Abstract
Free full text

Phosphoregulation of Spc105 by Mps1 and PP1 Regulates Bub1 Localization to Kinetochores
Summary
Kinetochores are the macromolecular complexes that interact with microtubules to mediate chromosome segregation [1]. Accurate segregation requires that kinetochores make bioriented attachments to microtubules from opposite poles. Attachments between kinetochores and microtubules are monitored by the spindle checkpoint, a surveillance system that prevents anaphase until every pair of chromosomes makes proper bioriented attachments [2]. Checkpoint activity is correlated with the recruitment of checkpoint proteins to the kinetochore [1]. Mps1 is a conserved protein kinase that regulates segregation and the spindle checkpoint, but few of the targets that mediate its functions have been identified. Here, we show that Mps1 is the major kinase activity that co-purifies with budding yeast kinetochore particles and identify the conserved Spc105/KNL-1/Blinkin kinetochore protein as a substrate. Phosphorylation of conserved MELT motifs within Spc105 recruits the Bub1 protein to kinetochores and this is reversed by protein phosphatase I (PP1). Spc105 mutants lacking Mps1 phosphorylation sites are defective in the spindle checkpoint and exhibit growth defects. Together, these data identify Spc105 as a key target of the Mps1 kinase and show that the opposing activities of Mps1 and PP1 regulate the kinetochore localization of the Bub1 protein.
Results and Discussion
The conserved Mps1, Bub1 and Aurora protein kinases regulate chromosome segregation and the spindle checkpoint, but few of the corresponding substrates that mediate these functions have been identified [3–6]. To elucidate the mechanism by which these kinases regulate these processes, we sought to identify novel kinetochore targets. We previously developed a method to purify native budding yeast core kinetochore particles and found that the Mps1 and Bub1 kinases co-purify [7]. Although the kinases cannot be visualized by silver staining of kinetochore particles, incubation of particles with radiolabeled-ATP resulted in the strong phosphorylation of several species (Figure 1A). To identify the kinase(s) responsible, we utilized temperature sensitive mutations in the mitotic kinases Cdk1 (cdc28-13), Aurora B (ipl1-321), and Mps1 (mps1-1), as well as a tetracycline-repressible Bub1 protein (Ubi-R-TetR-BUB1). Inactivation of any individual kinase did not alter the bulk composition of purified kinetochore particles as determined by silver-stained SDS-PAGE (Figure 1A). However, inactivation of Mps1 but not the other candidate kinases resulted in a loss of phosphorylation, indicating that Mps1 is the major kinase activity that co-purifies with native kinetochore complexes and that it does not require the activity of other mitotic kinases tested in the context of this assay (Figures 1A and S1). The effect does not appear to arise from major alterations in kinetochore particle composition in mps1-1 mutants because immunoblotting of kinetochore proteins that are not detectable by silver staining revealed normal levels of the subcomplexes analyzed (Figure 1B).

(A) WT (SBY8253), cdc28-13 (SBY8716), ipl1-321 (SBY8712) Ubi-R-TetR-BUB1 (SBY8920) and mps1-1 (SBY8726) strains were shifted to conditions to inactivate the kinases and kinetochores were purified by α-Flag immunoprecipitation of Dsn1-His-Flag. The kinetochores were treated with γ-32P-ATP and eluted with 3×-Flag peptide. Particle composition was analyzed by silver-stained SDS-PAGE (left) and radioactive incorporation was analyzed by autoradiography (right). (B) Wild type and mps1-1 kinetochore complexes prepared as in (A) were analyzed by immunoblotting with the indicated antibodies. The relative amount of sample loaded in each lane is indicated at top.
Mps1 activity is required for the localization of the Bub1 protein to kinetochores in other organisms [8–15], so we tested whether Mps1 also regulates its localization in budding yeast. WT and analog sensitive mps1-as1 mutant cells containing Bub1-3GFP were arrested in metaphase by depletion of the APC activator Cdc20, and were then treated with the microtubule-destabilizing drug nocodazole and the mps1-as1 inhibitor 1NM-PP1 [16]. While Bub1-3GFP co-localized with the core kinetochore protein Mtw1-mCherry in approximately 40% of wild type cells, this was reduced to 10% in the mps1-as1 cells treated with inhibitor (Figure 2A). Bub1 interacts with the Bub3 checkpoint protein and requires its function for kinetochore localization [17–23], and we found that Bub3 also requires Mps1 activity to localize to kinetochores (Figure S2A). We next tested whether Mps1 is required for Bub1 to associate with kinetochore particles in vitro. Bub1 co-purifies with native kinetochore particles but not with mps1-1 particles in the presence of phosphatase inhibitors (Figure 2B). Furthermore, Bub1 was released when native particles were treated with recombinant PP1 in vitro, suggesting that phosphorylation is directly required for Bub1 binding to kinetochores (Figure 2C). Phosphatase inhibitors preserved the interaction (Figure 2C). Importantly, PP1 treatment did not alter the core composition of the kinetochore particles or the associated Mps1 kinase activity (Figures S2B and S2C). To test whether phosphorylation of kinetochores by Mps1 could restore Bub1 binding to kinetochores, we incubated the phosphatase-treated kinetochore particles with ATP to specifically phosphorylate Mps1 sites. When the ATP-treated particles were incubated with cellular lysate, we observed robust Bub1 rebinding to the particles (Figure 2D). The effect requires Mps1-mediated phosphorylation because phosphatase-treated particles treated without ATP, as well as mps1-1 mutant control kinetochores treated with ATP, failed to recruit Bub1 (Figure 2D). These experiments also demonstrate that rebinding is not due to exchange of bead-bound kinetochore particles with those present in the lysate. Spc105 is phosphorylated by the Ipl1/Aurora kinase [24–26], but phosphorylation of kinetochores by Ipl1/Aurora did not promote Bub1 rebinding (Figure 2D). The Bub3 protein behaved similarly in the same assays (Figure S2D–F).
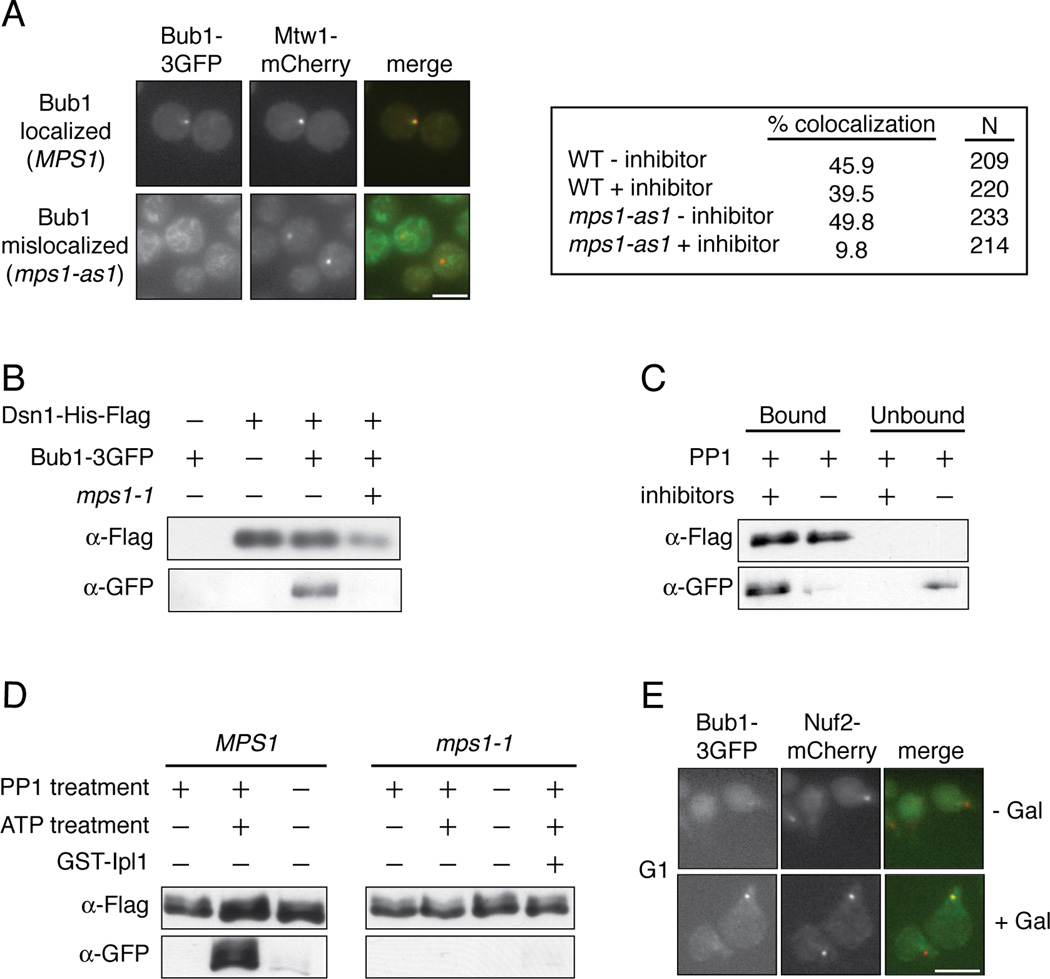
(A) pMet-Cdc20 BUB1-3GFP MTW1-mCherry cells containing MPS1 (SBY10318) or mps1-as1 (SBY10282) were arrested in mitosis by Cdc20 depletion and treated with nocodazole. They were then incubated with (+ inhibitor) or without (− inhibitor) 1NM-PP1 and analyzed for co-localization between Bub1 and Mtw1. Scale bar = 5.0 microns. (B) Kinetochore particles from strains containing Bub1-3GFP and MPS1 (SBY8502) or mps1-1 (SBY9348) were purified and analyzed by immunoblotting. (C) Kinetochore particles were purified from a strain containing Bub1-3GFP (SBY8502). The particles were treated with PP1 in the presence or absence of phosphatase inhibitors and the bound and unbound pools were analyzed by immunoblotting. (D) Kinetochore particles were purified from strains containing Bub1-3GFP and MPS1 (SBY8502) or mps1-1 (SBY9348). The particles were treated with PP1 and then incubated with +/− ATP. They were then incubated in a lysate from a WT strain (SBY8502) and analyzed by immunoblotting. (E) BUB1-3GFP NUF2-mCherry pGAL-MPS1 cells (SBY10271) were arrested in G1 and then treated with or without galactose. The co-localization of Bub1 with Nuf2 was analyzed by microscopy. Scale bar = 5.0 microns.
Because Mps1 phosphorylation is required for Bub1 to associate with kinetochores in vitro, we tested whether Mps1 activity is sufficient to recruit Bub1 to kinetochores in vivo. Bub1-3GFP foci co-localize with approximately 3% of kinetochores when cells are arrested in G1 (n=200, Figure 2E). However, Bub1 can be visualized on 22% of the kinetochore foci when Mps1 is overexpressed (n=206, Figure 2E), suggesting that Mps1 activity promotes Bub1 localization to kinetochores independently of upstream mitotic events. Similar results were previously demonstrated in fission yeast [15], as well as for Bub3 localization in budding yeast upon Mps1 overexpression [27]. Together, these data show that Mps1-mediated phosphorylation of kinetochores is required to promote the localization of the Bub1 protein and its interacting partner, Bub3.
We next wanted to identify the Mps1 substrate that mediates Bub1 localization to kinetochores. Bub1 interacts with the KNL-1/Blinkin protein in other organisms [8, 10, 19, 28, 29], making it a likely target. Consistent with this possibility, the molecular weight of the slowest migrating species phosphorylated by Mps1 on kinetochore particles corresponds to that of Spc105, the budding yeast homolog of KNL-1/Blinkin [30, 31](Figure 1A). This species migrated even slower when kinetochore particles with Spc105-3GFP were used in kinase assays, identifying it as Spc105 (Figure 3A). Similar experiments determined that the other two major substrates are Ndc80 and Dsn1 (data not shown). To ensure that Mps1 directly mediates this phosphorylation, we purified particles from an mps1-1 mutant strain and performed a kinase assay with recombinant GST-Mps1 (Figure 3A). Although the overall level of phosphorylation was lower with recombinant kinase, the pattern was identical to that of WT particles. Together, these data show that Mps1 is the major mitotic kinase that co-purifies with kinetochore particles and that it directly phosphorylates Spc105 and other kinetochore targets in vitro.
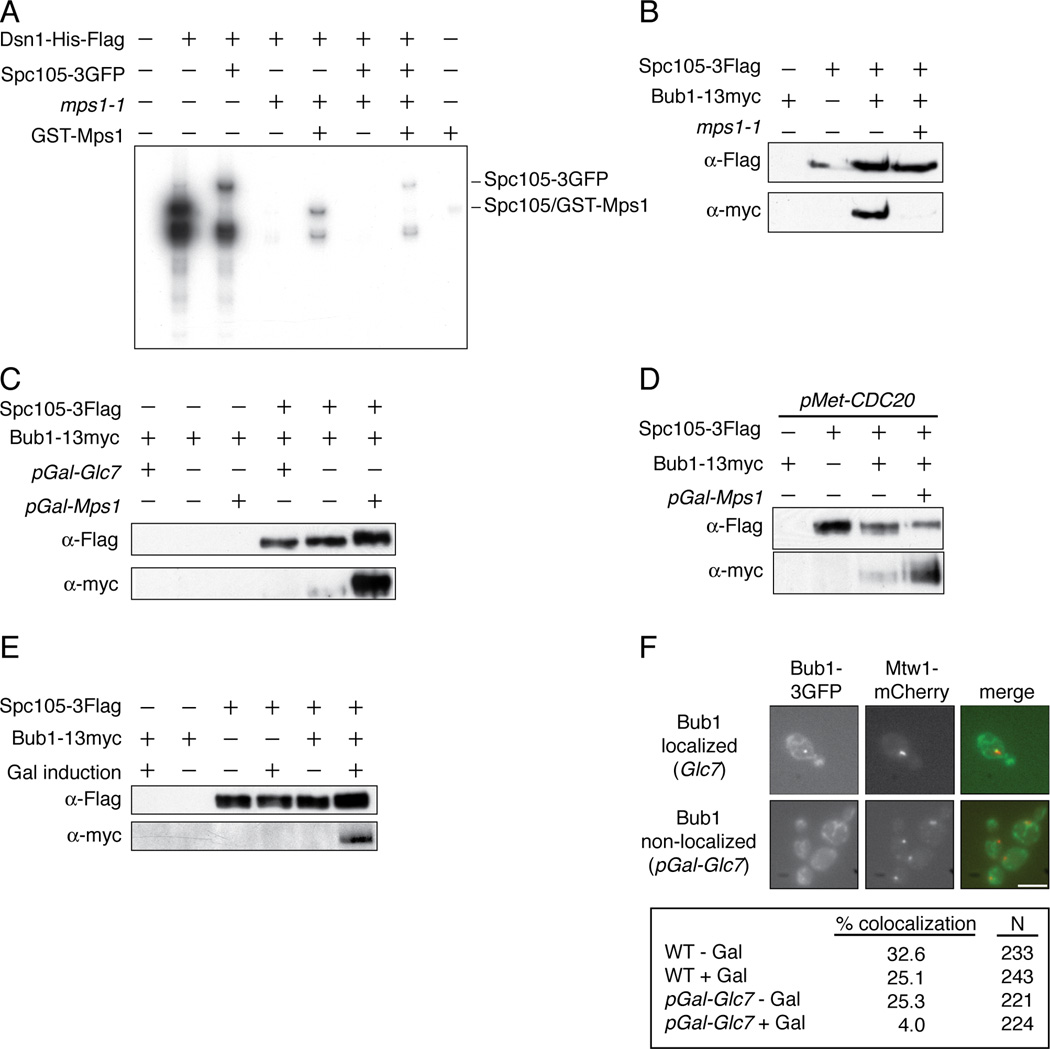
(A) Kinetochore particles from WT (SBY8253), SPC105-3GFP (SBY8750), mps1-1 (SBY8726), and SPC105-3GFP mps1-1 (SBY9099) strains were treated as in Fig 1A, as was a control purification from untagged strain SBY3. Recombinant GST-Mps1 was incubated with the mps1-1 samples. (B) Spc105-3Flag was immunoprecipitated from BUB1-13myc control cells (SBY3269) grown at 23 °C and from SPC105-3Flag (SBY7420), SPC105-3Flag BUB1-13myc (SBY9226) and SPC105-3Flag BUB1-13myc mps1-1 (SBY9476) cells shifted to 37 °C for 2 hours before harvest. Samples were analyzed by immunoblotting. (C) Spc105-3Flag was immunoprecipitated from WT (SBY9226), pGAL-MPS1 (SBY10100), and pGAL-GLC7 (SBY9971) cells containing BUB1-13myc and analyzed by immunoblotting. Control strains containing BUB1-13myc but lacking Spc105-3Flag are SBY4020 (pGAL-GLC7), SBY3269 (WT) and SBY4587 (pGAL-Mps1). (D) pMET-CDC20 cells containing Spc105-Flag and/or Bub1-13myc (SBY10464, SBY10364, SBY10465 and SBY10466) were arrested in metaphase and then galactose was added to induce Mps1 overexpression and cells were processed as in (C). (E) Spc105-3Flag was purified from cells arrested in G1 and treated with galactose for one hour to induce Mps1 overexpression. The strains used were BUB1-13myc (SBY3269), BUB1-13myc pGAL-MPS1 (SBY4587), SPC105-3Flag (SBY7492), SPC105-3Flag pGal-MPS1 (SBY9952), SPC105-3Flag BUB1-13myc (SBY9226), SPC105-3FLAG BUB1-13myc pGAL-MPS1 (SBY10100). (F) GLC7 (SBY9337) and pGAL-GLC7 (SBY10479) cells containing BUB1-3GFP and MTW1-mCherry were induced with or without galactose for 1 hour. The percentage of cells with Bub1-3GFP foci localizing to the kinetochore was quantified by microscopy. Scale bar = 5.0 microns.
We next tested whether Bub1 interacts with Spc105 in an Mps1-dependent manner. Bub1 co-purified with Spc105 that was immunoprecipitated from wild-type cells, and the interaction was abolished in mps1-1 mutant cells (Figure 3B). Consistent with phosphorylation being required for their association, Mps1 overexpression enhanced the co-purification of Bub1 with Spc105, whereas the overexpression of the budding yeast PP1 catalytic subunit, Glc7, inhibited it (Figure 3C). Mps1 overexpression also enhanced the interaction between Bub1 and Spc105 in metaphase-arrested and G1-arrested cells (Figures 3D and 3E). The phosphorylation state of Spc105 in vivo correlated with Mps1 activity because Mps1 overexpression reduced the relative mobility of Spc105 (Figures 3C and 3D). PP1 is known to interact with Spc105 [24, 26], so we tested whether it dephosphorylates Spc105. We found that Glc7 overexpression increased Spc105 mobility in vivo (Figure 3C) and PP1 treatment of kinetochore particles increased the migration of Spc105 in vitro (data not shown). Together, these data demonstrate that Spc105 mobility changes are due to changes in phosphorylation and confirm that there is a pool of phosphorylated Spc105 within kinetochore particles. Consistent with PP1 activity opposing Mps1 on kinetochores, Glc7 overexpression reduced Bub1 localization to kinetochores in vivo (Figure 3F) and reduced Bub1 binding to purified kinetochore particles (data not shown).
To identify potential Mps1 phosphorylation sites on Spc105 that might regulate its interaction with Bub1, we treated kinetochore particles with ATP and then analyzed Spc105 by mass spectrometry. We identified 17 sites that are phosphorylated (Table S1). Because an N-terminal region of Spc105 homologs has been reported to interact with Bub1 [8, 19, 28, 29], we focused on sites proximal to the conserved microtubule-binding N-terminus. Strikingly, we detected phosphorylation on three of six conserved threonines that match an expanded MELT-like motif [M/I][E/D][I/L/M][S/T] ([10, 19, 23, 28, 31, 32] and Figure 4A and Table S1). To test whether Mps1 phosphorylates these sites, we mutated the T residues to A in all six motifs to generate the spc105-6A allele. Kinetochore particles were then purified from cells containing WT Spc105 or the mutant Spc105-6A protein and incubated with radioactive ATP. There was a 54% reduction in phosphorylation on Spc105-6A relative to WT Spc105, indicating that Mps1 directly phosphorylates these sites on kinetochore particles (Figure 4B). We next tested whether Bub1 binding to Spc105 was altered by the mutation of the Mps1 phosphorylation sites. WT Spc105 co-purified with Bub1 whereas the Spc105-6A mutant did not (Figure 4C). Like Bub1, Bub3 also required Mps1 activity to interact with Spc105 (Figure S3A). Neither Bub1 nor Bub3 co-purified with kinetochore particles containing Spc105-6A (Figure S4D and S3B).
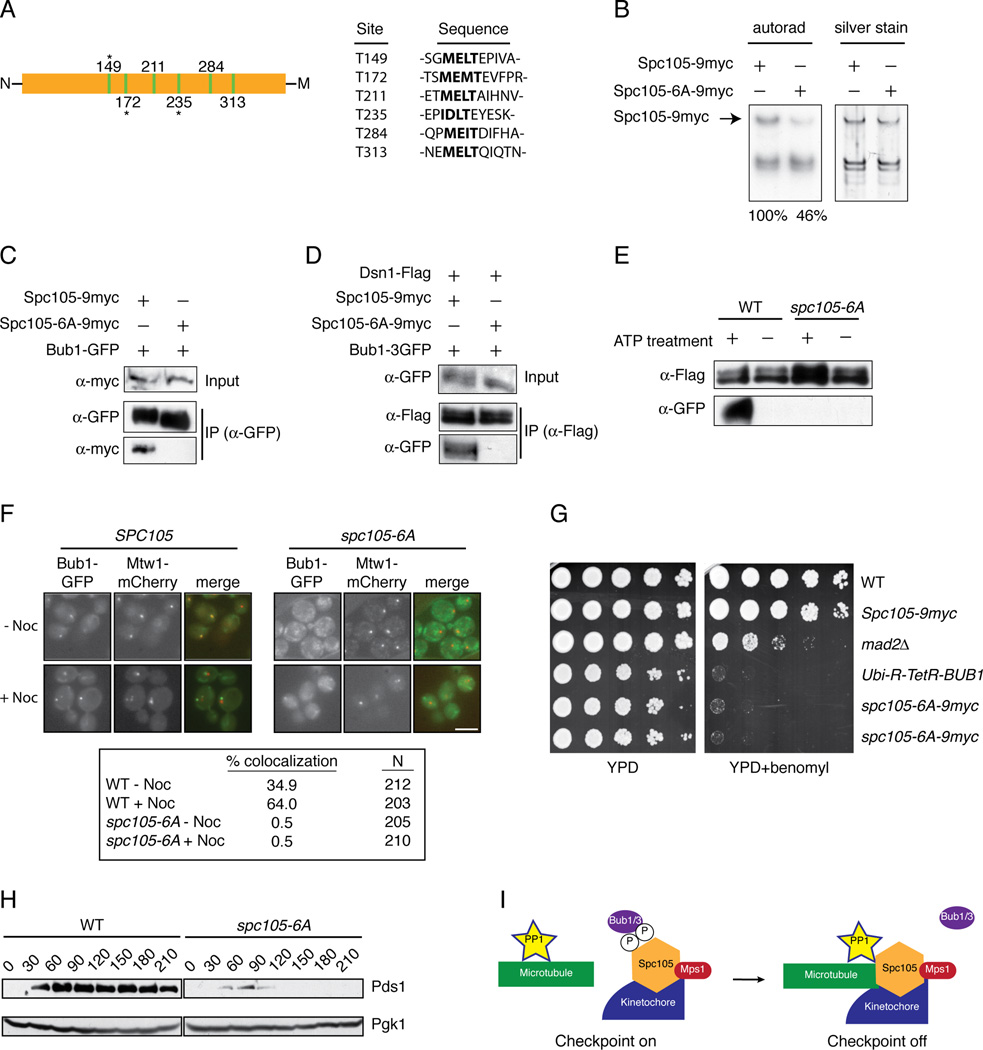
(A) A diagram of the N-terminal region of Spc105 shows the location of 6 MELT-like motifs illustrated as green bars. Residues that were detected as phosphorylated after Mps1 treatment are indicated with an asterisk. The N-terminus of Spc105 is denoted by “N” and the middle domain of the protein by “M”. Table shows MELT-like motifs (bolded) and surrounding sequence. (B) Kinetochores were purified (silver stain, right) from strains containing SPC105-9myc (SBY10267) or spc105-6A-9myc (SBY10265). They were incubated with radioactive ATP and the corresponding phosphate incorporation was quantified (autorad, left). Standard deviation of normalized Spc105 signal is +/− 5% (n=3). (C) Bub1-GFP was immunoprecipitated from strains containing Spc105-9myc (SBY10266) or Spc105-6A-9myc (SBY10264). (D) Dsn1-Flag was immunoprecipitated from strains containing SPC105-9myc Bub1-3GFP (SBY10426) or Spc105-6A-9myc Bub1-3GFP (SBY10422). (E) Kinetochore particles were purified from strains containing Bub1-3GFP and SPC105 (SBY10426) or spc105-6A (SBY10422). The particles were treated with PP1 and then incubated with +/− ATP. They were then incubated in lysates from WT strains (SBY10426) containing Bub1-3GFP and analyzed by immunoblotting. (F) Strains containing BUB1-GFP MTW1-mCherry and SPC105-9myc (SBY10266) or spc105-6A-9myc (SBY10264) were arrested in nocodazole for 60 minutes. The percentage of cells showing co-localization of Bub1 with Mtw1 was quantified. Note that Bub1 is enriched on kinetochores that have detached from microtubules and are no longer clustered together [18]. (G) Five-fold serial dilutions of WT (SBY3), SPC105-9myc (SBY10267), mad2Δ(SBY292), Ubi-R-TetR-BUB1 (SBY8920), or two independently generated spc105-6A-9myc strains (SBY10265, SBY10254) were analyzed for growth on YPD with doxycycline in the presence or absence of 10 µg/ml benomyl. (H) Pds1 levels were analyzed in SPC105-6His (SBY10373) or spc105-6A-6His (SBY10374) cells released from G1 into nocodazole. Pgk1 is a loading control. (H) Model for silencing of the spindle checkpoint. When kinetochores are not correctly attached to microtubules, Mps1 phosphorylates Spc105 to recruit Bub1/3. Proper microtubule attachment promotes PP1 interaction with Spc105, leading to dephosphorylation of Spc105 and release of Bub1/3 to promote checkpoint silencing.
To test whether phosphorylation of kinetochores by Mps1 could restore Bub1 binding to kinetochores containing the Spc105-6A mutant, we treated WT or Spc105-6A kinetochore particles with phosphatase, incubated them with ATP to allow phosphorylation of Mps1 sites, and then incubated them with cellular lysate, as in Figure 2D. Although Bub1 showed strong rebinding to ATP-treated particles from WT cells, the particles containing Spc105-6A failed to recruit Bub1 (Figure 4E). Consistent with this, Bub1 localization to kinetochores in vivo was abolished in cells containing spc105-6A (Figure 4F). Bub3 behaved the same in the rebinding assay (Figure S3C), indicating that Mps1 phosphorylation of one or more MELT motifs in Spc105 regulates the binding of Bub1 and Bub3 to kinetochores.
To analyze the phenotypes associated with mutation of the MELT motifs in Spc105, WT and spc105-6A strains were grown on media containing the microtubule-destabilizing drug benomyl. The spc105-6A mutant cells grew slowly compared to WT cells and exhibited hypersensitivity to benomyl media, similar to a Bub1 depletion (Figure 4G). Mutations in other Mps1 sites detected by mass spectrometry did not result in benomyl sensitivity or slow growth (data not shown). We attempted to analyze chromosome segregation by fluorescently marking a chromosome, but a high percentage of the spc105-6A cells exhibited aneuploidy of the marked chromosome, which prevented us from assaying biorientation or segregation (data not shown). To determine whether the spc105-6A cells were competent for the spindle checkpoint, we released them from G1 into nocodazole. Although WT cells arrested with high levels of the anaphase inhibitor Pds1, spc105-6A mutant cells did not stabilize Pds1 (Figure 4H). Consistent with this, 44% of the spc105-6A mutant cells exhibited rebudding 5 hours after release from G1 compared to 12% of WT cells. Together, these data show that Mps1-mediated phosphorylation of Spc105 contributes to accurate kinetochore and spindle checkpoint function.
The identification of targets of the conserved kinetochore kinases is key to fully understanding chromosome segregation and the spindle checkpoint, but progress has been slow due to the large number of potential kinetochore targets. Currently, Ndc80 is the only known Mps1 substrate at yeast kinetochores that regulates the spindle checkpoint [33]. We used a previously established kinetochore purification technique to identify Mps1 as the major kinase activity that associates with core kinetochore particles [7]. Although Bub1 also purifies with kinetochore particles, we did not detect any associated activity. This could be explained by low levels of Bub1 on kinetochore particles, a lack of active Bub1, or a low abundance of co-purifying substrates [34, 35].
Here, we identify Spc105 as a key Mps1 kinase target that recruits Bub1 to kinetochores via phosphorylation of the conserved MELT motifs. While a Bub1-binding KI motif was identified in human KNL-1/Blinkin that is not clearly conserved in yeast, this interaction is not sufficient for kinetochore recruitment of Bub1 in vivo [19, 28]. Therefore, there are additional unknown controls over the interaction that could be mediated by phosphorylation [19]. Given the strong conservation of the MELT motifs despite the generally strong divergence in much of the Spc105 homolog sequence, this mechanism may be conserved. Although the growth defects of the spc105-6A mutant are similar to a bub1 mutant, additional functions for the MELT motifs may also exist. Bub3 has been implicated in mediating Bub1 binding to kinetochores [18–20], and we found that Mps1 also regulates the association of Bub3 with Spc105. In the future, it will be important to clarify the dependency relationships between the Bub1 and Bub3 proteins and their interaction with Spc105.
We also show that Glc7 reverses Mps1-mediated phosphorylation on kinetochores to release Bub1, identifying a phosphatase activity that opposes the Mps1 kinase. This is consistent with work showing that Spc105 and PP1 are required to exit the spindle checkpoint [36–38]. Spc105 recruits PP1 to the kinetochore and requires its microtubule binding domain for checkpoint silencing [24, 26, 38], raising the possibility that PP1 disrupts Bub1 and Bub3 binding to Spc105 to turn off the checkpoint once kinetochores make proper bioriented attachments (Figure 4I). We showed that additional Mps1 phosphorylation sites exist on Spc105 and other kinetochore substrates, so an important line of future study will be to elucidate the associated functions. Together, these studies further our understanding of the molecular mechanisms by which Mps1 and PP1 regulate the complex signaling events that underlie spindle checkpoint function and chromosome segregation to maintain genomic stability.
Experimental Procedures
Yeast strains, plasmids and microbial techniques
Media and genetic and microbial techniques were essentially as described [39]. The construction of yeast strains and plasmids used in this study and the experimental growth conditions are described in Supplementary Experimental Procedures. Yeast strains are listed in Supplemental Table 2.
Biochemistry and microscopy techniques
Details of kinetochore purifications, immunoprecipitation conditions, mass spectrometry and microscopy assays are described in Supplementary Experimental Procedures.
Acknowledgments
We are very grateful to Nicole Duggan, Phil Gafken and Christian Nelson for technical assistance. We thank John Cannon, Trisha Davis, Arshad Desai, John Kilmartin, Andrew Murray, Kelly Tatchell and Mark Winey for reagents used in this work and Yoshi Watanabe for communicating unpublished results. We thank Bungo Akiyoshi and members of the Asbury and Biggins lab for discussions, and Chip Asbury, Gary Deyter and Erica Hildebrand for comments on the manuscript. This work was funded by an NIH center interdisciplinary training grant (T32 CA080416) to NL, an NIGMS grant (PM50 GM076547/Center for Systems Biology) to JAR and NIH grants (GM064386 and an associated ARRA supplement) to SB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cub.2012.03.052
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0960982212003399/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Dual TTK/PLK1 inhibition has potent anticancer activity in TNBC as monotherapy and in combination.
Front Oncol, 14:1447807, 09 Aug 2024
Cited by: 0 articles | PMID: 39184047 | PMCID: PMC11341980
Architecture of native kinetochores revealed by structural studies utilizing a thermophilic yeast.
Curr Biol, 34(17):3881-3893.e5, 09 Aug 2024
Cited by: 0 articles | PMID: 39127048 | PMCID: PMC11387133
A conserved site on Ndc80 complex facilitates dynamic recruitment of Mps1 to yeast kinetochores to promote accurate chromosome segregation.
Curr Biol, 34(11):2294-2307.e4, 21 May 2024
Cited by: 3 articles | PMID: 38776906
Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biol, 14(6):240025, 12 Jun 2024
Cited by: 3 articles | PMID: 38862021 | PMCID: PMC11286163
<i>FBXO42</i> activity is required to prevent mitotic arrest, spindle assembly checkpoint activation and lethality in glioblastoma and other cancers.
NAR Cancer, 6(2):zcae021, 20 May 2024
Cited by: 0 articles | PMID: 38774470 | PMCID: PMC11106029
Go to all (225) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Dual mechanisms regulate the recruitment of spindle assembly checkpoint proteins to the budding yeast kinetochore.
Mol Biol Cell, 27(22):3405-3417, 11 May 2016
Cited by: 13 articles | PMID: 27170178 | PMCID: PMC5221577
KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint.
Curr Biol, 21(11):942-947, 01 Jun 2011
Cited by: 149 articles | PMID: 21640906 | PMCID: PMC3109435
Killing two birds with one stone: how budding yeast Mps1 controls chromosome segregation and spindle assembly checkpoint through phosphorylation of a single kinetochore protein.
Curr Genet, 66(6):1037-1044, 06 Jul 2020
Cited by: 3 articles | PMID: 32632756
Review
Delineating the contribution of Spc105-bound PP1 to spindle checkpoint silencing and kinetochore microtubule attachment regulation.
J Cell Biol, 218(12):3926-3942, 24 Oct 2019
Cited by: 15 articles | PMID: 31649151 | PMCID: PMC6891095
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA015704
Grant ID: T32 CA080416
NIGMS NIH HHS (4)
Grant ID: PM50 GM076547
Grant ID: P50 GM076547
Grant ID: R01 GM064386
Grant ID: GM064386





