Abstract
Free full text

Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells
Abstract
Deficiency in C1q, the recognition component of the classical complement cascade and a pattern recognition receptor involved in apoptotic cell clearance, leads to lupus-like auto-immune diseases characterized by auto-antibodies to self proteins and aberrant innate immune cell activation likely due to impaired clearance of apoptotic cells. Here, we developed an autologous system using primary human lymphocytes and monocyte-derived macrophages (HMDMs) to characterize the effect of C1q on macrophage gene expression profiles during the uptake of apoptotic cells. C1q bound to autologous apoptotic lymphocytes modulated expression of genes associated with JAK/STAT signaling, chemotaxis, immunoregulation and NLRP3 inflammasome activation in LPS-stimulated HMDMs. Specifically, C1q sequentially induced type I interferons (IFNs), IL-27 and IL-10 in LPS-stimulated HMDMs and IL-27 in HMDMs when incubated with AL conditioned media. Co-incubation with C1q tails prevented the induction of type I IFNs and IL-27 in a dose dependent manner and neutralization of type I IFNs partially prevented IL-27 induction by C1q. Finally, C1q decreased procaspase-1 cleavage and caspase-1 dependent cleavage of IL-1β suggesting potent inhibitory effect of C1q on inflammasome activation. These results identify specific molecular pathways induced by C1q to suppress macrophage inflammation providing potential therapeutic targets to control macrophage polarization, and thus inflammation and autoimmunity.
Introduction
The complement system, a powerful effector of the innate immune system, consists of a group of proteins circulating as inactive precursors in the blood and in extracellular fluids. Upon activation through the classical, lectin or alternative pathway, a cascade of proteolytic cleavages and formation of central enzymatic complexes (C3 and C5 convertases) leads to the generation of active fragments resulting in the opsonization of invading pathogens (C1q, C3b and iC3b), release of pro-inflammatory chemotactic factors (C3a and C5a) which recruit leukocytes to the site of infection or injury, and finally formation of the membrane attack complex (C5b-9) and subsequent lysis of the pathogen (1,2). Complement functions as an important humoral defense system to sense danger by recognizing pathogen associated molecular patterns (PAMPs) but is also activated by damaged associated molecular patterns (DAMPs) or altered self tissues. Dysregulated complement activation has been associated with the development of various diseases including rheumatoid arthritis and Alzheimer’s disease (2,3). A causal link between complement deficiency and systemic lupus erythematosus (SLE) involves in part the role of complement in physiological waste disposal mechanisms, in particular clearance of dying cells (4). While activation by all three complement pathways can contribute to enhanced uptake of apoptotic cells by phagocytic cells (5–8), homozygous deficiency of any of the early complement components of the classical pathway (C1q, C1r, C1s, C4 and C2) predisposes to the development of SLE with over 90% of individuals with genetic deficiency of C1q developing severe SLE (9).
C1q is known to play a prominent non redundant tissue specific role in the clearance of apoptotic cells in vitro and in vivo (10–14). C1q binds to apoptotic cells and cellular debris through its globular heads (10,15) and to phagocytic receptors through its collagen tails (1,16). While at first thought to be primarily of liver origin, C1q is predominantly synthesized in vivo by peripheral tissue macrophages and dendritic cells (17,18) and by myeloid cells in vitro (8,19–21). While C1q is most often bound to C1r and C1s in the circulation (22), this local synthesis of C1q is hypothesized to be the major source of C1q for the rapid opsonization of dying cells in tissue before recruitment of plasma-derived components such as C1r and C1s and subsequent activation of the complement cascade. In addition, induced synthesis of C1q has been detected in several injury models in vivo and in vitro ((23,24) and reviewed in (3)), suggesting that the induction of C1q synthesis in tissue may be a response to injury that promotes rapid clearance of apoptotic cells and concomitant suppression of inflammation. For example, interaction of C1q with human monocytes or dendritic cells results in the down-regulation of pro-inflammatory cytokines upon TLR4 stimulation by LPS (25,26). Recently, we showed that C1q enhances uptake of apoptotic Jurkat T cells by human monocytes but has no effect on the basal clearance level of these apoptotic cells by human monocyte-derived macrophages (HMDMs) and dendritic cells (8). In addition, although C1q influences the induction of cytokines in all myeloid cell types tested in this study, both the degree and direction of modulation depend on the state of differentiation of the phagocytic cell (8). However, because several C1q receptors have been identified and none has been shown to specifically mediate C1q-enhancement of phagocytosis of apoptotic cells (1,12,27), the intracellular signaling pathways engaged upon interaction of C1q with phagocytic cells remain to be fully elucidated. In addition, since characterization of macrophage activation in response to C1q has been limited to the study of few candidate cytokines, chemokines and/or signaling molecules, the extent of the effect of C1q on macrophage polarization and inflammatory responses during uptake of apoptotic cells remains largely uncharacterized. In this study, we developed a unique system using primary human autologous lymphocytes and HMDMs to characterize the effect of C1q on macrophage gene expression profiles during the uptake of autologous apoptotic cells, a more physiologic system than transformed cell lines as a source of apoptotic cells. Our results show that C1q bound to autologous apoptotic lymphocytes (AL) significantly modulates the response of HMDMs to LPS by increasing expression of cytokines, chemokines and effector molecules associated with immunoregulation and by directly suppressing caspase-1 dependent cleavage of IL-1β, in absence of any other complement proteins.
Material and Methods
Media and reagents
RPMI 1640, penicillin/streptomycin, trypsin-EDTA and L-Glutamine were from InVitrogen. HL-1 medium was from BioWhittaker and defined FCS from HyClone. Recombinant human (rh) M-CSF and IL-2 were from PeproTech. ATP was from Sigma-Aldrich. Mouse IgG1 antibodies were from R&D Systems and anti-human IFNα and β antibodies were from PBL Biomedical Laboratories. Human serum albumin (HSA) used for elutriation was obtained from Talecris Biotherapeutics. Ultra-pure LPS was from List Biological Laboratories Inc. C1q was isolated from plasma-derived normal human serum by ion-exchange chromatography, followed by size-exclusion chromatography according to Tenner et al. (28) and modified as described (29). C1q tails were prepared as described (30). All C1q preparations showed equivalent purity (determined by SDS-PAGE and Coomassie staining) and have less than 0.03 EU/ml endotoxin by Limulus Amoebocyte Lysate clot assay (Lonza).
Cell isolation and culture
All blood samples were collected into CPDA1 at the UCI Institute for Clinical and Translational Science in accordance with guidelines and approval of the University of California Irvine (UCI) Institutional Review Board. Human peripheral blood lymphocytes and monocytes were isolated by counter flow elutriation using a modification of the technique of Lionetti et al. (31) as described previously (32). Cell purity was determined by standard flow cytometry on a FACS calibur (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Tree Star). About 80% of the cells of the lymphocyte fraction were CD3+ (Fig. S1A) and greater than 90% of the monocyte fraction was CD11b+. Lymphocytes were maintained for 7 days in RPMI1640, 10% FCS, 2 mM L-Glutamine and 1% penicillin/streptomycin (complete media) containing 50 U/ml rhIL-2 and then γ-irradiated (10 Gy) and maintained overnight in complete media or media without FBS to generate EAL (Annexin V+/PI−) or LAL (Annexin V+/PI+), respectively (Fig. S1B). In some experiments, lymphocytes were pre-labeled with a red membrane cell tracker (PKH26 from Sigma-Aldrich) according to manufacturer’s instructions. HMDMs were generated from monocytes by culture for 8 days in complete media containing 25 ng/ml rhM-CSF. For every experiment, apoptosis (apoptosis detection kit from BioVision) and HMDMs phenotype (Fig. S2A) were assessed by flow cytometry.
C1q binding assay
EAL and LAL were incubated with 150 µg/ml purified human C1q for 1 h in PBS/1% HSA at 37°C. Binding of C1q was assessed for every experiment by flow cytometry using a monoclonal antibody against C1q (Quidel) and FITC-anti mouse IgG (Jackson ImmunoResearch Laboratories). For each experiment, C1q binding was greater than 50% for EAL or LAL.
Uptake assay
PKH26-labeled or unlabeled EAL and LAL, pre-coated or not with C1q were incubated with HMDMs at a 5:1 ratio for 1 h (optimal ratio and time determined in preliminary experiments, see Fig. S2B) in phagocytosis buffer (RPMI1640, 25 mM HEPES and 5 mM MgCl2). For uptake quantification, cells were washed, harvested with Trypsin/EDTA and stained with CD11c-FITC antibodies for flow cytometry analysis or fixed with 3.7% paraformaldehyde and stained with FITC-phalloidin (InVitrogen) according to manufacturer’s instructions. For confocal imaging, cells were analyzed using the Nikon Ti microscope and the EZ C1 software. Images were analyzed using Adobe Photoshop CS and ImageJ.
RNA extraction and microarray analysis
After uptake, HMDMs were stimulated with 10 ng/ml LPS (each condition performed in triplicate) for 3 h in serum free HL-1 media. Total RNA was extracted using the Illustra RNA spin Mini Isolation Kit (GE Healthcare). Gene expression profiles were studied using the Human Gene 1.0 ST array (Affymetrix). RNA labeling and hybridization were performed according to manufacturer’s instructions by the UCI Genomics High Throughput Facility at University of California, Irvine. Data processing and analysis were performed using JMP Genomics 5.0 software (SAS Institue Inc.). Briefly, inter-array median correction was used to normalize signal intensities. Significant differences in gene expression compared to unstimulated HMDMs were identified by ANOVA test using Holm multiple testing method and a false positive rate (alpha error) of 0.05 (see Table S1 for a complete list of modulated genes). Hierarchical clustering (Pearson correlation coefficient-based heat map using complete linkage method) was performed using JMP genomics and TMeV (33). Functional classification was performed using DAVID software (http://david.abcc.ncifcrf.gov/) (34) and pathway network analysis and visualization was performed using Cytoscape (35). All data were submitted to the MIAME-compliant (36) database Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE30177).
Reverse transcription and quantitative real-time PCR (qRT-PCR)
The cDNA synthesis was carried out with 100 ng of total RNA and the M-MLV reverse transcriptase RT (Invitrogen) as previously described (37). Quantitative PCR was performed using the Maxima SYBR/Green Master Mix (Thermo Fisher Scientific), the iCycleriQ and the iQ5 software (Bio-Rad). The fold-change was determined as follows: FC = 2−ΔΔCt, where ΔΔCt = (CtTarget − CtGAPDH) test − (CtTarget − CtGAPDH) unstimulated. Ct values were defined as the number of cycles at which the fluorescence signals were detected (38).
Cytokine secretion assays
Human IFNα detection kit was from Mabtech and IL-27 and IL-10 human detection kits were from BioLegend. ELISAs were performed according to manufacturer’s instructions.
Detection of cleaved caspase-1
After uptake, HMDMs were washed and stimulated with 10 ng/ml LPS in HL-1 media for 6 h. ATP (1 mM) was added to the cell culture 90 min before the end of LPS stimulation. HMDMs were then incubated with Green FLICA™ Caspase-1 probes (ImmunoChemistry Technologies) and Cell Tracker™ Blue CMF2HC (InVitrogen) 1 h before the end of the stimulation. HMDMs were then washed, fixed and analyzed immediately by confocal microscopy as described above.
Western blot
For detection of inflammasome components, HMDMs were stimulated with 10 ng/ml LPS for 6 h and 1 mM ATP was added for the last 1 h of stimulation. For detection of IL-1β, HMDMs were stimulated with LPS for 18 h and ATP was added during the last 3 h of LPS stimulation. HMDMs culture supernatants were concentrated using 10 kDa Amicon columns (Millipore). HMDMs were harvested in RIPA lysis buffer. Proteins were separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes (GE Healthcare). Immunoblots were performed using primary antibodies against NLRP3 (NLR family pyrin domain containing 3, Enzo Life Sciences), caspase-1 (Cell Signaling), ASC (apoptosis-associated speck-like protein, Medical & Biological laboratories), IL-1β (clone 3ZD form the NCI Biological Resources Branch) and β-actin (Sigma-Aldrich) and secondary HRP-conjugated anti-mouse or rabbit IgG antibodies (Jackson ImmunoResearch Laboratories). The blots were developed using enhanced chemiluminescence plus (ECL+, GE Healthcare) and analyzed using the Nikon D700 digital SLR camera and the ImageJ software as described (39).
Statistical analysis
Results were calculated as means ± s.d. and compared with two-way ANOVA followed by Bonferroni post hoc test, alpha error = 0.05, using GraphPad Prism (unless otherwise stated, all conditions are compared to unstimulated HMDMs). Differences were considered significant when p was < 0.05.
Results
C1q binds to human apoptotic lymphocytes and enhances their ingestion by HMDMs
To determine how C1q modulates human macrophage responses during the uptake of apoptotic cells we developed a unique autologous system (lymphocytes and monocytes being isolated from the same donor) where HMDMs were incubated with C1q-coated apoptotic lymphocytes. Lymphocytes were irradiated to induce apoptosis and maintained overnight in complete media or under serum deprivation to generate early (EAL, AV+PI−) and late (LAL, AV+PI+, secondary necrosis stage) apoptotic lymphocytes, respectively (Fig. S1B). In preliminary experiments (Fig. S1D–E), the optimal C1q concentration (150 µg/ml) and incubation time (1 h) to obtain the highest percentage of C1q binding to AL while maintaining C1q concentration at near physiological levels was determined (C1q serum concentration is about 113 ± 40 µg/ml, ranging from 56 to 276 µg/ml depending on the studies and methods (40,41)). We found that C1q binds to human EAL and LAL with the same efficiency since about 55 to 60% of both EAL and LAL were C1q positive and have comparable MFI (Fig. 1A and B). Pre-labeling of EAL and LAL with the red cell membrane tracker PKH26 did not affect the induction of apoptosis (Fig. S1C) or C1q binding to those cells (Fig. S1F). The effect of C1q on the uptake of EAL and LAL by HMDMs was then determined by immunocytochemistry and flow cytometry (Fig. S2B–C and Fig. 1C–1D). It is of note that all uptake assays and subsequent stimulation were performed in serum free medium so no other complement proteins than C1q are present in this system in order to mimic tissue environment (early in injury or during homeostatic apoptotic cell clearance) before the recruitment of plasma-derived complement proteins. C1q significantly (p < 0.05) increased the percentage of HMDMs that have ingested at least one EAL but did not increase the percentage of HMDMs that have ingested at least one LAL (Fig. 1C–D). However, C1q increased the number of EAL and LAL per HMDMs, particularly the percentage of HMDMs that have ingested 3 or more EAL (p < 0.01, Fig. 1C) or LAL (p < 0.05, Fig. 1C). All together these results showed that C1q binds with the same efficiency to EAL and LAL and enhances their uptake by HMDMs.

(A and B) EAL and LAL were incubated without (gray peak) or with 150 µg/ml C1q (black lane) for 1 h, washed and stained for C1q. Representative FACS plot of multiple experiments are shown. (B) Percentage of C1q binding and anti-C1q MFI determined by flow cytometry. Results represent means ± s.d. (n = 5). (C–D) HMDMs were incubated with PKH26-labeled EAL and LAL, pre-incubated or not with C1q, at a 5:1 ratio for 1 h, washed and fixed. Cells were stained with FITC-phalloidin and analyzed by confocal microscopy (Fig. S2) to determine the percentage of phagocytosis and the number of targets per HMDMs (C) or stained with CD11c-FITC antibodies and analyzed by flow cytometry (D).Results represent means ± s.d. (n = 3 different donors), two-way ANOVA, * p < 0.05 and ** p < 0.01. (E–F) PKH26-prelabeled EAL and LAL (red) were incubated with C1q for 1 h, washed and then added to HMDMs at a 5:1 ratio for 1 h. Cells were fixed and stained with anti-C1q antibodies (blue) and FITC-phalloidin (green) and analyzed by confocal microscopy. Representative micrographs of 3 independent experiments are shown in E. Scale bar = 10 µm. (F) Percentage of HMDM-bound/ingested EAL or LAL bound or not to C1q. Results represent means ± s.d. (n = 3), two-way ANOVA, * p < 0.05 and ** p < 0.01.
To determine if HMDMs preferentially ingest those EAL and LAL bound to C1q, we quantified the number of C1q positive cells after uptake by immunostaining. (Fig. 1E–F). C1q was detectable on EAL or LAL during the uptake by HMDMs (Fig. 1E) and more than 70% of macrophage-bound EAL (72.1 ± 17.6%, p < 0.05) or LAL (74.1 ± 12.3%, p < 0.01) were C1q positive (Fig. 1F). All together these results suggest that C1q remained on EAL and LAL surfaces acting as a powerful “eat-me” signal to enhance the uptake and therefore may directly signal macrophages to modulate their responses during phagocytosis.
C1q modulates expression of genes associated with chemotaxis, inflammation, signaling and NLRP3 inflammasome activation in LPS-stimulated HMDMs during the uptake of apoptotic lymphocytes
To delineate the C1q-modulated pathways in macrophages during the uptake of AL, HMDMs were incubated with C1q-bound EAL or LAL for 1 h and then stimulated for 3 h with a low dose of LPS (10 ng/ml). LPS is used as a tool in this system to mimic local inflammation induced by DAMPs such as through activation of TLRs (such as TLR4) by HMGB1 or HSP that are normally intracellular but are released when the cells die (42).. Global transcriptional gene expression profiles of HMDMs were analyzed using the Gene 1.0 ST array from Affymetrix (Fig. 2 and S3) and validated by qRT-PCR (Fig. S3). Hierarchical clustering analysis highlighted groups of genes specifically modulated by the uptake EAL and LAL as compared to LPS alone and also by C1q when bound to EAL or LALas compared to EAL or LAL in the absence of C1q (Fig. 2A). Using Gene Ontology (GO) annotation, genes modulated by C1q showed enriched GO biological processes related to signal transduction, G protein-coupled receptor (GPCR) signaling pathway, immune response, homeostasis and macromolecule biosynthesis (Fig. 2B). Network pathway analysis of these biological processes using Cytoscape showed that C1q modulated genes associated with chemotaxis, inflammation/cytokines, JAK/STAT signaling and NLRP3 inflammasome activation (Fig. 2C–F and Table S1), some of those genes being differentially modulated when C1q is bound to EAL or LAL suggesting that the apoptotic cell stage influences the C1q effect on macrophage response. For example, C1q increased the expression of the chemokine CX3CL1 (fractalkine) and the chemokine receptor CCR3 in HMDMs only when bound to EAL (Fig. 2C). When bound to LAL, C1q significantly increased the expression of versican (VCAN), IL-13, a typical M2-driven cytokine, and IL1F7, also known as IL-37, while decreasing the expression of the M1-asssociated chemokine CXCL9 in HMDMs (Fig. 2C–D). C1q bound to either EAL and LAL decreased the expression of VEGF-C and increased the expression of immunoregulatory and immunosuppressive cytokines such as IL-33, IL-27 and the type I IFNs receptor IFNAR2 in HMDMs (Fig. 2C–D). It is important to note than no IL-27 mRNA, the most up-regulated gene by C1q, was detectable in C1q-bound EAL or LAL thus eliminating any possibilities of RNA contamination from AL in our microarray results (Supplementary Fig. 3G). In addition, C1q bound to EAL and LAL modulated the expression of several signaling molecules of the JAK/STAT pathway (Fig. 2E). Finally, C1q bound to EAL and LAL increased the expression of NLRP12, a negative regulator of NF-κB and inflammasome activation (Fig. 2F). When bound to LAL, C1q decreased the expression of NLRP3/NALP3 (also down-regulated by EAL alone by 2-fold with no further effect of C1q on this down-regulation, see Table S1), one of the main components of the NLRP3 inflammasome, a cytosolic protein complex formed by the association of NLRP3, procaspase-1, and ASC (apoptosis-associated speck-like protein containing a CARD domain) that cleaves procaspase-1 to generate active caspase-1. At the same time, C1q bound to LAL increased the expression of POP1/ASC2 (Fig. 2F), a pyrin-domain containing protein that associates with ASC and may destabilize the NLRP3 inflammasome. All together these results suggest that C1q promotes the expression of potent immunoregulatory and immunosuppressive cytokines and negatively regulates NLRP3 inflammasome activation.
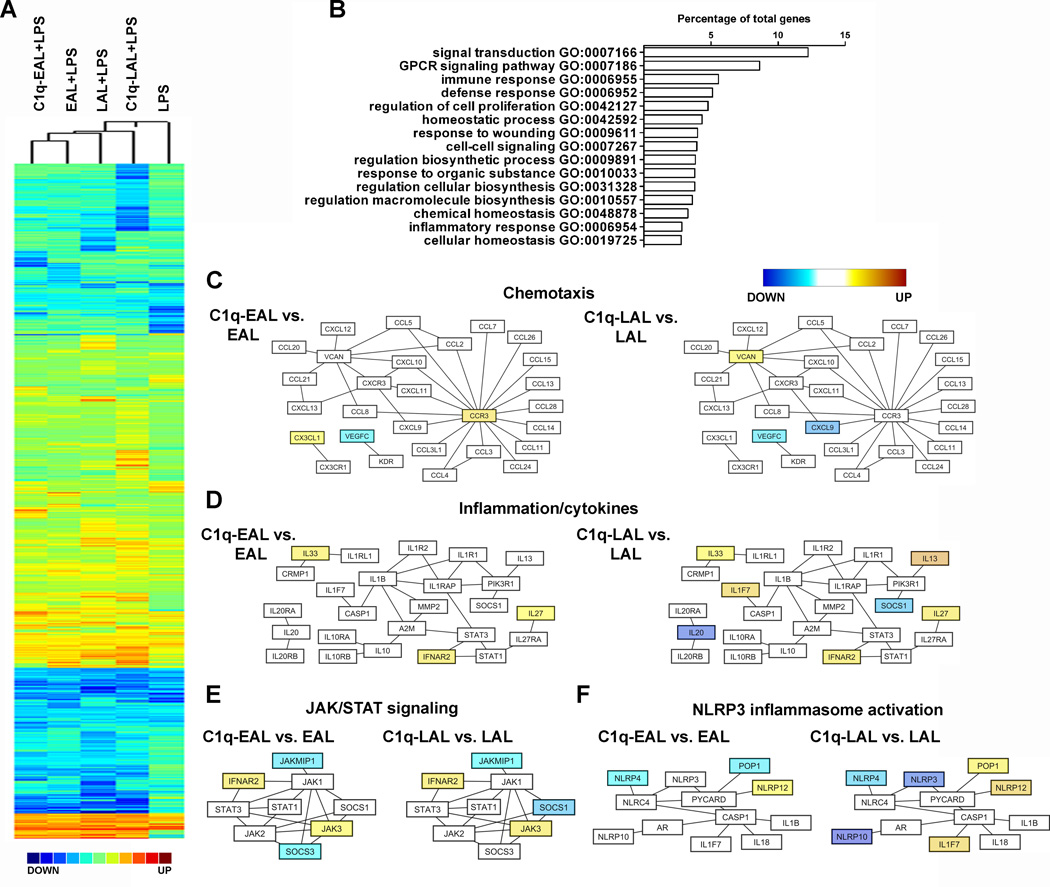
(A) Pearson correlation coefficient-based heat map (complete linkage method) representation of the Log2 fold-change (all conditions performed in triplicate) of HMDMs incubated with EAL, C1q-EAL, LAL and C1q-LAL and then stimulated with LPS for 3 h over unstimulated HMDMs. (B) GO-based functional annotation of genes modulated by C1q in HMDMs. Major biological processes are shown as the percentage of differentially expressed annotated genes (redundancy is due to the involvement of individual genes in multiple biological processes). (C–F) Network diagrams of chemotaxis (C), inflammation/cytokines (D), JAK/STAT signalling (E) and NLRP3 inflammasome activation (F) pathways modulated by C1q in HMDMs. Node colors represent changes in gene expression in C1q-EAL vs. EAL or C1q-LAL vs. LAL, shown using a color gradient (blue = down-regulated, white = not modulated and yellow = up-regulated by C1q).
C1q increases type I IFNs, IL-27 and IL-10 secretion by LPS-stimulated HMDMs during the uptake of AL
LPS stimulation of mouse macrophages induces IL-10 production through the sequential induction of type I IFNs and IL-27 (43). Since IL-27 was one of the most up-regulated cytokine by C1q bound to EAL and LAL in HMDMs (about 4-fold-increase compared to unstimulated HMDMs and 2 fold-increase compared to EAL or LAL alone, Fig. 2D and Table S1) and IFNA6 mRNA was slightly up-regulated by C1q in HMDMs (Table S1), we investigated if C1q modulates type I IFNs, IL-27 and IL-10 expression in human macrophages. C1q transiently increased IFNα and IFNβ mRNA levels (Fig. 3A–D) in HMDMs, and the secretion of IFNα by HMDMs was significantly (p < 0.001) increased by C1q bound to EAL or LAL after 6 h of LPS stimulation (Fig. 3A and B, insets). C1q bound to EAL and LAL significantly increased IL-27 mRNA levels after 3, 6 and 12 h of LPS stimulation (Fig. 3E and F) and the IL-27 protein levels were significantly (p < 0.001) increased by C1q-EAL and C1q-LAL after 18 h of LPS stimulation (Fig. 3E and F, insets). Interestingly, the levels of IL-27 were decreased by LPS in a dose dependent manner and the effect of C1q on IL-27 expression was totally inhibited after stimulation with higher dose of LPS (Fig. 3I). Finally, C1q bound to EAL and LAL significantly (p < 0.05) increased IL-10 mRNA levels in HMDMs after 18 h of LPS stimulation (Fig. 3G and H). The protein levels of IL-10 were also increased by C1q after 18 h of LPS stimulation when compared to LPS (p < 0.05) and EAL or LAL (Fig. 3G and H, insets).
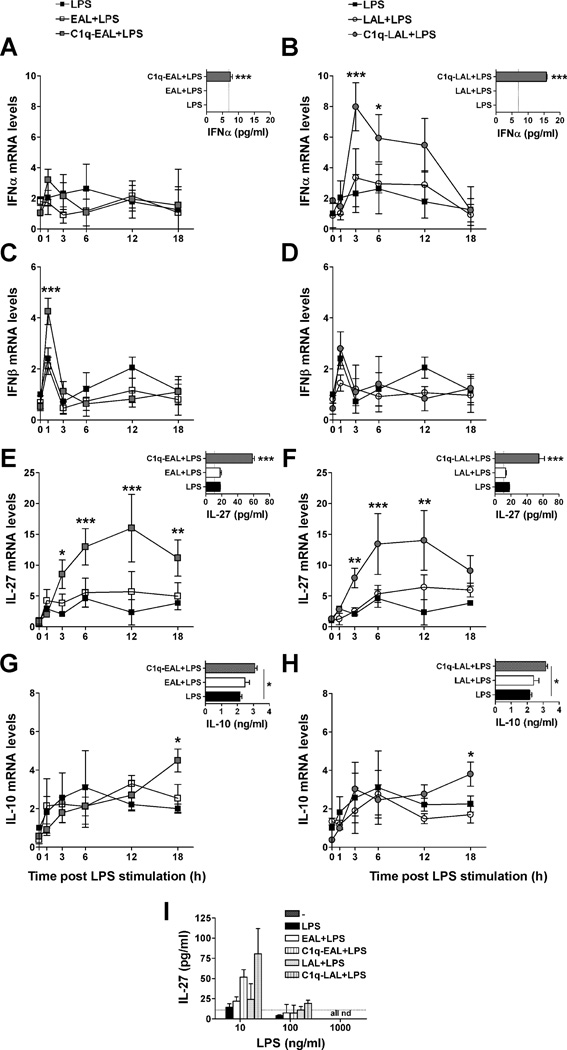
HMDMs were incubated with EAL (left panels) or LAL (right panels), pre-incubated or not with C1q, at a 5:1 ratio for 1 h and then stimulated with 10 ng/ml LPS (A–H) or 10–1000 ng/ml (I) in HL-1 for up to 18 h. Changes in mRNA levels for IFNα (A and B), IFNβ (C and D), IL-27 (E and F) and IL-10 (G and H) were determined by qRT-PCR. Protein levels of IFNα (A and B, insets) were detected 6 h after LPS stimulation and IL-27 (E and F, insets and I) and IL-10 (G and H, insets) after 18 h of LPS stimulation. (A–H) LPS data (black squares) are identical between left and right panels (single LPS control experiment performed for both EAL and LAL simultaneously). Results represent means ± s.d. (n = 2–3 different donors), two-way ANOVA, * p < 0.05, ** p < 0.01 and *** p < 0.001.
C1q bound to apoptotic cells synergizes with apoptotic cell signals in inducing IL-27 in HMDMs during the uptake of AL
We next investigated whether C1q modulates type I IFNs, IL-27 and/or IL-10 in resting HMDMs (i.e. non LPS-stimulated). HMDMs were incubated with C1q-bound EAL or LAL for 1 h and then further cultured for 3 h in absence of LPS (time point corresponding to the 3 h LPS stimulation done in our microarray). While C1q-bound to AL did not modulate expression of type I IFNs or IL-10 at 3 h of incubation, C1q-bound to LAL significantly (p < 0.01) increased IL-27 mRNA levels compared to LAL alone, although this 2-fold increase in IL-27 was lower than the induction observed in presence of LPS (Fig. 3F).
Dying cells release numerous DAMPs that can activate TLR4, similarly to LPS (44). To model the more physiologic condition in which this release of DAMPs by EAL and LAL might affect the modulation of HMDMs response by C1q, HMDMs were incubated with C1q-bound EAL or LAL for 1 h and then incubated with EAL or LAL conditioned media for 3 h. While again at this time point, no induction of type I IFNs or IL-10 was detectable, the presence of EAL conditioned media induced a 3-fold increase in IL-27 expression in HMDMs ingesting C1q-bound EAL compared to ingestion of EAL alone (Fig. 4B). Moreover, in presence of LAL conditioned media, C1q-bound LAL induced a 6-fold increase in IL-27 expression in HMDMs compared to LAL alone (Fig. 4B), induction similar to what is observed in the presence of LPS without the conditioned media (Fig. 3F). These results suggest that C1q sequentially induced type I IFNs, IL-27 and IL-10 expression in LPS-stimulated HMDMs and may act in synergy with soluble factors released by dying cells to induce IL-27 in resting HMDMs.
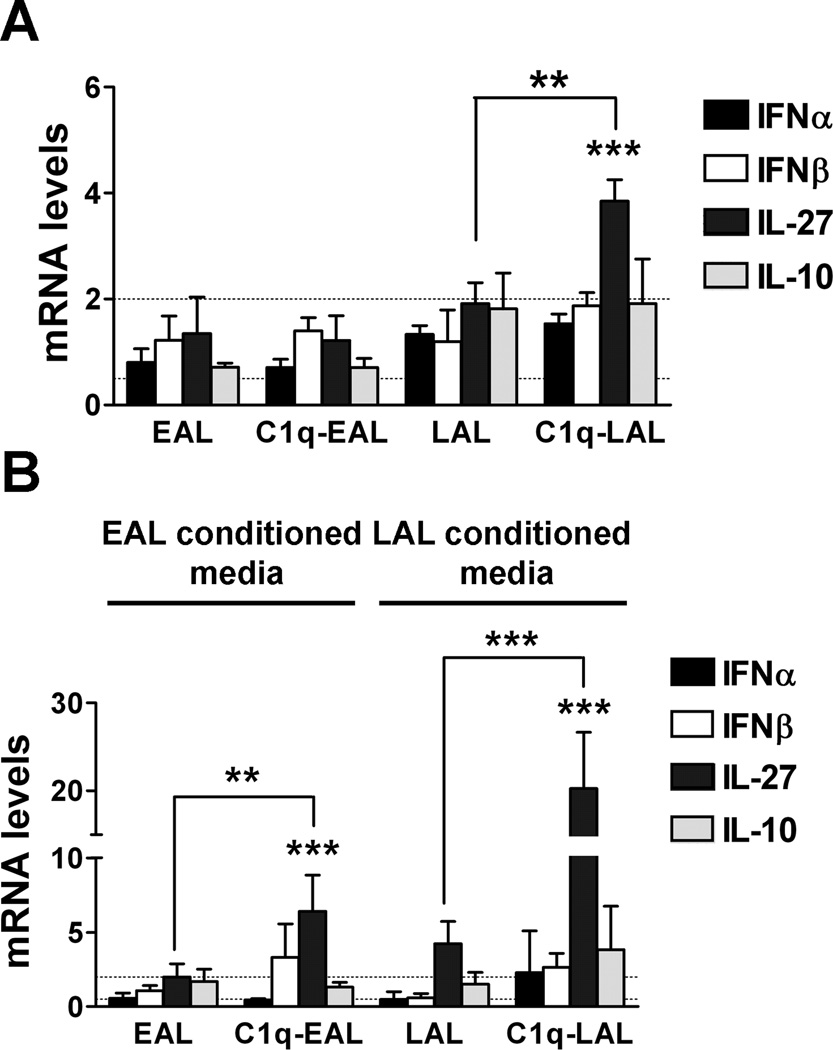
HMDMs were incubated with EAL or LAL, pre-incubated or not with C1q, at a 5:1 ratio for 1 h. HMDMs were then cultured in HL-1 for 3 h in absence of LPS (A) or in presence of EAL or LAL conditioned media (B). Changes in type I IFNs, IL-27 and IL-10 were determined by qRT-PCR. Results represent means ± s.d. (n = 3 different donors), two-way ANOVA, ** p < 0.01 and *** p < 0.001.
Co-incubation with C1q tails or neutralization of type I IFNs suppresses IL-27 induction by C1q in LPS-stimulated HMDMs
C1q has been shown to bind apoptotic cells through its globular heads and can interact with receptors on phagocytic cells through its tail region. Thus, to determine the specificity of C1q signaling in the induction of cytokines, HMDMs were incubated with C1q-bound EAL or LAL for 1 h in presence of increasing doses of C1q tails, which should compete with C1q bound to LAL to bind HMDMs without inducing C1q receptor clustering on HMDMs and thus no signaling. After washing to remove unbound targets and C1q tails, HMDMs were stimulated with LPS for 3 h and mRNA levels of IFNα and IL-27 were determined by qRT-PCR (Fig. 5A and B). When HMDMs were incubated with C1q-bound EAL or LAL in presence of C1q tails, the C1q-induced expression of IFNα and IL-27 is significantly decreased in a dose dependent manner by C1q tails (Fig. 5A and B), suggesting a competition between C1q tails and C1q bound to AL to induce cytokines in HMDMs.

HMDMs were incubated with EAL, C1q-EAL, LAL or C1q-LAL at a 5:1 ratio for 1 h (A–B) in presence of C1q tails and then stimulated with 10 ng/ml LPS for 3 h or (C–E) stimulated with LPS for 18 h in presence of 1 µg/ml control mouse IgG1 or neutralizing antibodies against IFNα and/or IFNβ (C–D) or goat IgG or neutralizing antibodies against IL-27 (E). mRNA levels were determined by qRT-PCR and proteins levels by ELISAs. Results represent means ± s.d. (n = 2–3 different donors, performed in duplicates), two-way ANOVA, * p < 0.05, ** p < 0.01 and ***, p < 0.001.
To determine the contribution of type I IFNs in the induction of IL-27 and IL-10 by C1q in LPS-stimulated macrophages, HMDMs were incubated with C1q-bound EAL or LAL for 1 h and then stimulated with LPS for 18 h in presence of control mouse IgG or type I IFNs or IL-27 neutralizing antibodies (Fig. 5C–E). Incubation with anti-IFNα antibodies, anti-IFNβ antibodies or both significantly decreased the IL-27 secretion induced by C1q bound to EAL to levels of EAL alone (Fig. 5C). The induction of IL-27 in HMDMs by C1q bound to LAL was also significantly decreased by anti-IFNα antibodies and/or anti-IFNβ antibodies, however the levels of IL-27 after neutralization of type I IFNs remained significantly higher than IL-27 levels induced by LAL alone (Fig. 5C). In addition, no change was observed in IL-10 levels after neutralization of type I IFNs (Fig. 5D) and neutralization of IL-27 slightly decreased IL-10 protein levels but the differences did not reach statistical significance (Fig.5E). This suggests that induction of IL-27 by C1q is partially dependent on type I IFNs in LPS-stimulated HMDMs while induction of IL-10 likely involves multiple pathways.
C1q suppresses NLRP3 inflammasome activation and IL-1β cleavage
Our microarray data suggest that EAL, C1q-EAL and C1q-LAL may decrease NLRP3 inflammasome activity through decreased NLRP3 mRNA levels and increased expression of negative regulators of inflammasome activity such as POP1/ASC2 or NLRP12 (Fig. 2 and Table S1). To test this hypothesis, PKH26-prelabeled EAL and LAL (red) with or without bound C1q were added to HMDMs at a 5:1 ratio for 1 h. HMDMs were then stimulated with LPS for 6 h with 1 mM ATP added during the last 90 min of LPS stimulation to activate the inflammasome. Cleavage of procaspase-1 was assessed by immunostaining using a green fluorescent probe specific to cleaved caspase-1, and HMDMs were stained using a blue cell tracker (Fig. 6A). About 30–40% of LPS-stimulated HMDMs and HMDMs that had ingested LAL showed cytoplasmic cleaved caspase-1 (Fig. 6A and B). However, HMDMs that have ingested EAL, C1q-EAL or C1q-LAL showed almost no cleaved caspase-1 (the only cleaved caspase-1 signal detected in these HMDMs was associated with AL themselves) (Fig. 6A and B). This decrease was slightly enhanced when counting only those HMDMs that had ingested C1q-LAL (3-fold decrease) vs. cleaved caspase-1 in HMDMs with and without evidence of ingested C1q-LAL (2-fold decrease) (Fig. 6A and B). It is of note that while NLRP3 mRNA levels were down-regulated in HMDMs that have ingested EAL, C1q-EAL or C1q-LAL, the protein levels of NLRP3, as well as procaspase-1 and ASC, appear similar in all conditions at 6 h post LPS stimulation (Fig. 6C).
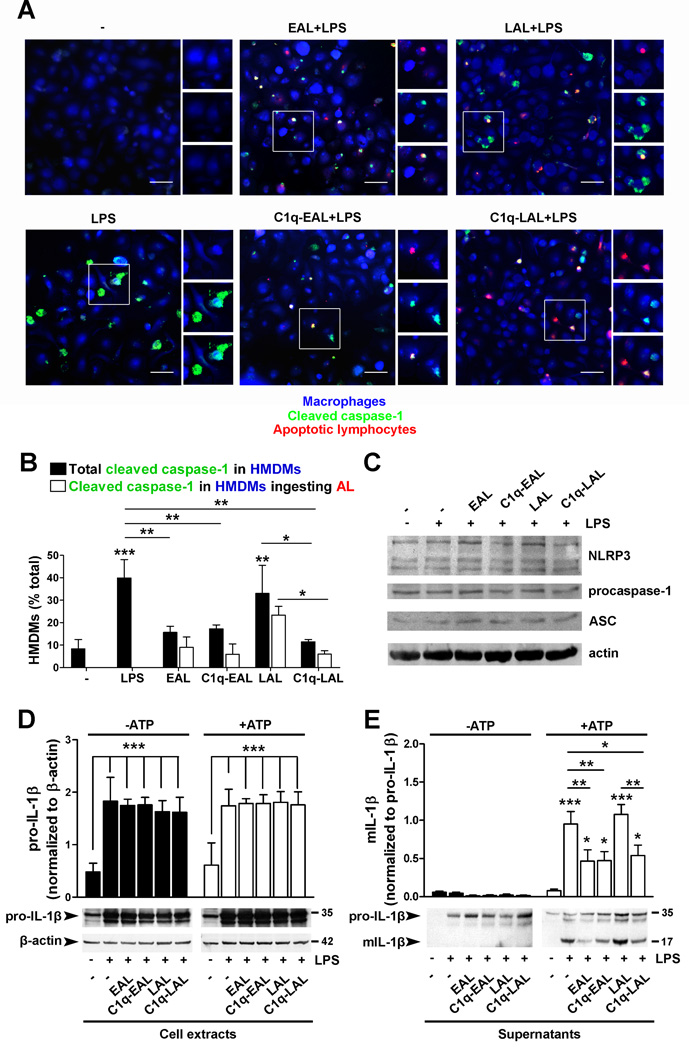
HMDMs were incubated with PKH26-labeled EAL and LAL, pre-incubated with C1q, at a 5:1 ratio for 1 h and then stimulated with 10 ng/ml LPS. (A–B) HMDMs were stimulated with LPS for 6 h. ATP (1 mM) was added 90 min before the end of the stimulation. Cleaved caspase-1 was detected by FITC fluorescent caspase-1 probes and HMDMs were stained with a blue cell tracker. Representative merged micrographs of 3 independent experiments (from 3 different donors) are shown. Scale bar = 50 µm. Areas in white boxes were enlarged to show PKH26-AL (top, red), cleaved caspase-1 (middle, green) and the merge (bottom). (B) Quantification of cleaved caspase-1 in HMDMs. (C) NLRP3, procaspase-1, ASC and actin expression in HMDM cell extracts. Representative blots of 2 independent experiments are shown. (D–E) Levels of pro-IL-1β relative to β-actin levels (D, cell extracts) and mIL-1β relative to pro-IL-1β (E, supernatants) in HMDMs stimulated with LPS for 18 h with ATP added during the last 3 h of stimulation. Representative blots of 3 independent experiments are shown. All results represent means ± s.d. (n = 3 different donors), two-way ANOVA, * p < 0.05, ** p < 0.01 and ***, p < 0.001.
Next, we examined whether the inhibition of caspase-1 cleavage by C1q resulted in decreased mature IL-1β levels released by macrophages. HMDMs were incubated with C1q bound to EAL or LAL for 1 h and stimulated with LPS for 18 h with ATP added during the last 3 h. Upon LPS stimulation, pro-IL-1β levels were similarly increased in all conditions (Fig. 6D), observations in accordance with gene expression data showing similar increased in IL-1β mRNA levels in all conditions (Table S1). No mature IL-1β levels were detected in HMDM supernatants in absence of ATP (Fig. 6E). In presence of ATP, amount of mature IL-1β released by HMDMs was significantly (p < 0.001) increased after LPS stimulation and in LPS-stimulated HMDMs that have ingested LAL compared to unstimulated HMDMs (Fig. 6E). Consistent with the observed decreased in caspase-1 cleavage, HMDMs that had ingested EAL or C1q-bound EAL or LAL showed a significant decrease (50–60%, p < 0.01) in the amount of mature IL-1β released by HMDMs (Fig. 6E). All together these results demonstrate that C1q inhibited cleavage of caspase-1 and subsequent IL-1β cleavage.
Discussion
It is now well established that C1q can play a prominent role in the clearance of apoptotic cells and facilitates the rapid removal of damaged cells, thereby avoiding the release of potentially damaging intracellular components. Data presented here show that in addition to enhancing phagocytosis of autologous apoptotic lymphocytes, C1q bound to apoptotic cell “cargo” significantly influences the responses of HMDMs in a more physiologic model than our previous studies. C1q modulated several signaling pathways, increased the expression of immunoregulatory cytokines including IL-10, IL-27, IL-33 and IL-37 and inhibited NLRP3-dependent cleavage of caspase-1 and subsequent IL-1β cleavage, potentially through increased expression of negative regulators of inflammasome activity such as NLRP12 and/or POP1/ASC2 (Fig. 7).
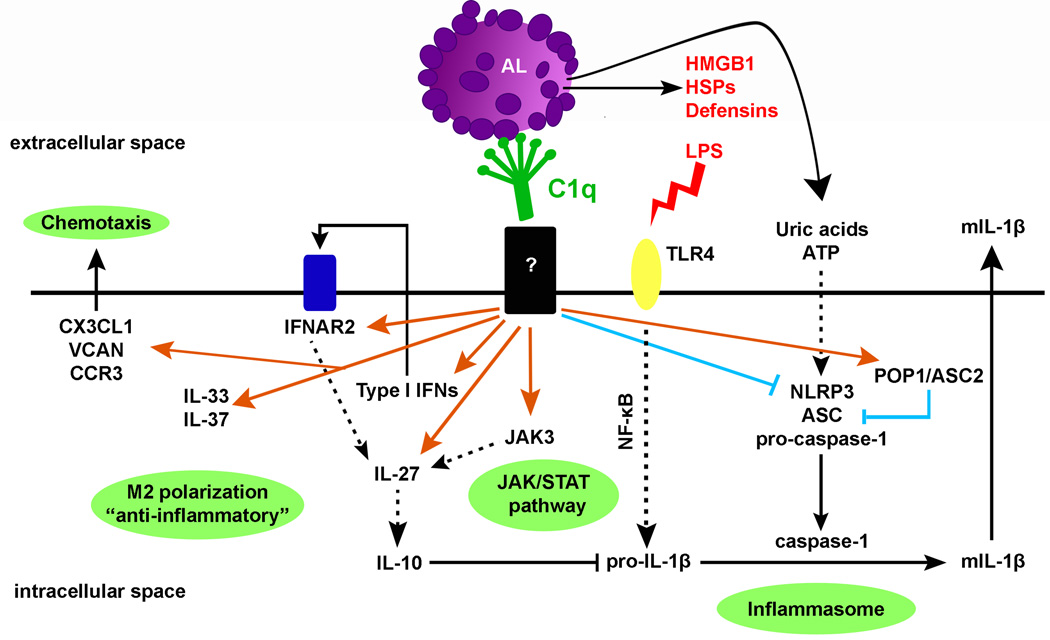
C1q increases the expression of type I IFNs and IL-27, known to act sequentially to stimulate expression of IL-10 (also up-regulated by C1q), IL-33, known to promote alternative activation of macrophages, IL-37, a potent natural suppressor of innate inflammatory responses and JAK3 that may be involved in IL-27 up-regulation. C1q also suppresses procaspase-1 and pro-IL-1β cleavage and subsequent mIL-1β release through possibly increase expression of negative regulators of inflammasome activity and indirectly (at later times) through increase IL-10 expression, which is known to decrease pro-IL-1β mRNA levels. C1q may thus prevent excessive and dysregulated inflammasome activation induced by the release of DAMPs (HMGB1, HSPs, ATP) from apoptotic cells during secondary necrosis that can activate TLR4 and the NLRP3 inflammasome. Orange arrows represent up-regulated genes and blue lines represent down-regulated genes by C1q.
Previous studies using apoptotic lymphocytic cell lines such as Jurkat cells have shown that C1q binds stably to late apoptotic cells but much less to early apoptotic cells (8,45). Our data showed that C1q, at physiological concentrations, binds directly, and to the same extent, to primary human EAL and LAL, suggesting differences between primary and transformed apoptotic cells in this regard. The fact that C1q binds strongly and stably to EAL in our system supports a central role of C1q in the rapid removal of apoptotic cells to avoid auto-immunity.
C1q enhanced the uptake of EAL and LAL by HMDMs, in accordance with numerous previous reports showing an enhancement of uptake of apoptotic cells bound to C1q by different subsets of phagocytes (5,8,45,46). While previously assumed, we report that C1q remained on apoptotic cells during the phagocytosis process, suggesting that C1q can directly interact with and signal the macrophages. C1q bound to EAL and LAL indeed profoundly affected the HMDM response to LPS, this effect being sometimes dependent on the stage of the apoptotic cell (early vs. late). It is likely that C1q engaged multiple receptors on HMDMs in addition to the engagement of other phagocytic receptors, such as Mer, SR-A or CD36 (47), by the apoptotic cells themselves thus triggering a complex signaling cascade, which remains to be fully characterized, leading to the modulation of the several different pathways identified in this study. C1q bound to EAL and LAL regulated the expression of several cytokinesin HMDMs. Particularly, C1q increased the expression of IL-33, a newly described member of the IL-1 family that can amplify M2 (alternative) polarization of macrophages induced by IL-13 (48,49), which is also induced by C1q bound to LAL. In addition, C1q bound to LAL increased the expression of IL-37 (IL1F7), a natural suppressor of innate inflammatory responses (50). In previous studies, C1q has been shown to enhance IL-10 production after LPS stimulation (8,25,46). Among myeloid cells, macrophages are the main source of IL-10, which is produced in response to TLR stimulation to limit and resolve inflammation (51). Recently, it has been shown that induction of IL-10 in LPS-stimulated murine macrophages results from the sequential induction of type I IFNs and IL-27 (43). Our data showed that C1q sequentially induces type I IFNs, IL-27 and IL-10 secretion in LPS-stimulated human macrophages. However, the induction of IL-27 by C1q was totally abolished after stimulation with higher dose of LPS, suggesting that the immunoregulatory effect of C1q occurs only when limited inflammation is present (i.e. during sterile inflammation). However, in presence of higher dose of LPS (i.e. higher TLR stimulation such as during an infection) the effect of C1q to dampen the inflammatory response is overwhelmed, a result which would be beneficial to promote the resolution of an infection. Moreover, addition of C1q tails inhibited in a dose dependent manner the C1q-induced expression of type I IFNs and IL-27, consistent with the reported binding of C1q to apoptotic cells via the globular heads, and thus orienting the “tail” domain to interact with the macrophage and induce cytokine expression in HMDMs. Finally, neutralization of type I IFNs only partially decreased the induction of IL-27 and neutralization of IL-27 moderately affected the induction of IL-10. In addition, IL-27 was also up-regulated by C1q in resting HMDMs while no increase in type I IFNs was observed in these cells. These results suggest that type I IFNs participate in the induction of IL-27 and possibly IL-10 in HMDMs stimulated with low dose of LPS, but that these pathways likely involve other partners in resting HMDMs that are still to be identified.
C1q has been shown to inhibit IFN-α production by plasmacytoid dendritic cells (pDCs) in response to stimulation by SLE immune complexes in vitro and C1q deficiency has been associated with defective regulation of IFN-α production by pDCs in SLE patients (52). This apparent opposite effect of C1q on regulation of type I IFNs production can be due to differences in the source of type I IFNs between these two systems (HMDMs and pDCs), the transient and very local induction of type I IFNs in our system vs. a systemic dysregulation effect with immune complexes and pDCs and/or the differences in the stimulus (apoptotic cells/PAMPs/DAMPs vs. immune complexes). These observations reinforce the premise that depending on the context, the stimulus and the cell type it interacts with, the outcome of the C1q response may be widely different. These differences must be considered in the development of therapeutics targeting these pathways.
Importantly, our data provides the first demonstration that C1q bound to “self-cargo” directly inhibits inflammasome activation, cleavage of caspase-1 and subsequent IL-1β processing in HMDMs. Interestingly, late apoptotic cells that exhibit decreased membrane integrity (“secondary necrosis”) release numerous DAMPs that can trigger inflammasome activation (ATP and uric acids) and/or activate TLR4 such as high mobility group box 1 (HMGB1), heat shock proteins (HSPs) and defensins (44). By directly regulating inflammasome activation, C1q may prevent excessive and dysregulated inflammasome activation induced by these DAMPs (Fig. 7). The mechanism by which C1q is modulating inflammasome activation still under investigation but one possible hypothesis is that C1q bound to LAL increases negative regulators of inflammasome activation such as POP1/ASC2 mRNA levels. POP1 (which is not encoded in the mouse genome (53)) has been shown to bind ASC and modulate NF-κB activation and procaspase-1 cleavage (54) in human macrophages. Others have reported that inhibition of inflammasome activation by autocrine IL-10 is dependent on signaling through JAK3 (55), suggesting that C1q may induce both an early direct effect on the inflammasome (at 3–6 h through regulation of ASC2 and potentially other negative regulators), and a long lasting effect via upregulation of type I IFNs (56), JAK3 and IL-10. Finally, C1q bound to EAL and LAL increased the expression of NLRP12, an important inhibitor of inflammatory gene expression in human myeloid cells through suppression of NF-κB activation (57). Taken together, these data indicate that more than one pathway is induced by C1q to direct macrophage polarization and inhibit inflammasome activation.
In summary, C1q enhanced phagocytosis of autologous apoptotic lymphocytes and significantly modulated gene expression profile and inflammasome activity of HMDMs (Fig. 7). Importantly, the predominant effect of C1q on HMDM inflammatory responses was observed when C1q is bound to late apoptotic cells since early apoptotic cells themselves seem to have direct suppressive effects especially on inflammasome activation. This suggests that C1q, in the absence of other complement proteins, is a potent immunoregulatory molecule, which contributes to containing the inflammatory response induced by secondary necrosis by both immediate effects and through induction of regulatory cytokines. This study extends our initial understanding of the consequences of C1q-macrophage interactions and identifies specific and potentially novel molecular pathways induced by C1q that suppress macrophage inflammation. These results there by identify candidate therapeutic targets to control inflammation, suppress autoimmunity (SLE) and promote host defense and/or vaccine design.
Acknowledgements
We are very grateful to healthy volunteers for donating blood and the staff of the UCI Institute for Clinical and Translational Science for obtaining blood. We thank Dr. Michael Buchmeier (University of California, Irvine) for use of confocal microscope, Michelle Yu, Nathan Yu, Elsie Jang, Anthony Chen and Sophie Chu for excellent technical help and Dr. Edward Nelson, Dr. Melissa Lodoen and members of the laboratory for engaging and helpful discussions.
This work was supported by Grant Number UL1 RR031985 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and the NIH Roadmap for Medical Research, NIH grants AI 41090 and AG 00538 and the Cypress College STEM Summer Bridge Program.
Abbreviations used
| ASC | apoptosis-associated speck-like protein |
| DAMPs | damaged associated molecular patterns |
| EAL | early apoptotic lymphocytes |
| HMDMs | human monocyte-derived macrophages |
| LAL | late apoptotic lymphocytes |
| NLRP | NLR family pyrin domain containing |
| PAMPs | pathogen associated molecular patterns |
| POP1 | PAAD-only protein-1 |
References
Full text links
Read article at publisher's site: https://doi.org/10.4049/jimmunol.1103760
Read article for free, from open access legal sources, via Unpaywall:
https://www.jimmunol.org/content/jimmunol/188/11/5682.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4049/jimmunol.1103760
Article citations
Augmenting fibronectin levels in injured adult CNS promotes axon regeneration in vivo.
Exp Neurol, 379:114877, 27 Jun 2024
Cited by: 0 articles | PMID: 38944331
Association of Complement Factors With Disability Progression in Primary Progressive Multiple Sclerosis.
Neurol Neuroimmunol Neuroinflamm, 11(4):e200270, 21 Jun 2024
Cited by: 0 articles | PMID: 38912898 | PMCID: PMC11226316
Single-cell RNA profiling reveals classification and characteristics of mononuclear phagocytes in colorectal cancer.
PLoS Genet, 20(2):e1011176, 26 Feb 2024
Cited by: 0 articles | PMID: 38408082 | PMCID: PMC10919852
Immune receptors and aging brain.
Biosci Rep, 44(2):BSR20222267, 01 Feb 2024
Cited by: 0 articles | PMID: 38299364 | PMCID: PMC10866841
Review Free full text in Europe PMC
Cellular and transcriptome signatures unveiled by single-cell RNA-Seq following ex vivo infection of murine splenocytes with Borrelia burgdorferi.
Front Immunol, 14:1296580, 08 Dec 2023
Cited by: 1 article | PMID: 38149246 | PMCID: PMC10749944
Go to all (146) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE30177
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells.
J Immunol, 183(10):6175-6185, 28 Oct 2009
Cited by: 98 articles | PMID: 19864605 | PMCID: PMC2843563
Binding of von Willebrand Factor to Complement C1q Decreases the Phagocytosis of Cholesterol Crystals and Subsequent IL-1 Secretion in Macrophages.
Front Immunol, 10:2712, 21 Nov 2019
Cited by: 10 articles | PMID: 31824501 | PMCID: PMC6881245
Aloe vera downregulates LPS-induced inflammatory cytokine production and expression of NLRP3 inflammasome in human macrophages.
Mol Immunol, 56(4):471-479, 01 Aug 2013
Cited by: 74 articles | PMID: 23911403
Caspase-11 non-canonical inflammasome: a critical sensor of intracellular lipopolysaccharide in macrophage-mediated inflammatory responses.
Immunology, 152(2):207-217, 31 Jul 2017
Cited by: 115 articles | PMID: 28695629
Review
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000153
NCRR NIH HHS (2)
Grant ID: UL1 RR031985
Grant ID: UL1 RR031985-02
NIA NIH HHS (3)
Grant ID: P01 AG000538
Grant ID: AG 00538
Grant ID: P01 AG000538-33
NIAID NIH HHS (3)
Grant ID: R01 AI041090
Grant ID: AI 41090
Grant ID: R01 AI041090-14





