-self targets containing the correct PAM sequence was not investigated. To test this, we measured the binding affinity of Cascade to a 71-bp dsDNA target containing the correct PAM (5′-CAT-3′) and proto-spacer sequences (O1—O2 duplex, Fig. S1a) by adding increasing amounts of purified E. coli CasA to a constant concentration of the CasBCDE—crRNA subcomplex (Fig. 1b). The CasBCDE-crRNA subcomplex alone displays no dsDNA binding activity (Fig. 1b, first lane), while dsDNA binding increases over the course of the CasA titration, indicating that specific dsDNA binding depends on incorporation of CasA into the complex.
The apparent dissociation equilibrium constant (Kd) of DNA binding (5 ± 1 nM) as measured by CasA titration is similar to the affinity of the full Cascade complex for the same target (Fig. 1b and S1b). Additionally, the Kd of CasA binding to the CasBCDE-crRNA complex as measured directly by isothermal titration calorimetry (8 ± 4 nM, Fig. 1c) is indistinguishable from the Kd of Cascade binding dsDNA. Based on these observations, we hypothesized that CasA may dissociate from Cascade at low concentrations of the complex, resulting in an inflation of the apparent dissociation constant for dsDNA. To test this, we added 2 μM CasA to a titration of Cascade or the CasBCDE-crRNA subcomplex to ensure that CasA remains bound even at low concentrations of the complex. Addition of saturating amounts of CasA increases the affinity for a 71 bp dsDNA target from a Kd of 8 nM to 1.5 nM, indicating that CasA dissociation limits dsDNA binding (Fig. S1b).
The observation that Cascade binds dsDNA with higher affinity than the interaction between CasA and the CasBCDE-crRNA subcomplex suggests that cellular CasA concentrations could regulate Cascade targeting function. CasA may require higher expression levels than the rest of Cascade in order to ensure high affinity binding of invasive dsDNA. Consistent with this idea, previous studies have shown that CasA mRNA is present in relatively high abundance compared to other Cascade transcripts (Hernandez-Lucas et al., 2008; Shimada et al., 2009; Westra et al., 2010).
Structural insights into CasA function
To further understand the critical role that CasA plays in the targeting step of CRISPR immunity, we determined the structure of CasA from T. thermophilus at 2.3 Å resolution using X-ray crystallography (Fig. 1d, Table 1). The structure reveals two separate domains: an N-terminal domain (NTD, residues 1-364) that adopts a fold not found in the Protein Data Bank when searched using the Dali server (Holm and Rosenstrom, 2010); and a C-terminal four-helix bundle (CTD, residues 365-502). Two helices of the CTD bundle pack against the NTD, maintaining the relative orientation of the domains in the two molecules present in the asymmetric unit. Each domain contains a loop (L1, residues 130-143 and L2, residues 401-410) that was disordered in the crystal, indicating flexibility of these regions.
Table 1
Data collection and refinement statistics
| Native | SeMet | ||
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | |
| Cell dimension | |||
 a, b, c (Å) a, b, c (Å) | 94.36, 48.11, 129.78 | 94.54, 47.82, 129.26 | |
| α, β, γ (°) | 90.0, 97.0, 90.0 | 90.0, 97.3, 90.0 | |
| Peak | Remote | ||
|
| |||
| Wavelength (Å) | 1.11587 | 0.97968 | 0.95704 |
| Resolution (Å)* | 46.83–2.30 (2.36–2.30) | 46.89–2.90 (2.98–2.90) | 46.89–2.90 (2.98–2.90) |
| Rsym (%)* | 5.6 (45.6) | 11.5 (52.7) | 12.0 (55.1) |
| I/δI* | 18.9 (2.9) | 10.3 (2.7) | 10.0 (2.5) |
| Completeness (%)* | 99.6 (96.8) | 99.8 (100.0) | 99.8 (99.9) |
| Redundancy* | 3.9 (3.6) | 3.8 (3.8) | 3.8 (3.8) |
| Refinement | |||
| Resolution (Å) | 46.8–2.3 | ||
| No. reflections | 51896 | ||
| Rwork/Rfree | 0.196/0.247 | ||
| No. atoms | |||
 Protein Protein | 7323 | ||
 Water/ligands Water/ligands | 360 | ||
| B-factors | |||
 Protein Protein | 44.4 | ||
 Water/ligands Water/ligands | 53.4 | ||
| R.m.s deviations | |||
 Bond lengths (Å) Bond lengths (Å) | 0.009 | ||
 Bond angles (°) Bond angles (°) | 1.1 | ||
Outside the context of Cascade, purified CasA is incapable of binding to DNA (Fig. 1b) (Jore et al., 2011b); consistent with this observation, no prominent positively charged regions are present in the electrostatic surface map for CasA (Fig. S1c). To better understand the function of CasA in DNA binding by Cascade, we docked the high-resolution structure of CasA into cryo-EM reconstructions of Cascade from E. coli (Fig. 1e–g) (Wiedenheft et al., 2011a). The T. thermophilus and E. coli CasA sequences are ~50% similar, and both are predicted to have very similar secondary structures based on PSIPRED prediction (Jones, 1999). The crystal structure fits well into the EM density for CasA, indicating that the tertiary architecture of CasA is unaltered in Cascade (Fig. 1e). In the structure of Cascade with no target bound, CasA L1 appears to contact the 5′ end of the crRNA (Fig. 1f). The EM density in the region of L1 is contiguous with the density for the crRNA, suggesting that L1 becomes ordered upon interaction with the RNA.
Upon target binding, Cascade undergoes a concerted conformational change in which the CasA, CasB1-2 and CasE subunits shift and rotate along the CasC—crRNA backbone (Wiedenheft et al., 2011a). The shifted position of CasA in the target-bound structure of Cascade places L1 just below the 5′ end of the crRNA-target duplex (Fig. 1g). In contrast to the non-target bound EM density, no density is present for L1 in this reconstruction, suggesting that the loop no longer interacts with the crRNA and becomes flexible. We note that no PAM sequence is present in the target-bound Cascade reconstruction, but the trajectory of the crRNA—target duplex suggests that a longer target would come into close proximity of L1. Based on these observations, we hypothesized that L1 may contact the PAM sequence when a non-self target is bound.
CasA L1 is required for viral resistance
Our structural data indicate that CasA L1 could interact with both the crRNA and the PAM, suggesting that this sequence may be very important for Cascade function and viral resistance. We wished to probe the importance of the L1 sequence in a lambda phage (λ) challenge experiment using a previously described over-expression system that relies on arabinose for the tunable expression of cas genes under control of a T7 promoter. Previously, this system has been shown to confer >107-fold resistance against λ infection, even when the spacers targeted λ sequences with incorrect PAMs (Brouns et al., 2008). We hypothesized that overexpression of Cascade could allow the system to overcome the stringent PAM requirements, as high concentrations of Cascade can bind targets with incorrect PAM sequences (Brouns et al., 2008; Semenova et al., 2011). To avoid masking defects in viral resistance upon mutation of L1, we attenuated expression of the immune system by decreasing the amount of induction reagent by 10-fold (0.01 mM IPTG and 0.02% L-arabinose). Reduction of the amount of inducer weakened the immune system relative to previous reports (Brouns et al., 2008; Semenova et al., 2011) but permitted distinction between the WT immune system, mutation of CasA or the role of the PAM in CRISPR-mediated defense (Fig 2a). Although these results suggest that overexpression of the CRISPR system can overcome the requirement for a correct PAM sequence, the CRISPR-bearing plasmid seems to escape destruction under these conditions based on continued antibiotic resistance and immunity against phage. While we are unsure how the plasmid is preserved, it is possible that complete complementarity between the crRNA and target may inhibit Cascade binding or subsequent cleavage by Cas3.
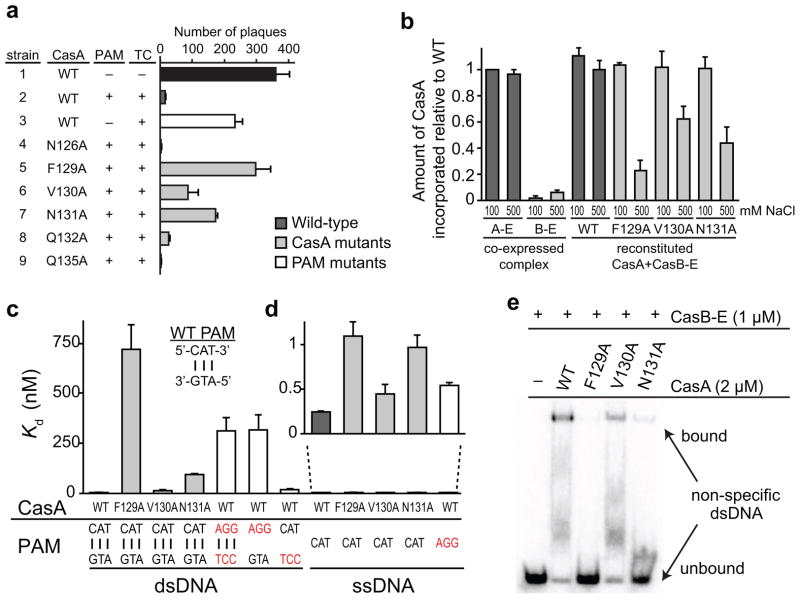
CasA L1 is important for Cascade assembly and target binding. (a) Phage sensitivity of E. coli strains expressing a CRISPR/Cas system with either WT CasA or the indicated CasA mutant. Cas genes were co-expressed with a CRISPR containing spacers that are not complementary to lambda phage, a CRISPR containing spacers complementary to the coding strand of lambda phage genes J,O, R, and E but no PAM; or a CRISPR containing spacers complementary to the coding strand of lambda phage genes J,O, R, and E, and including a PAM. The presence (+) or absence (−) of a PAM and/or a targeting CRISPR (TC) is indicated. See also Table S1. The number of plaques were counted and values from three replicates were averaged. Error bars represent one standard deviation. (b) Incorporation defects for CasA L1 mutants. The amount of CasA relative to CasC was determined from denaturing protein gel band intensities and normalized against relative amount of CasA in low salt Cascade immunoprecipitation. Aggregate data from three replicates are shown, with error bars representing one standard deviation. For WT, F129A, V130A and N131A CasA, reconstituted CasA + CasBCDE-crRNA (CasB-E) complex was formed and washed in the presence of low (100 mM) or high (500 mM) concentrations of NaCl. (c–e) Target binding defects for CasA L1 and PAM mutants (PAM mutations labeled in red). (c–d) Bar graph plotting Kd values for (c) dsDNA or (c–d) ssDNA targets. Average values from three replicates are shown, with error bars representing one standard deviation. Targets according to nomenclature in Figure S1a are as follows: Bars 1–4: O1—O2 duplex; Bar 5: O3—O4 duplex; Bar 6: O3—O2 duplex; Bar 7: O1—O4 duplex; Bars 8–11: O1; Bar 12: O3. (e) Gel shift assay for non-specific O5—O6 dsDNA (Fig. S1a) target binding by WT and mutant CasA + CasBCDE-crRNA (CasB-E).
Using this CRISPR/Cas expression system, we probed the importance of the L1 sequence by mutating several similar amino acids present in both the T. thermophilus and E. coli L1 CasA sequences and tested the effect of these mutations on resistance to λ infection. For cells expressing a fully functional CRISPR immune system, we observed a ~95% reduction in the sensitivity to phage infection compared to a strain containing a non-targeting CRISPR (Fig. 2a, S2a–c and Table S1). In contrast, strains containing CasA mutations in a three amino acid sequence of L1 (Phe129-Asn131) were far more sensitive to phage. In particular, cells expressing CasA with mutations at either F129A or N131A showed a ~80% and ~45% increases in phage sensitivity relative to wild-type (WT), respectively. In comparison, a strain expressing crRNAs that target λ phage at sites containing incorrect PAM sequences showed similar levels of phage sensitivity as observed with CasA F129A and N131A mutations (~60% more sensitive than WT), indicating that these residues are as important for viral resistance as the PAM itself. Because our structural data suggested that CasA L1 might interact with both the target and the crRNA (Fig. 1f–g), we hypothesized that the observed loss of viral resistance may be caused by defects in Cascade assembly, target binding, or both.
CasA L1 is important for incorporation into Cascade
To determine whether the three amino acid motif in L1 is important for CasA incorporation into Cascade, we incubated WT or mutant CasA with the CasBCDE-crRNA subcomplex and performed pull-down assays using an affinity tag on the CasB subunits (Fig. 2b and S2d). In comparison to co-expressed Cascade, all four CasA constructs were incorporated at the same levels when the complex was formed at low salt concentrations (100 mM NaCl). By contrast, the amount of incorporated mutant CasA decreased by 40–80% in the presence of 500 mM NaCl, while incorporation of WT CasA was unaffected. These data indicate that residues in L1 (Phe129-Asn131) are important, but not essential, for CasA binding to the CasBCDE-crRNA subcomplex.
Importantly, the CasA mutants remain stably associated with the complex at low salt concentrations, and the stoichiometry and size exclusion chromatography elution profiles are indistinguishable from purifications of the complex that include the WT CasA protein (Fig. S2e–f). We predict that mutations in L1 may disrupt interactions with the crRNA, but that contacts with other protein subunits of Cascade are sufficient for incorporation of CasA into the complex. At cellular protein concentrations, it is possible that the instability of the interaction between L1 mutant CasA and the CasBCDE-crRNA subcomplex could be at least partially responsible for the observed loss of viral resistance.
CasA L1 is important for dsDNA binding
We next asked whether CasA L1 plays a role in target binding by measuring the affinities of Cascade containing WT or mutant CasA for the 71-bp dsDNA with correct PAM and proto-spacer sequences (O1—O2 duplex, Fig. S1a). DNA binding experiments were performed using excess CasA (2 μM) in a low salt buffer to ensure that the WT and mutant CasA subunits were all stably associated with the CasBCDE-crRNA complex. While the V130A mutation has little effect on DNA binding, mutation of Phe129 or Asn131 to alanine causes substantial defects in dsDNA binding (Fig. 2c and Table S2), with ~500- and ~60-fold increases in Kd for F129A and N131A, respectively. In contrast, we observe only a 2–4-fold increase in Kd for ssDNA binding (O1, Fig. S1a) for all three mutants (Fig. 2c,d and Table S2). These data indicate that the L1 sequence is far more important for binding dsDNA than ssDNA. This difference suggests that CasA L1 may destabilize dsDNA targets, allowing the crRNA to access the seed and nucleate crRNA—target duplex formation.
An additional factor that could affect the affinity of Cascade for dsDNA is the ability of the complex to localize on DNA by non-specific binding prior to crRNA base pairing to a target sequence. Indeed, both F129A and N131A CasA mutations substantially reduce the amount of binding observed for a 71-bp dsDNA target containing a non-complementary crRNA binding sequence (O5—O6 duplex, Fig. S1a), indicating that L1 is the source of the previously observed CasA-dependent non-specific binding activity of Cascade (Fig. 2e) (Jore et al., 2011b).
Notably, similar to the CasA L1 sequence, we observe a difference in the importance of the PAM sequence for double-stranded versus single-stranded target binding (Fig. 2c–d and Table S2). Mutations in the PAM cause only a ~2-fold defect in binding to ssDNA targets (O3, Fig. S1a), while binding to dsDNA with an incorrect PAM (O3—O4 duplex, Fig. S1a) is significantly impaired. The observed binding of this target likely occurs mainly through non-specific interactions, as binding is reduced substantially in the presence of a non-specific competitor DNA (Fig. S2h). Importantly, mutation of the PAM sequence on only the target strand (O3—O2 duplex, Fig. S1a) has a similar effect as mutation of both strands, while mutation of the PAM sequence on the non-target strand (O1—O4 duplex, Fig. S1a) causes only a ~15-fold decrease in binding affinity (Fig. 2c and Table S2). These data suggest that only the target strand of the PAM is actively recognized by Cascade, while the non-target strand may indirectly promote PAM recognition by positioning the target strand through base pairing interactions.
Mapping CasA L1 position on dsDNA target
The relative importance of both CasA L1 and the PAM sequence for double-stranded target binding supports our structure-based hypothesis that L1 is involved in PAM recognition. Alternatively, mutations in L1 may affect the ability of Cascade to properly undergo conformational changes upon target binding (Wiedenheft et al., 2011a), resulting in the observed dsDNA binding defects. To test these two possibilities, we performed hydroxyl radical cleavage on the 71-bp dsDNA target (O1—O2 duplex, Fig. S1a) bound by Cascade containing WT, F129A or N131A CasA (Fig. 3). Overall, the cleavage patterns for all three Cascade variants are very similar, indicating that mutations in L1 do not significantly alter the overall architecture of Cascade. Together with the observation that L1 mutation does not significantly affect ssDNA target binding, it is likely that the concerted conformational change observed upon target binding still occurs when L1 is mutated.
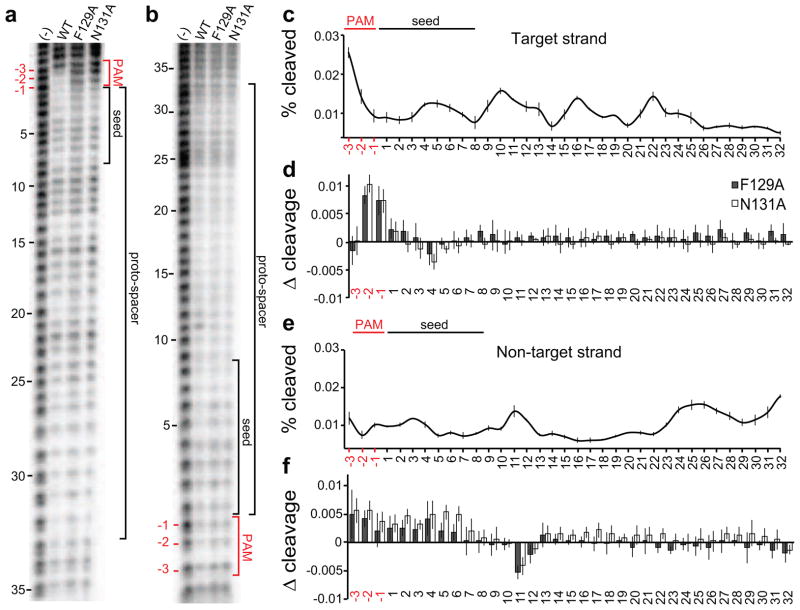
CasA L1 interacts directly with the PAM. (a,b) Representative sequencing gels for hydroxyl radical cleavage of O1—O2 dsDNA target in the absence (−) or presence of Cascade with either WT CasA, F129A or N131A. (a) Target (O1) strand or (b) non-target (O2) strand was 5′ end labeled with 32P and annealed to cold complement (Fig. S1a). (c) Percent cleaved at each position of the PAM and proto-spacer on the target strand for WT CasA + CasBCDE-crRNA bound to dsDNA. For each band, the peak intensity from three replicates were averaged and plotted versus sequence position, with error bars representing standard deviations. (d) Difference plot of percent of the target strand cleaved for mutant versus WT CasA + CasBCDE-crRNA bound to dsDNA. Positive values indicate regions of decreased protection upon mutation of L1. Aggregate data from three replicates are plotted, with error bars representing propagated standard deviations for average mutant and WT peak intensity. (e) Percent cleaved at each position of the PAM and proto-spacer on the non-target strand for WT CasA + CasBCDE-crRNA bound to dsDNA, plotted as in (c). (f) Difference plot of percent of the non-target strand cleaved for mutant versus WT CasA + CasBCDE-crRNA bound to dsDNA, plotted as in (d).
On the target strand (Fig. 3a,c, O1 Fig. S1a), a periodic 6-nt oscillation between protection and cleavage marks the gaps between the CasC2-C6 subunits, while CasC1 and C2 appear to fully protect the target from cleavage (Fig. S3). Strikingly, the only major difference in protection between WT and mutant Cascade occurs in the PAM region, where the −2 and −1 positions of the target are protected by Cascade containing WT CasA but not by Cascade containing F129A or N131A CasA (Fig. 3a,d). The loss of protection at such a localized region of the target strongly suggest that these differences are due to a loss of interaction between L1 and the PAM sequence, and that the position of CasA is likely not significantly altered outside of the L1 region.
Footprinting of the non-target strand (O2, Fig. S1a) reveals two main regions of protection, positions 5–8 and 13–22 (Fig. 3e), which may correspond to interactions with the CasA CTD and the two CasB subunits. The CasB subunits have been previously proposed to stabilize the displaced strand in the R-loop, consistent with this observed protection pattern (Wiedenheft et al., 2011a). Similar to the target strand, we observe reduced protection in the PAM and seed region of the non-target strand when F129A or N131A CasA is bound, although the effect is not as pronounced as on the target strand (Fig. 3f). In addition, position 11 is less protected in the presence of mutant CasA. These data suggest that the loss of interaction between L1 and the target strand PAM may result in a rearrangement of interactions between CasA and the displaced strand.
Model for CasA-dependent dsDNA target selection
Collectively, our data suggest that the CasA L1 motif is important for incorporation of CasA into Cascade, non-specific interactions with DNA, and specific double-stranded target binding, likely through interactions with the PAM. The apparent dual role of L1 in both non-specific DNA binding activity of Cascade and in contacting the PAM suggests that the two functionalities may be linked. In addition, the far greater importance of both the L1 and PAM sequences for dsDNA versus ssDNA binding suggests that PAM binding may destabilize the dsDNA helix, facilitating duplex unwinding by crRNA-seed base pairing. We hypothesize that Cascade may search cellular DNA for potential target sites by initially scanning for PAM sequences through CasA L1-mediated non-specific contacts (Fig. 4a). Once the complex encounters a PAM sequence, L1 may locally destabilize the dsDNA helix, enabling the crRNA to access the adjacent target sequence (Fig. 4b). Intriguingly, an aromatic residue is present at nearly identical locations in highly divergent CasA L1 sequences (Fig. S4), consistent with the superlative importance of Phe129 within the L1 motif. This aromatic residue may participate in DNA intercalation and helical distortion, similar to mechanisms observed in the Type II restriction endonuclease HinP1I and the DNA repair enzyme MutM. Both enzymes insert Phe residues into the DNA helix, causing substantial distortions of the B-form helix (Banerjee et al., 2006; Horton et al., 2006). In Cascade, similar helical distortions could lead to destabilization of the duplex, triggering seed nucleation (Fig. 4b).
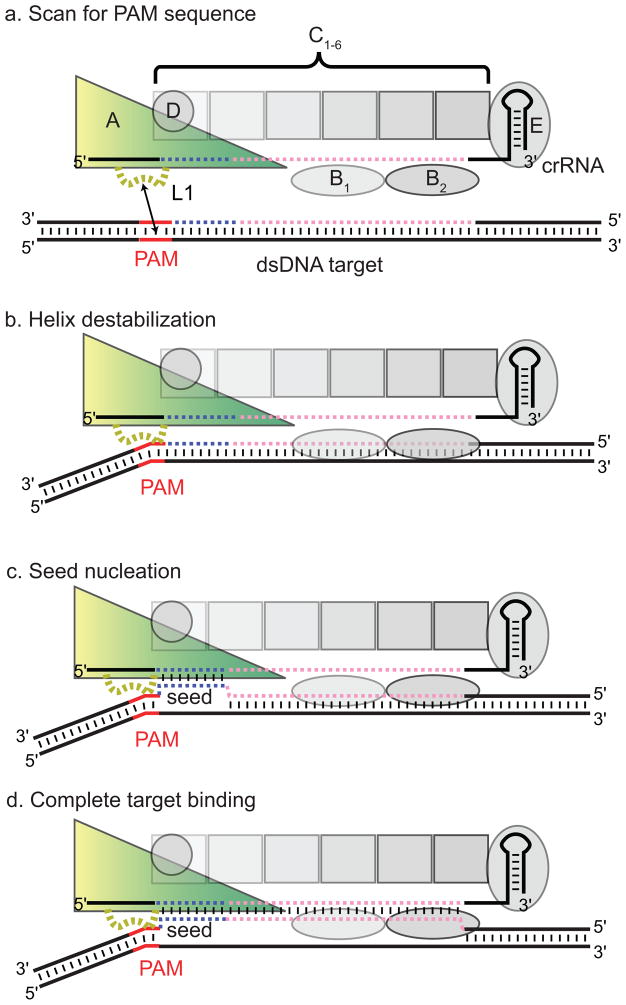
Model for CasA L1-dependent non-self target recognition by Cascade. (a) CasA L1-dependent non-specific binding localizes Cascade on target DNA. (b) Upon encountering a PAM sequence, CasA L1 binding destabilizes the duplex. (c) In the presence of a proto-spacer, L1-mediated PAM destabilization facilities strand invasion by the seed sequence. (d) Upon duplex nucleation at the seed, full spacer—proto-spacer binding can be completed. CasA is shown as a yellow-green triangle, CasB1-2 as horizontal ellipses, CasC1-6 as squares, CasD as a circle, and CasE as a vertical ellipse. The PAM is shown in red, the seed in blue, and the rest of the crRNA and proto-spacer in pink.
Because the tri-nucleotide PAM sequence will be abundant in any target genome, PAM scanning must occur transiently in the absence of an adjacent seed sequence. A similar strategy is required for MutM, which must search for rare occurrences of 8-oxoguanine (oxoG) lesions without invasively interrogating every undamaged guanine (Banerjee et al., 2006). In this case, helix intercalation by the probe residue distorts each interrogated base pair, but bases are only fully extruded in the presence of an oxoG. When Cascade encounters a bona fide target, helical distortion at the PAM would be stabilized by base pairing between the seed and crRNA, pausing Cascade at this site and initiating nucleation of the full crRNA—target duplex (Fig. 4c) and complete non-target strand displacement (Fig. 4d). This mechanism would allow Cascade to probe for a complementary seed match, while also facilitating efficient by-pass of PAM sequences that do not have an adjacent target sequence.
CasA sequences are not conserved in all CRISPR systems where PAMs have been identified. In particular, CasA homologs are not part of the CRISPR-associated targeting complexes isolated from Sulfolobus solfataricus and Pseudomonas aeruginosa (Lintner et al., 2011; Wiedenheft et al., 2011b). This observation suggests that functional orthologs of CasA may exist in other systems, or that some of these systems may employ alternative mechanisms for PAM recognition. In the future, it will be interesting to explore the importance of PAM sequences for target binding in other CRISPR systems, and to compare the strategies of various crRNA-protein complexes for non-self target selection.
Experimental Procedures
Construct cloning and preparation
CasA genes from T. thermophilus and E. coli K12 were cloned into the expression vector pSV272, encoding an N-terminal His6-MBP-tag followed by a tobacco etch virus (TEV) protease recognition site (Table S1). CasBCDE-crRNA with or without CasA were co-expressed using previously described expression vectors (Jore et al., 2011b; Wiedenheft et al., 2011b) (Table S1). Mutations were introduced using site-directed mutagenesis and confirmed by DNA sequencing. All constructs were expressed in BL21(DE3) cells, grown to 0.5 OD600, and induced overnight at 16°C with 1mM isopropyl β-D-1 thiogalactopyranoside (IPTG). Selenomethionine-incorporated CasA was expressed using methionine pathway inhibition (Doublie and Carter, 1992). Cells were harvested and lysed by sonication.
His6-MBP-tagged CasA was initially purified using Ni-NTA affinity resin (Qiagen), then incubated with TEV protease at room temperature for 4 hours (T. thermophilus) or at 4°C overnight (E. coli). T. thermophilus CasA was incubated at 65°C for 10 minutes to precipitate most of the released His6-MBP and TEV protease, then centrifuged to remove the precipitate. The protein was then concentrated to ~1 mL, followed by a second 65°C incubation for 10 minutes to remove the remaining contaminating proteins. The protein was finally purified by size exclusion chromatography using a Superdex 75 16/60 column (GE Life Sciences) in a buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 5% glycerol, and 1 mM TCEP. E. coli CasA was dialyzed against a buffer containing 50 mM HEPES, pH 7.0, 50 mM NaCl, 5% glycerol, and 1 mM TCEP during TEV protease cleavage, then bound to a 5 mL HiTrap HP SP (GE Healthcare Life Sciences) ion exchange column equilibrated with dialysis buffer. The column was then washed with 8% elution buffer (50 mM HEPES, pH 7.0, 1 M NaCl, 5% glycerol, and 1 mM TCEP) to remove contaminants, followed by a gradient from 8–30% elution buffer over 25 mL to elute CasA. The eluent was concentrated to ~1 mL, then finally purified by size exclusion chromatograph using a Superdex 200 16/60 column (GE Life Sciences) in a buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 5% glycerol, and 1 mM TCEP.
Cascade and the CasBCDE-crRNA subcomplex were initially purified using Strep-Tactin Superflow Plus resin (Qiagen) using an N-terminal Strep-II tag on the CasB subunit as previously described (Wiedenheft et al., 2011a). The complex was then concentrated to ~1 mL and purified by size exclusion chromatograph using a Superose 6 10/30 column (GE Life Sciences) in a buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 5% glycerol, and 1 mM TCEP.
Crystallization and structure determination of CasA
Native CasA crystals were grown in 2.1 M sodium acetate and 0.1 M MES pH 6.5, and SeMet CasA crystals were grown at 2.25M sodium acetate and 0.1 M Tris, pH 8.0 at 18°C using hanging drop vapor diffusion by mixing 15 mg/mL CasA and reservoir solutions at 2:1 ratio. Native crystals diffracting to 2.3 Å grew after ~1 month, while selenomethionine crystals diffracting to 2.9 Å grew after ~3 months. For data collection, crystals were flash frozen in liquid nitrogen using a cryoprotectant containing 2.8 M sodium acetate and 30% glycerol.
Diffraction data were collected at 100 K on beamline 8.3.1 of the Advanced Light Source (Lawrence Berkeley National Laboratory). Two-wavelength multiple anomalous dispersion (MAD) data were collected using SeMet crystals at wavelengths of 0.97938 Å and 0.95704 Å. All datasets were processed using XDS(Kabsch, 2010). Eight selenium sites (four from each of two molecules in the asymmetric unit) were located using the Hybrid Substructure Search (HySS) module of the Phenix package(Adams et al., 2010). Substructure refinement, phasing and density modification were performed using autoSHARP (Vonrhein et al., 2007). The resulting electron density map displayed clear secondary structural elements, allowing for manual model building of ~60% of the protein. This model was completed by iterative rounds of refinement against the native dataset in Phenix.refine, followed by manual building in COOT (Emsley and Cowtan, 2004). The final model has a Rfree of 24.7% and Rwork of 19.6%, with 97% of torsion angles falling in the Ramachandran favored region. Each molecule in the asymmetric has one Ramachandran outlier (Arg184), but the electron density for this amino acid and the surrounding region is well defined, indicating that the geometry in this region is correct. The rmsd between the two molecules in the asymmetric unit is 0.28 Å. Rigid-body docking of crystal structures into segmented cryo-EM densities was performed using the “Fit in Map” module in UCSF CHIMERA (Goddard et al., 2007). Cross-correlation values of 0.53 and 0.55 were calculated between the atomic model of CasA and the apo and target-bound versions of Cascade, respectively. All structure figures were also generated using UCSF CHIMERA.
Viral resistance
Host sensitivity to Lambda (λvir) phage was performed using E. coli BL21-AI (Invitrogen) cells, essentially as described previously (Brouns et al., 2008), with the exceptions noted below. All strains (Table S1) were grown in 2YT-Lambda (2YTL) media (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, 10 mM MgSO4, 0.2 % maltose) until the optical density (OD600nm) reached 0.3. Cas protein and pre-crRNA production was induced for 45 min by adding L-Arabinose (Sigma-Aldrich) and IPTG to a final concentration of 0.02% and 0.01 mM, respectively. Following induction, the cells were pelleted by centrifugation and resuspended in 10 mM MgSO4. Plaque assays were performed in triplicate according to standard procedures (Sambrook et al., 1989), using a synthetic CRISPR locus containing four spacers that do not match the Lambda phage genome (pWUR477) (Brouns et al., 2008), that target proto-spacers located within essential genes (genes J, O, R and E) but that are not adjacent to a PAM sequence (pWUR478) (Brouns et al., 2008), or that target proto-spacers located within genes J, O, R and E that are adjacent to PAM sequence (Fig. S2a–c, Table S1). The new CRISPR construct was cloned into pACYCDuet-1 vector (Novagen) between NcoI and XhoI restriction sites. Error-bars represent one standard deviation.
Isothermal titration calorimetry
ITC experiments were measured on a nanoITC (TA Instruments) containing a 190 μL cell. All samples were purified by size exclusion chromatography using identical buffer (20 mM Tris, pH 7.5, 100 mM KCl, 1 mM TCEP, 5% glycerol), then concentrated to the indicated concentration. 10 μM CasA was titrated into the cell containing 80 μM CasBCDE-crRNA complex with 25–30 1.5 μL injections at 22°C. Concentrations of CasA and CasBCDE-crRNA complex were determined by UV absorbance (CasA ε280: 59,525 M−1cm−1; CasBCDE-crRNA complex ε280: 654,620 M−1cm−1). Data were baseline corrected, integrated and fit using NanoAnalyze software (TA Instruments). Titrations were performed in triplicate. Reported values are average of three replicates with errors bars reflecting one standard deviation.
Cascade pull-down assays
500 nM CasA and 250 nM CasBCDE-crRNA complex were incubated in 20 mM Tris, pH 8.0 and either 100 mM or 500 mM NaCl at 37°C for 10 minutes, then incubated with 15 μL Strep-Tactin Superflow Plus resin (Qiagen) for 15 minutes at 4°C. The resin was washed two times with either low or high salt buffer, then 40 μL of SDS loading dye was added to the resin. Samples were boiled and resolved by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue dye and scanned using a Typhoon scanner, and densitometry was performed using ImageQuant software (GE Life Sciences). Pull-downs were performed in triplicate, and the average relative amount of CasA versus CasC is reported with error bars representing one standard deviation.
DNA binding assays
DNA oligomers were ordered from Integrated DNA Technologies (Fig. S1a). Duplexes were formed by mixing 40 μM of each strand in 40 mM Tris pH 8.0, 38 mM MgCl2, and 1 mM Spermidine, heating at 95°C for 2 minutes, and cooling at RT for 10 minutes. All DNAs were purified on native gels containing 6% 29:1 polyacrylamide and 1X TBE. DNA bands were visualized by UV light, excised, and eluted by soaking gel pieces in deionized H2O. Gel pieces were removed, and DNA was ethanol precipitated and resuspended in deionized H2O. DNA samples were 5′ labeled with γ-[32P]-ATP using polynucleotide kinase (PNK, New England Biolabs) for 30 minutes at 37°C. PNK was heat denatured at 65°C for 20 minutes, and excess ATP was removed using a G-25 spin column (GE Healthcare).
All binding assays were performed in buffer containing 25 mM Tris, pH 8.0, 130 mM KCl, 1 mM TCEP and 5% glycerol. CasA and CasBCDE-crRNA complex were pre-incubated for 10 minutes at 37°C to ensure complex formation prior to addition of DNA. CasA or CasBCDE-crRNA complex were titrated with constant final concentrations of 1 μM CasBCDE-crRNA complex (Fig. 1b) or 2 μM CasA (Fig. 2c,d, S1b and S2h), respectively. DNA was added to a final concentration of 5–20 pM, and samples were incubated at 37°C for 30 minutes, then resolved at 4°C on a 6% 29:1 polyacrylamide gel containing 1X TBE. Gels were dried and DNA was visualized by phosphorimaging and quantified using ImageQuant software (GE Life Sciences). The fraction of DNA bound (amount of bound DNA divided by sum of free and bound DNA) was plotted versus concentration of titrated species (CasA or CasBCDE-crRNA complex) and fit to a binding isotherm using Kaleidagraph software. Reported Kd values are the average of three replicates, and errors are the standard deviations between the three values.
Hydroxyl radical cleavage assays
~10 nM 5′-end 32P-labeled target or non-target DNA was annealed with 50 nM unlabeled complement in 5 mM Tris, pH 7.5 and 50 mM KCl at 95°C for 3 minutes, then cooled at room temperature for 10 minutes. Cascade containing WT, F129A or N131A CasA was formed by incubating 5 μM CasA with 2.5 μM CasBCDE-crRNA complex in the annealing buffer at 37°C for 10 minutes. 10 μL of annealed DNA was added to 40 μL of Cascade and incubated at 37°C for 20 minutes. An aliquot of each complex was run on a 6% native polyacrylamide gel to ensure complex formation. Hydroxyl radicals were formed by pipetting 1 uL each of Fe(II)-EDTA (100 mM ammonium iron(II) sulfate, 200 mM EDTA, pH 8.0), 500 mM sodium ascorbate, and 0.3% hydrogen peroxide onto separate spots on the side of a 1.5 mL tube containing 10 μL of Cascade—DNA complex. Negative controls contained 3 μL deionized H2O instead of hydroxyl-radical-generating reagents. All components were mixed by brief centrifugation then incubated at 37°C for 2 minutes. Radicals were quenched by addition of 1 μL 100 mM thiourea. Samples were phenol:chloroform extracted, ethanol precipitated, resuspended in 5 μL formamide loading dye and resolved on a 10% polyacrylamide sequencing gel containing 8 M urea and 1X TBE. Gels were dried, and DNA was visualized by phosphorimaging. Intensities were measured by drawing lines through each lane and plotting counts versus pixels using ImageQuant software. Intensities were normalized for variations in the amount of DNA loaded by calculating the overall fraction cleaved (pixel intensity divided by sum of all pixel intensities). Peak intensities were determined for each band and plotted versus nucleotide position. All experiments were performed in triplicate, and peak intensities were averaged with error bars representing one standard deviation.
Acknowledgments
We thank Kaihong Zhou for purification of Cascade and CasBCDE-crRNA complexes; Stan J. J. Brouns and John van der Oost for providing Cascade expression and viral resistance plasmids; Syed Habib Tahir Bukhari for help designing CRISPRs; members of the Doudna lab for critical reading of the manuscript; J. Holton and G. Meigs (Beamline 8.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory) for assistance with X-ray data collection; and C. Quinn, M. Golinar and J. Berger for use of the NanoITC. This project was funded by a National Science Foundation grant to J.A.D.. D.G.S. is supported by the Damon Runyon Cancer Research Foundation. B.W. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. J.A.D. is a Howard Hughes Medical Institute investigator.
Footnotes
Author contributions: D.G.S. determined the CasA structure, performed CasA incorporation, DNA binding and hydroxyl radical footprinting assays. D.G.S. and B.W. performed crystal structure-EM density modeling, viral resistance assays and ITC experiments. D.G.S., B.W. and J.A.D. designed experiments and wrote the manuscript.
Accession Number
Coordinates and structure factors for CasA have been deposited to the Protein Data Bank under accession code 4AN8.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. [Europe PMC free article] [Abstract] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. [Abstract] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. [Abstract] [Google Scholar]
- Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. [Europe PMC free article] [Abstract] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. [Abstract] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. [Abstract] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. [Europe PMC free article] [Abstract] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. [Europe PMC free article] [Abstract] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. [Europe PMC free article] [Abstract] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. [Europe PMC free article] [Abstract] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. [Abstract] [Google Scholar]
- Doublie S, Carter CWJ. Preparation of selenomethionyl protein crystals. In: Ducruix A, Giegg R, editors. Crystallization of Nucleic Acids and Proteins A Practical Approach. Oxford University Press; 1992. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. [Abstract] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. [Abstract] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. [Abstract] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. [Abstract] [Google Scholar]
- Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, et al. Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Mol Cell 2012 [Europe PMC free article] [Abstract] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. [Europe PMC free article] [Abstract] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. [Europe PMC free article] [Abstract] [Google Scholar]
- Hernandez-Lucas I, Gallego-Hernandez AL, Encarnacion S, Fernandez-Mora M, Martinez-Batallar AG, Salgado H, Oropeza R, Calva E. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J Bacteriol. 2008;190:1658–1670. [Europe PMC free article] [Abstract] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. [Europe PMC free article] [Abstract] [Google Scholar]
- Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. [Europe PMC free article] [Abstract] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. [Abstract] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. [Abstract] [Google Scholar]
- Jore MM, Brouns SJ, van der Oost J. RNA in Defense: CRISPRs Protect Prokaryotes against Mobile Genetic Elements. Cold Spring Harb Perspect Biol 2011a [Europe PMC free article] [Abstract] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011b;18:529–536. [Abstract] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. [Europe PMC free article] [Abstract] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal CASCADE complex for CRISPR-mediated viral defense. J Biol Chem 2011 [Europe PMC free article] [Abstract] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. [Europe PMC free article] [Abstract] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. [Europe PMC free article] [Abstract] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. [Europe PMC free article] [Abstract] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. [Abstract] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. [Abstract] [Google Scholar]
- Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–31903. [Europe PMC free article] [Abstract] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8 [Abstract] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. [Abstract] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. [Europe PMC free article] [Abstract] [Google Scholar]
- Shimada T, Yamamoto K, Ishihama A. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J Bacteriol. 2009;191:4562–4571. [Europe PMC free article] [Abstract] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. [Europe PMC free article] [Abstract] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. [Europe PMC free article] [Abstract] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. [Abstract] [Google Scholar]
- Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–1393. [Abstract] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–489. [Europe PMC free article] [Abstract] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. [Abstract] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011b;108:10092–10097. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Mol Cell. 2012. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2012.03.020
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276512002274/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
CRISPR-Cas12a exhibits metal-dependent specificity switching.
Nucleic Acids Res, 52(16):9343-9359, 01 Sep 2024
Cited by: 2 articles | PMID: 39019776 | PMCID: PMC11381342
Unity among the diverse RNA-guided CRISPR-Cas interference mechanisms.
J Biol Chem, 300(6):107295, 18 Apr 2024
Cited by: 1 article | PMID: 38641067 | PMCID: PMC11127173
Review Free full text in Europe PMC
Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol Cell, 84(3):463-475.e5, 18 Jan 2024
Cited by: 6 articles | PMID: 38242128
Cas4/1 dual nuclease activities enable prespacer maturation and directional integration in a type I-G CRISPR-Cas system.
J Biol Chem, 299(9):105178, 20 Aug 2023
Cited by: 0 articles | PMID: 37607619 | PMCID: PMC10504553
The biology and type I/III hybrid nature of type I-D CRISPR-Cas systems.
Biochem J, 480(7):471-488, 01 Apr 2023
Cited by: 1 article | PMID: 37052300 | PMCID: PMC10212523
Review Free full text in Europe PMC
Go to all (159) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(1 citation)
PDBe - 4AN8View structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural basis for CRISPR RNA-guided DNA recognition by Cascade.
Nat Struct Mol Biol, 18(5):529-536, 03 Apr 2011
Cited by: 356 articles | PMID: 21460843
CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference.
Proc Natl Acad Sci U S A, 111(18):6618-6623, 18 Apr 2014
Cited by: 133 articles | PMID: 24748111 | PMCID: PMC4020112
Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding.
J Biol Chem, 287(27):22445-22449, 23 May 2012
Cited by: 23 articles | PMID: 22621933 | PMCID: PMC3391111
Cascade-mediated binding and bending of negatively supercoiled DNA.
RNA Biol, 9(9):1134-1138, 01 Sep 2012
Cited by: 25 articles | PMID: 22954644 | PMCID: PMC3579878
Review Free full text in Europe PMC
Funding
Funders who supported this work.





