Abstract
Free full text

Antiviral therapies: focus on Hepatitis B reverse transcriptase
Abstract
Hepatitis B virus (HBV) is the etiologic agent of mankind’s most serious liver disease. While the availability of a vaccine has reduced the number of new HBV infections, the vaccine does not benefit the approximately 350 million people already chronically infected by the virus. Most of the drugs approved by the FDA for the treatment of hepatitis B target the reverse transcriptase (RT or P gene product) and are nucleoside RT inhibitors (NRTIs) that suppress viral replication. However, prolonged monotherapies directed against a single target result in the emergence of viral resistance. HBV genotypic differences affect NRTI resistance, and because the reading frames of the S (surface antigen) and P genes partially overlap, genomic differences that affect the surface of the virus may also alter the viral polymerase sequence, function and drug susceptibility. The scope of this review is to assess the effects of HBV genotypic variation on the development of drug resistance to NRTIs. Some RT residues that vary among different genotypes are in the vicinity of residues that mutate and give rise to NRTI resistance. Interactions between these amino acids can help explain the effect of HBV genotype on the development of NRTI resistance during antiviral therapies, and might help in the design of improved therapeutic strategies.
1. Introduction
Hepatitis B virus (HBV) infection is a global health problem, with approximately a third of the world’s population exposed and up to 5% chronically infected (ca. 350 million people). The prevalence is highest in Africa, Asia, and in the Western Pacific. HBV is transmitted through blood and other bodily fluids, sexual contact, and through perinatal mother-to-child transmission, similar to hepatitis C virus (HCV) and human immunodeficiency virus (HIV). Co-infections by these viruses are frequent and may result in significant co-morbidities (Soriano et al., 2006). In acute HBV infection the main symptoms are liver inflammation and jaundice that may lead to chronic hepatitis, especially in younger children. The immune response causes hepatocellular damage and may eventually lead to liver cirrhosis and cancer. According to the World Health Organization, an estimated 600,000 persons die each year due to acute or chronic HBV infection. Currently, there are two FDA-approved treatment options for chronic HBV infection: interferon alpha (IFNα), and nucleos(t)ide analogs using one or more of seven approved drugs. IFNs work directly by inhibiting the synthesis of viral DNA and by activating antiviral enzymes. They also act indirectly by increasing the cellular immune responses against HBV-infected liver cells. The antiviral activity of NRTIs is based on the inhibition of the synthesis of either the negative strand or the positive strand or both strands (Figure 1).
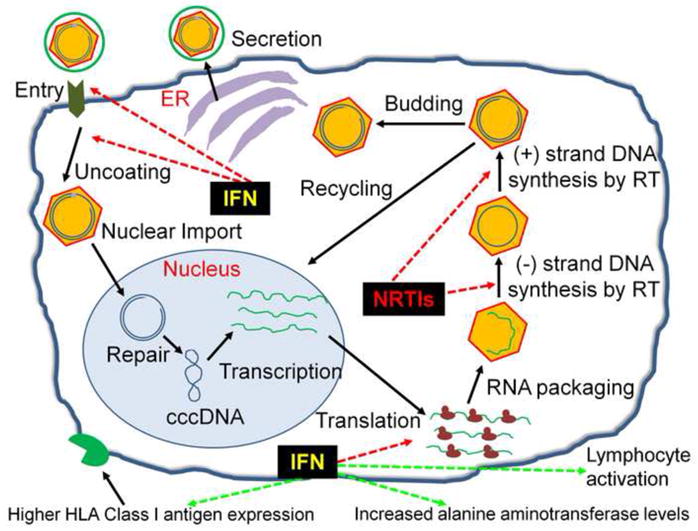
The different steps of the life cycle of HBV are represented in a simplified way. IFNs either inhibit indirectly the viral DNA synthesis (red dotted lines) or activate cellular enzymes and immune responses (green dotted lines). The NRTIs inhibit the negative and positive strand DNA synthesis.
2. HBV genome organization
HBV is the prototype member of Hepadnaviridae. It has a partially double-stranded circular DNA genome of ~3,200 bases with four overlapping open reading frames (ORFs): P (DNA polymerase), pre-C/C (pre-core/core), pre-S/S (surface proteins), and X (transcriptional co-activator) (Figure 2). The pre-S/SORF encodes the three viral surface proteins that surround the nucleocapsid (HBV surface antigen - HBsAg). The pre-S/S ORF is contained completely within the P ORF but is translated in a different reading frame. ORFs C and X overlap ORF P by 1/4 and 1/3 of their respective sequence lengths (Miller et al., 1989). HBV replicates through RNA intermediates used by protein P to produce complementary DNA (cDNA). Protein P consists of four domains: a terminal protein (TP) that is covalently linked to the DNA primer during negative-strand DNA synthesis, a spacer domain that is tolerant to mutations, the reverse transcriptase (RT) domain, and the ribonuclease H (RNase H) domain. The RT and RNase H domains have sequences highly conserved among proteins with similar enzymatic functions, including HIV reverse transcriptase (Das et al., 2001, Bartholomeusz et al., 2004). The high error rate of HBV RT results in many genomic substitutions during viral replication, as also occurs for HIV.
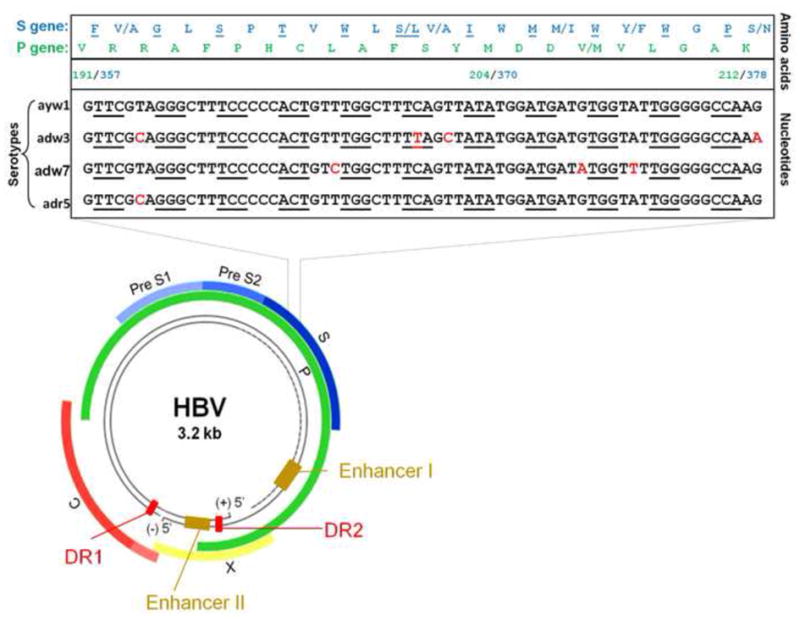
The genome of approximately 3,200 nucleotides is divided into four partially overlapping open reading frames (ORFs). Colored boxes represent protein coding regions. These are abbreviated as C for the core, P for the polymerase gene, X for the x gene, and for the envelope genes pre-S1 and pre-S2 and surface S. The positions of the two enhancers, as well as the positions of the direct repeats, DR1 and DR2, are indicated. The magnified area represents part of the overlapping region of the P and S genes. The letters in red indicate nucleotide variations among different serotypes. Every other codon of the S gene overlapping with the P gene is underlined. The amino acid sequences encoded by the S and P genes are shown in blue and green, respectively.
3. HBV serotypes and genotypes
Mutations affecting the antigenic characteristics of the virus have given rise to four major serotypes (adw, adr, ayw, ayr) (Le Bouvier et al., 1972, Norder et al., 1992a). Part of the S protein is the immunological “a” determinant, which is conserved in all the serotypes and is the target of vaccine-elicited antibodies. While serotypic classification groups viruses based on differences in their surface proteins (amino acid sequence), genotypic classification categorizes viruses based on their DNA sequence divergence throughout the entire genome. Because of technical advances in sequencing and bioinformatic technologies as well as increasing availability of viral genomic sequences, genotypic classification of viruses has now become routine. Hence, based on whole-genome sequences, at least ten major HBV genotypes have been defined (A through J), which have variations from each other in at least 8% of their whole genomic sequences (Okamoto et al., 1988, Norder et al., 1992a, Norder et al., 1994, Schaefer, 2007). Each genotype has numerous sub-genotypes. The relation between serotypes and genotypes has been studied extensively (Norder et al., 1992a, Norder et al., 1992b, Ohba et al., 1995, Stuyver et al., 2000a, Liu et al., 2002, Devesa et al., 2004, Norder et al., 2004, Sugauchi et al., 2004, Kay and Zoulim, 2007). Although serotypes adr and ayr are generally encountered in genotype C, serotype ayw is very rare in genotype C but present in all other genotypes. Finally, serotype adw is found in all genotypes except D and E (Shiina et al., 1991, Kay and Zoulim, 2007).
Figure 3 illustrates the geographical distribution of the main HBV genotypes. HBV genotypes have been associated with variable clinical outcomes and different responses to IFNα and NRTI treatments that are discussed below (Chien et al., 2003, Hsieh et al., 2009, Chen et al., 2011, Lin and Kao, 2011). Since the S and P gene sequences partially overlap with each other but are translated in different reading frames, single nucleotide changes among different HBV genotypes may or may not affect the amino acid composition of both gene products (Figure 2) (Mizokami et al., 1997). The importance of HBV genotypic differences in the mechanism of viral DNA synthesis or for NRTI resistance has been elusive and is discussed in the last section of this review.
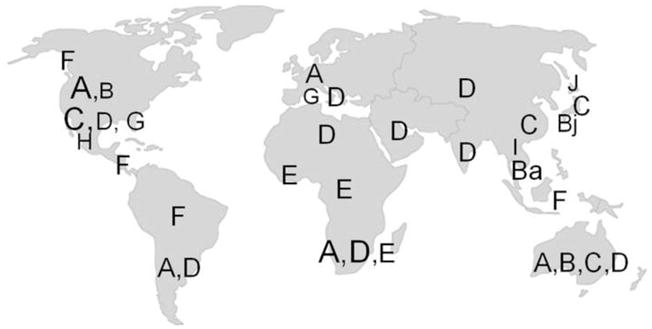
The predominant genotypes of regions of the world are shown in larger font sizes.
Furthermore, due to the partial overlap of P and S ORFs, NRTI-induced mutations on the polymerase gene may result in sequence and structural changes in the surface antigen (HBsAg) (Figure 2) (Torresi, 2002, Kamili et al., 2009). At the same time some of the changes in the surface genes may alter critical functions of the HBV envelope proteins, thus affecting the replication ability and infectivity of the virus (Villet et al., 2009). These events may be linked to the emergence of drug-resistant variants during antiviral therapy (Litwin et al., 2005, Villet et al., 2009, Billioud et al., 2012). Recently, Svicher et al. reported the synergistic effect of the genetic barrier and the S/P overlap on the development of drug resistance and immune escape (Svicher et al., 2011). The selection of a long-term therapy with a high barrier to resistance can determine the success of this therapy (Gish et al., 2012).
3.1 HBV genotypes and treatment with interferon alpha
Several studies have shown that differences in HBV genotype affect the response to IFNα-based treatment. Zhang et al. reported that the response to IFNα treatment was higher in patients infected with genotype A relative to patients infected with genotypes D or E (70% vs. 40%) (Zhang et al., 1996). Two other studies reported that response to IFNα was higher in patients infected with genotype B rather than genotype C (41% vs. 15%) (Kao et al., 2000, Wai et al., 2002). Other studies have shown that interferon-treated patients infected with genotype A HBV had higher rates of HBeAg seroconversion (a marker that is usually associated with a decline in viral replication) than patients infected with genotypes D or C (Erhardt et al., 2000, Janssen et al., 2005). Hou et al. reported recently that genotype A has a better response to short 16-week IFNα therapy than other HBV genotypes and also represents an independent predictor of sustained HBeAg clearance. The higher response rate of genotype A has been reported to be based on its molecular characteristics, particularly the ability to develop core promoter mutations (A1762T/G1764A) and to have the lowest variations in the core gene (Hou et al., 2007). It is likely that genotypic differences in the HBsAg surface protein may have an impact on immune recognition and virulence. The molecular basis of how genotypic differences in HBV affect the response to IFNα therapy requires further investigation.
4. Viral Reverse Transcriptases
The viral reverse transcriptase (RT) has two activities: a DNA polymerase activity that uses either RNA or DNA as a template and an RNase H activity that degrades RNA in RNA/DNA hybrids. RTs do not possess proof-reading activity and during viral replication their error-prone DNA synthesis results in enhanced mutation frequency and the production of virus variants. Over twenty years of research has established RT as the main target of antivirals for the treatment of HIV-1 and HBV. FDA-approved drugs that target HIV-1 RT are either a) nucleoside (or nucleotide) reverse transcriptase inhibitors (NRTIs) that mimic the natural nucleosides for incorporation to the elongating DNA chain, or b) non-nucleoside RT inhibitors (NNRTIs) that bind to a hydrophobic pocket close to the RT active site. Because NRTIs and NNRTIs bind at different sites their resistance profile rarely overlaps and can thus be used in combination therapies called highly active antiretroviral therapies (HAART) that also include inhibitors of other steps of the HIV-1 life cycle, such as protease, integrase, entry, or fusion inhibitors. In the case of HBV, combination therapies which use different NRTIs with and without INFα are under investigation. Moreover, combination of different NRTIs is an important option for rescue therapy. In addition, combination therapy is used successfully in HIV therapy. Clearly, new antivirals that block other functions of HBV or that target novel binding sites are needed to improve the existing therapeutic regimens.
5. NRTIs
Of all the FDA-approved drugs for treatment of HBV infection, IFNα-2b was the first licensed IFNα. The pegIFNα-2a and pegIFNα-2b are also now available for the treatment of HBV. IFNα boosts the antiviral immune response and has been reviewed elsewhere (Perrillo, 2009, Bhattacharya and Thio, 2010, Dienstag, 2008). The others are NRTIs that suppress viral replication and are the focus of this review, including lamivudine (LVD; also known as 3TC, the [–]-β-L-stereoisomer of 2′,3′-dideoxy-3′-thiacytidine), adefovir (ADV), entecavir (ETV), telbivudine (LdT), and TDF (Figure 4). Other β-L-NRTIs that inhibit HBV are emtricitabine (FTC), a fluorinated analog of LVD, valtorcitabine (LdC), also an analog of deoxycytidine, and LdT.
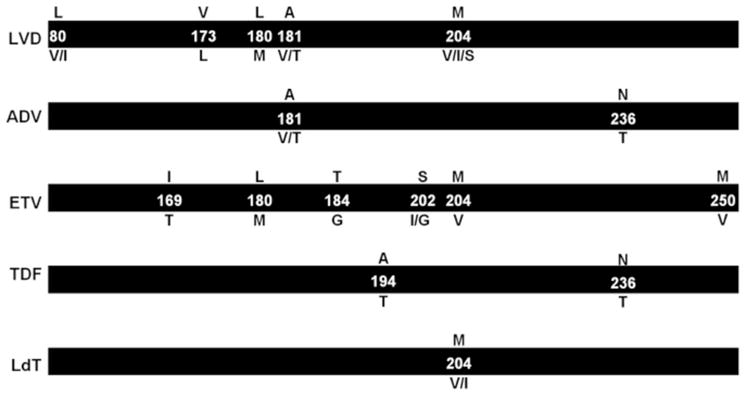
The major resistance mutations for each inhibitor are shown in boxes.
Most NRTIs are administered as unphosphorylated prodrugs and are phosphorylated to their active triphosphate form by cellular kinases. ADV and TDF are both acyclic nucleotides and do not have a sugar or pseudosugar ring but contain a phosphonate group replacing the α-phosphate, and hence require the addition of only two phosphate groups for activation. The activated NRTIs compete with natural nucleotide substrates for incorporation into the growing DNA chain. Once incorporated, they act as chain-terminators due to their lack of a 3′-OH (Mitsuya et al., 1987). An exception is ETV, which has a 3′-OH and appears to act as a delayed chain-terminator by being incorporated into the DNA primer (DNAETV) and by causing subsequent dissociation of DNAETV from the polymerase (Seifer et al., 1998, Langley et al., 2007, Tchesnokov et al., 2008).
Prolonged use of NRTIs leads to the emergence of HBV strains with RT mutations that confer resistance and eventually lead to treatment failure (Lok et al., 2007, Zoulim and Locarnini, 2012). Typically, resistance mutations selectively decrease the incorporation of NRTIs without significantly affecting the incorporation of natural dNTPs. Currently there is a universally accepted numbering nomenclature for the HBV RT mutations which is genotype-independent (Stuyver et al., 2001). The NRTIs used for treatment of hepatitis B and the associated resistance mutations in the HBV polymerase are discussed below and summarized in Figures 4 and and55 (Ghany and Liang, 2007, Tenney et al., 2007, De Clercq et al.).
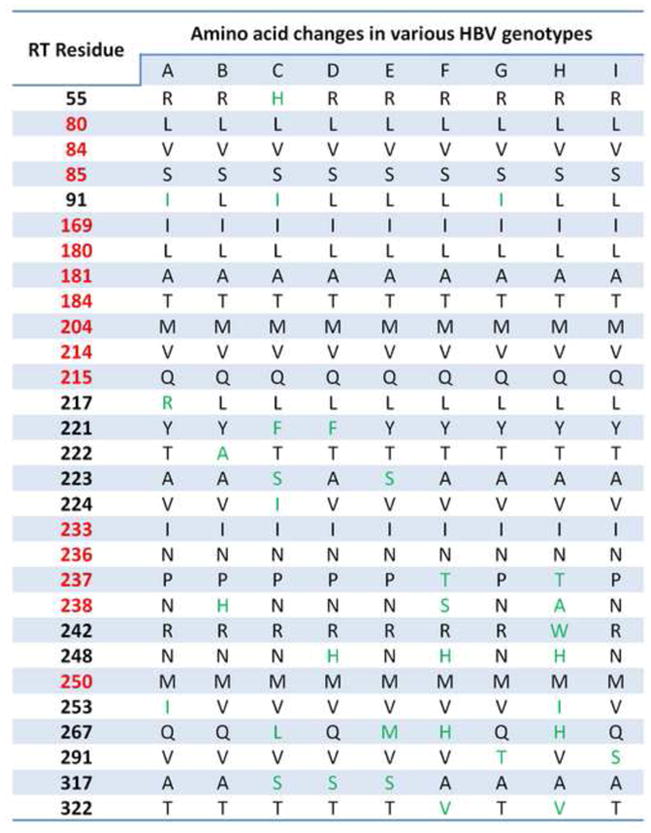
DNA consensus sequences of genotypes A, B, C, D, E, F, G, H, and I were constructed according to the HBV RT Stanford Database (Rhee et al., 2010). Residues in green are genotypically variable in treatment-naïve patients and may have an effect on NRTI drug resistance because of three dimensional proximity to residues that are mutated in response to NRTI treatment (shown in red) (Michailidis et al., 2011).
5.1 Lamivudine (LVD)
LVD was the first safe, effective, and well-tolerated oral medication for the treatment of HBV infection (Dienstag et al., 1995, Dienstag et al., 1999, Lai et al., 1998, Liaw et al., 2000, Liu and Schinazi, 2002). Both LVD and TDF are approved for use in HBV and HIV co-infected individuals. Similar to LVD, FTC also is an L-nucleoside analog of dC with a 5-fluoro substitution. FTC has been approved for the treatment of HIV and is in clinical trials for the treatment of HBV infection. LVD significantly suppresses HBV viral load, reduces liver inflammation, and improves certain laboratory markers. However, prolonged treatment with LVD leads to the emergence of resistant HBV viruses (and resistant HIV viruses, in the case of co-infected patients).
LVD resistance occurs in approximately 20% of HBeAg-positive patients after one year and up to 70% after five years (Lok et al., 2003, Pawlotsky et al., 2008). Resistance is conferred by a single mutation, M204V/I/S, within the highly conserved YMDD motif at the catalytic center of the polymerase (Locarnini and Mason, 2006, Bozdayi et al., 2003). M204I can be found by itself, whereas M204V/S mutations are always associated with other changes, in particular with mutation L180M (Figures 4–6) (Bartholomew et al., 1997, Ling et al., 1996, Das et al., 2001, Delaney et al., 2001, Shaw et al., 2006, Ono et al., 2001), which has been reported to partially restore replication fitness of M204V/I/S viruses and to enhance resistance in in vitro studies (Das et al., 2001, Langley et al., 2007). In addition, mutation A181T can confer resistance to LVD (Yeh et al., 2000). Substitutions L80V/I and V173L have a compensatory role (Ogata et al., 1999, Delaney et al., 2003). Specifically, the V173L mutation is associated with resistance to LVD and is invariably found in the background of L180M and M204V (Delaney et al., 2003). It is a compensatory mutation that does not affect the sensitivity of HBV RT to LVD or ADV (Delaney et al., 2003). Similar to HBV, HIV acquires resistance to LVD and FTC in the presence of the M184V/I at the equivalent YMDD motif of the HIV reverse transcriptase, and the mechanism of resistance involves steric hindrance between the oxathiolane group of the inhibitor and the beta branched side chain of Val or Ile at position 184 (Sarafianos et al., 1999, Chong et al., 2003).
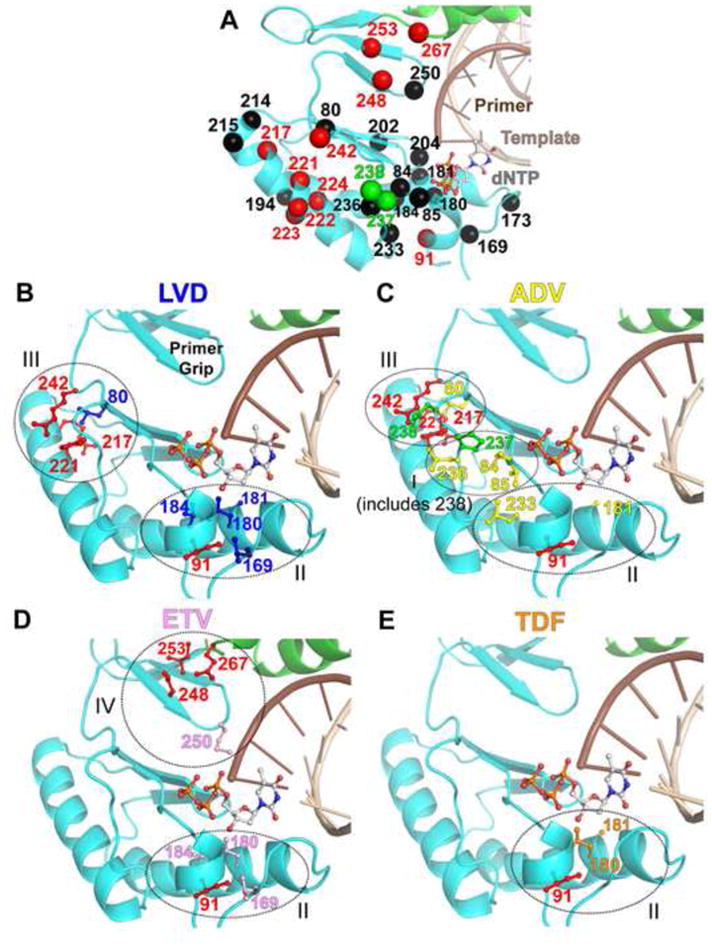
Homology models were built as described in Das et al. and Michailidis et al. (A) Overview of genotypic variability (red) or NRTI resistance (black) residues at the HBV RT active site. Residues that vary among different genotypes and appear also as NRTI resistance residues (either primary or secondary mutations) are shown in green. (B–E) The positions that are different among various genotypes and are close to NRTI-resistance residues are shown in red sticks. In panels B, C, D, and E residues involved in resistance to LVD, ADV, ETV, and TDF are shown in blue, yellow, pink, and orange, respectively. The model for LdT is similar to TDF and therefore has been omitted. These resistance residues may be at interacting distances from residues that vary among different genotypes. In panel C residues 237 and 238, which are both ADV-resistant mutations and genotypic variants, are shown in green. Other residues are numbered accordingly. Circles indicate the general location of clusters I–IV. In panel C, cluster I includes residue 238 and cluster III includes residues 217, 221, 242, and 80 (Michailidis et al., 2011). NRTI resistance residues not in proximity to genotypic variability residues are omitted from this figure for clarity, but are shown in panel A.
5.2 Adefovir (ADV)
At pharmacologically tolerated levels, ADV is a potent inhibitor of HBV replication but is only a weak inhibitor of HIV replication. Consequently, ADV has been approved by FDA only for the treatment of HBV infection. Early studies suggested ADV suppressed wild-type and LVD-resistant HBV (Benhamou et al., 2001, Delaugerre et al., 2002, Westland et al., 2003a, Westland et al., 2003b, Westland et al., 2005) and that resistance mutations were rare (Villeneuve et al., 2003). After a five-year period an estimated 29% of treated patients were reported to develop ADV resistance as compared to 70% for LVD (Borroto-Esoda et al., 2007). However, other studies revealed that as many as 50% of patients on ADV therapy fail to achieve adequate viral suppression (Villeneuve et al., 2003) and that high levels of ADV resistance occur after 1–2 years of treatment (Fung et al., 2006, Lee et al., 2006b). Liu et al. reported that patients with LVD-resistant mutations treated for 2–5 months with ADV and LVD achieved improved rates of viral suppression but did not improve biochemical indicators of liver health (Liu et al., 2006). A more recent study demonstrated that virological suppression by ADV is not ideal in the majority of LVD-resistant patients. However, early treatment by ADV when HBV DNA is low is important to maintain virological suppression (Chan et al., 2007).
The main resistance mutations in HBV polymerase associated with ADV resistance occur at codons 181 and/or 236 (A181V, A181T, and N236T, Figures 4–6) (Lacombe et al., 2006, Osiowy et al., 2006). Interestingly, despite the seemingly strong structural similarity between HIV and HBV polymerases (Das et al., 2001) resistance to ADV is conferred by mutations at structurally distinct parts of the two enzymes. The HBV resistance mutations are located in the palm subdomain of the enzyme, whereas HIV resistance mutations are positioned in the fingers subdomain (Das et al., 2001, Foli et al., 1996). When ADV is administered in combination with LVD to patients with preexisting LVD-resistant viral strains, resistance to both drugs develops (Yim et al., 2006). Cell culture studies indicate that N236T and A181V mutations do not confer cross-resistance to ETV (Figure 4) (Villeneuve et al., 2003, Langley et al., 2007).
5.3 Tenofovir (TDF)
TDF has been approved for the treatment of HIV infections and is known also to inhibit HBV polymerase. Jain et al. showed that combined LVD/TDF therapy suppresses synthesis of HBV DNA more effectively than either alone (Jain et al., 2007). More patients infected with HBV genotype A responded to TDF-based therapy better than the patients infected with non-A genotype HBV, regardless of therapeutic regimen or compliance, or prior antiretroviral treatment for those with HIV co-infection (Jain et al., 2007). In vitro drug combination studies have revealed that TDF has an additive effect when combined with LVD, ETV, or LdT (Zhu et al., 2009). However, in Jain et al. the patients were HBV/HIV co-infected and so far LVD/TDF combination is not recommended as first-line therapy in HBV mono-infected patients.
The HIV resistance profile of TDF (single codon mutation at fingers subdomain residue 65 or 70 of HIV RT) is distinct from the HIV resistance profile of LVD and FTC (M184V/I mutation at the YMDD motif of HIV RT). Hence, the TDF/FTC (Truvada) combination has been used successfully to treat HIV. Similarly, because of the differences in the resistance profiles of TDF and LVD, the TDF/LVD combination is being tested in HBV-infected patients and has been reported to be effective against wild-type and LVD-resistant HBV (Nelson et al., 2003, de Vries-Sluijs et al.). Moreover, the selection of HBV TDF resistance mutations in HBV/HIV co-infected patients treated with TDF was rare in the first 12 months. However, after 12 months a novel mutation appeared at position 194 of HBV RT, a site distal to the catalytic site of the polymerase. The A194T mutation confers reduced susceptibility to TDF in the presence of LVD-associated mutations and may affect DNA polymerization by altering the position of the DNA template relative to the dNTP binding site (Sheldon et al., 2007).
5.4 Entecavir (ETV)
ETV has several distinct advantages over LVD and ADV: it is the most potent inhibitor of HBVRT; it inhibits both wild-type and LVD-resistant HBV; it is not associated with any major adverse effects; and it has limited potential for development of resistance. Unlike LVD and ADV, ETV has a 3′ OH and appears to stop elongation after the incorporation of a few nucleotides (Langley et al., 2007). In clinical trials, ETV was superior to LVD in all primary endpoints in both nucleoside-naïve and LVD-refractory HBeAg-positive and HBeAg-negative patients. Long-term monitoring showed that ETV resistance in nucleoside-naïve patients is rare through 5 years of therapy (Tenney et al., 2009). However, in LVD-experienced patients who switched to ETV treatment, the 5-year cumulative probability of genotypic ETV resistance and genotypic ETV resistance associated with breakthrough were 51% and 43%, respectively (Tenney et al., 2009).
Although ETV is structurally different from LVD and LdT, ETV-resistant HBV viruses have combinations of mutations that include LVD resistance mutations M204V, L180M, I169T, and V173L (Figures 4–6). As shown in Figure 7, residue 204 is affected by a network of interactions of a region under the β-sheet adjacent to residue 204. This also has been observed in HIV-1 RT where changes in 182–184 interactions are observed in different complexes (Hachiya et al., unpublished). Such changes are likely to affect the position of residue 204 in the YMDD motif, which is known to change conformation during the course of DNA polymerization and recognition of NRTIs and NNRTIs (Sarafianos et al., 2002, Sarafianos et al., 2009). In ETV-resistant viruses, residues 180, 184, 202, and 204 are primarily mutated to hydrophobic residues (L180M, T184L, S202G/I, M204V), which may stabilize the YMDD motif in a conformation that disfavors ETV binding (Figure 7A) (Langley et al., 2007). Interestingly, a mutation to cysteine at position 202 may afford additional stability to the YMDD motif through a hydrogen bond with the main chain of V204. The S202C mutation does not appear to cause viral breakthrough in ETV-treated HBV patients in the background of LVD resistance (Han et al., 2011, Mukaide et al., 2010).
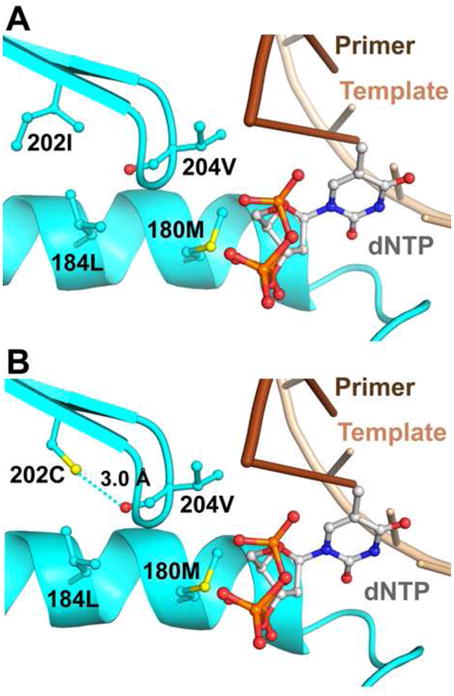
ETV-resistance mutations at 180, 184, 202, and 204 are shown as cyan sticks. In panel A, mutation of 202 to the hydrophobic residue I (or also to G, not shown) results in no interaction with residue 204. Panel B shows the hydrogen bond that forms between C202 and the main chain of V204, creating additional stability of the YMDD motif.
ETV is not recommended for HIV/HBV co-infected patients because it can lead to the emergence of the HIV M184V mutation (McMahon et al., 2007, Jain and Zoellner, 2007).
5.5 Telbivudine (LdT)
LdT is an orally administered nucleoside analog, approved for the treatment of chronic hepatitis B, with good tolerance, lack of mitochondrial toxicity, and no dose-limiting side effects. As an L-nucleoside, its resistance profile is similar to LVD although only the M204I mutation emerged in clinical trials (Ghany and Liang, 2007, Fung et al., 2006) (Figure 4).
Similarly, in vitro studies have shown that unlike LVD, M204V does not impart resistance to LdT; however, LdT is inactive against viruses that contain M204I or M204V/L180M mutations (Seifer et al., 2009). Although it is more potent than LVD and ADV, LdT is cross-resistant with LVD and engenders a high rate of drug resistance. A 2-year phase III GLOBE trial with chronically infected hepatitis B patients indicated that LdT is more effective than LVD (Liaw et al., 2009).
6. Structural basis of HBV resistance
6.1 HBV genotype and treatment with NRTIs
There is increasing evidence that HBV genotypic differences affect patient response to NRTI-based therapies. In a study that followed LVD-treated patients for 5 years or longer, there was virological and biochemical response in 67% of patients infected with genotype B and in 54% of patients infected with genotype C (Kumada, 2003). In a separate study, Kao et al. reported significant differences in the response to LVD treatment among patients infected with genotype B compared to those infected with genotype C (23% vs. 11%) (Kao et al., 2002). Similarly, Chien et al. reported that the sustained response to LVD was much higher in patients infected with genotype B compared with genotype C (61% vs. 20%) (Chien et al., 2003). On the other hand, Akuta et al. did not find significant differences in appearance of YMDD mutations in patients infected with genotype A, B, or C. However, they reported significantly more YMDD mutations in HBV Ba, rather than Bj (Akuta et al., 2003). Fung et al. found that patients with genotype B HBV were more likely to sustain their response when treatment was discontinued (Fung et al., 2004). A study of 78 German patients reported that LVD-resistant mutants emerged more rapidly in those with genotype A than D (54% vs.8%) (Zollner et al., 2004). A recent study showed that genotype B was significantly associated with development of LVD resistance within the first 12 months of LVD therapy compared with genotype C (Hsieh et al., 2009). A four-year study showed recently that LVD-ADV combination therapy was more effective in HBV patients with genotype B (Inoue et al., 2011).
Due to the partial overlap of the polymerase and surface ORFs, in some cases NRTI treatment results in the development of RT mutations that are often associated with substitutions in the “a” determinant (amino acids 124–147 on the small HBsAg protein) with serious implications for the public health (Locarnini and Yuen, 2010, Teo and Locarnini, 2010). Specifically, the triple LVD-resistant RT mutant V173L/L180M/M204V, also causes 2 amino acid substitutions in the overlapping S gene (E164D and I195M), which are responsible for decreasing binding to anti-HBs (Torresi et al., 2002). In addition, Sheldon et al. reported that mutations in the surface protein selected by LVD therapy are more common in genotype A than genotype D (Sheldon et al., 2007).
Recently, Damerow et al. demonstrated that HBV genotypes are significantly different in their mutation pattern of lamivudine resistance (Damerow et al., 2010).
Hence, genotypic differences clearly affect the outcome of HBV therapies by mechanisms that are under investigation and not completely understood.
6.2 Correlation of NRTI resistance and genotype
The crystal structure of HBV polymerase has not yet been solved. However, based on the similarities between the HIV and HBV enzymes, we constructed a three-dimensional homology model (Das et al., 2001) that helped rationalize the molecular basis of NRTI resistance in HBV by extrapolating the extensive structural and biochemical studies with NRTIs that target HIV. With this model, we proposed that resistance to LVD and FTC is caused by steric conflict between the Cγ2-methyl group of V204 (or I204) in HBV polymerase and the sulfur atom of the oxathiolane ring of these drugs (Das et al., 2001, Sarafianos et al., 1999). The effects of the L180M mutation, which also occurs near the HBV polymerase active site, appeared to be less direct, potentially involving rearrangement of the deoxynucleoside triphosphate binding pocket residues (Das et al., 2001).
Similarly, Langley et al. predicted that the exocyclic alkene moiety of ETV is accommodated in a hydrophobic pocket close to the dNTP binding site of HBV RT. This pocket is formed by residues A87, F88, P177, L180, and M204, along with the nucleotide at the 3′ end (+1 position) of the primer DNA strand. It was also suggested that ETV is inactive against human polymerase β as the same hydrophobic pocket is not present in this enzyme. LVD resistance mutations M204V and L180M, in addition with changes in T184, S202, and M250, that affect the positioning of the YMDD motif result in a reduction of susceptibility to ETV (Langley et al., 2007).
We have recently reported that HBV RT residues involved in NRTI resistance are proximal in primary and in some cases in tertiary structure to residues that vary among HBV genotypes. This observation offers novel insights into possible molecular interactions that may cause differences in viral resistance to NRTIs among various HBV genotypes (Michailidis et al., 2011).
In Figure 6 we show the side chains of residues involved in resistance to individual NRTIs (Michailidis et al., 2011). We also highlight in each case the positions that vary among different HBV genotypes (Michailidis et al., 2011). These data are based on consensus sequences of each genotype from the HBV RT Stanford database (http://hivdb.stanford.edu/HBV/DB/cgi-in/MutPrevByGenotypeRxHBV.cgi) (Rhee et al., 2010). Remarkably, several of the resistance mutation residues are in the proximity (in primary and/or tertiary structure) of residues that vary among different genotypes (Figure 6). Such proximity may contribute to the observed variability in the outcome of NRTI therapies among patients infected with various HBV genotypes. Below, we briefly discuss four regions of HBV RT where residues involved in NRTI resistance are clustered with residues that vary among genotypes (Michailidis et al., 2011).
Cluster I comprises residues 221–224, 236–238, and 84–85
Residues 236–238 (Figure 6, panel C)
The primary ADV mutation at position 236 (N236T) (Angus et al., 2003, Villeneuve et al., 2003), together with residues P237 and N238 which have been described in some patients as ADV-resistance mutations P237H (Bartholomeusz and Locarnini, 2006), and N238H (Bartholomeusz and Locarnini, 2006) define a hotspot of genotypic variability. Specifically: a) residue 237, is proline (P) in all genotypes except in F and H, where it is a more flexible threonine (T); b) residue 238, is an asparagine (N) in all genotypes except for genotype B where it is a histidine (H), genotype F where it is a serine (S), and genotype H, where it is an alanine (A); and c) residue 242, which is an arginine (R) in all genotypes except for genotype H where it is a tryptophan (W). However, some of these substitutions are not widely accepted as resistance mutations but as polymorphisms like N238H (Zhong et al., 2012).
Four other residues vary among different genotypes and are relatively close to residue 236 in the molecular model: a) residue 224, which is usually an isoleucine (I) in genotype C, and valine (V) in all the other genotypes; b) residue 223, which is usually a serine (S) in genotypes C and E, and an alanine (A) in the other genotypes; c) residue 222, which is an alanine (A) in genotype B and threonine (T) in all other genotypes; and d) residue 221, which is a phenylalanine (F) in genotypes C and D and a tyrosine (Y) in all other genotypes. In the molecular model, the hydroxyl group of Y221 forms a hydrogen bond with the carboxamide side chain of residue 236. This interaction would not be present in F221 (genotypes C and D). Because residue 221 has the potential to interact with residues from clusters I and III, we discuss it as part of both clusters. For clarity, residues 222, 223, and 224 are not shown in Figure 6. Secondary mutations that have been also detected in patients receiving ADV treatment are mutations V84M (Bartholomeusz and Locarnini, 2006) and S85A (Bartholomeusz and Locarnini, 2006, Mitsui et al., 2010). These mutations are in the vicinity of residues 237–238 that vary among genotypes (see above) and also close to the I233V (Liu et al., 2010, Schildgen et al., 2006, Chang and Lai, 2006) ADV-resistance site, which is close to residue 91 that varies among different genotypes (233 and 91 are in cluster II).
Cluster II comprises residues 169, 180, 184, 233, and genotypically variable residue 91
Residue 169 (Figure 6, panels B, D)
Mutation of this residue to a threonine (I169T) causes resistance to ETV and LVD (Tenney et al., 2004). 169 is close to residue 91 which is an isoleucine (I) in genotypes A, C, and G and a leucine (L) in all other genotypes.
Residues 180, 181, and 184 (Figure 6, panels A, B, C, D, E)
The L180M, A181V/T, and T184G/L mutations affect resistance to multiple NRTIs (Figures 4–6). They are close to each other and also close to residue 91, which also may interact with the ETV resistance residue 169, as local structural changes may affect their function.
Cluster III comprises the LVD compensatory mutation L80V/I which has also been associated with poor response to ADV (Lee et al., 2009), and genotypically variable residues 217, 242, 221 and 214–215
Residue 80 (Figure 6, panels B, C)
The L80V/I mutation residue is close to two residues that are different in various genotypes: a) residue 217 is a leucine (L) in all genotypes except for genotype A where it is an arginine (R); b) residue 242 is an arginine (R) in all genotypes except for genotype H where it is a tryptophan (W); c) residue 221, which was also discussed in Cluster I.
Residue 214 (not shown)
Substitution of this residue to an Alanine (V214A) is a secondary ADV and TDF mutation. It is close to residue 217 which is a leucine (L) in all genotypes except for genotype A where it is an arginine (R). Residue 215 (not shown). The LVD, ADV, and TDF secondary mutation Q215S is also close to residue 217. Therefore, genotypic differences in residues of this cluster potentially could alter the role of compensatory mutation L80V/I in resistance to LVD and ADV. However, like other RT substitutions Q215S has been also reported to be a polymorphism rather than a drug-resistance mutation (Amini-Bavil-Olyaee et al., 2009).
Cluster IV comprises ETV-resistance residue 250 and genotypically variable residues 248, 253, and 267
Residue 250 (Figure 6, panel E)
Mutation of this residue to a valine (M250V) causes resistance to ETV (Tenney et al., 2004, Baldick et al., 2008). This residue is part of the “primer grip”, a structural element that helps position the primer strand at the polymerase active site, and is also proximal to residues that differ among various genotypes: a) residue 248 is an asparagine (N) in most genotypes except for genotypes D, F, and H where it is typically a histidine (H); b) residue 253 is usually an isoleucine (I) in genotype A and H and a valine (V) in all other genotypes; c) residue 267 is a glutamine (Q) in genotypes A, B, D, G, and I, a leucine (L) in genotype C, a methionine (M) in genotype E, and a histidine (H) in genotypes F and H. The general proximity of residues 248, 253, and 267 to 250 opens the possibility that their interactions differ among genotypes, thereby having the potential to affect the resistance function of 250.
In addition to these residues, that are proximal to those involved in drug resistance, we have also identified several additional residues that may indirectly contribute to drug resistance by having an effect on the binding of the nucleic acid (Michailidis et al., 2011). Specifically, residue 55 is part of the palm subdomain of RT and is usually an arginine (R) except in genotype C where it is a histidine (H). Other residues are located in the thumb subdomain and may affect nucleic acid binding: a) residue 267 that we described earlier, b) residue 291 which is a valine (V) except for genotypes G and I where it is a threonine (T) or serine (S), respectively, c) residue 317 which is usually an alanine except for genotypes C, D and E where it is a serine (S); residue 322 is usually a threonine (T) except for genotypes F and H where it is a valine (V).
Despite the importance of the interaction between residues of drug resistance and residues that vary among different genotypes it is important to highlight the fact that many resistance mutations appear as secondary or compensatory mutations and they are not selected alone.
7. Detection of genotypic polymorphisms
There are several methods to study HBV genotypic polymorphisms and their effect on NRTI drug resistance. Specifically, direct DNA sequencing provides accurate and complete information of the HBV DNA sequences but it is time-consuming and labor intensive (Sablon and Shapiro, 2005, Clarke and Bloor, 2002). In addition, this method is not very sensitive for the identification of drug resistance mutations in mixtures of wild-type and mutant viruses (Aberle et al., 2001). In order to overcome this problem clonal analysis by sequencing multiple clones from a given sample has been used. However, a large number of clones must be analyzed for the identification of minor subpopulations (Zoulim, 2002, Stuyver et al., 2000b). Other methods that are being used to identify HBV drug resistance mutations involve DNA hybridization techniques (Cane et al., 1999, Whalley et al., 2001), fluorescence (Hall et al., 2000, Bai et al., 2003) and MALDI-TOF (Murray, 1996, Hong et al., 2004) technologies, oligonucleotide microarrays (Song et al., 2006), flow-through reverse dot-blot (Zhang et al., 2007), PCR invader assay (Tadokoro et al., 2006), and PCR-Luminex assay (Liu et al., 2011).
Some of the limitations of the above techniques can be circumvented by using the restriction fragment mass polymorphism (RFMP) assay (Kim et al., 2005b, Han et al., 2011). The assay is based on PCR ampli cation and mass detection using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry of oligonucleotides excised by type IIS restriction enzyme digestion (Kim et al., 2005b). The use of a type IIS restriction enzyme makes the assay independent of restriction sites within the HBV genome and universal to different genotypes because these enzymes cleave DNA at a fixed distance from the recognition sites incorporated into the amplification primers. The RFMP approach has been shown to be an accurate and reliable assay for genotyping hepatitis C virus and human papilloma virus, identifying antiviral drug resistant hepatitis B virus, as well as human genomic variations (Kim et al., 2005a, Ahn et al., 2006, Lee et al., 2006a, Paik et al., 2006, Yeon et al., 2006, Hong et al., 2008). RFMP was shown to be more sensitive and to have superior quantitation of mixtures containing multiple viral genotypes without the need for population-based cloning and subsequent sequencing (Lee et al., 2006a, Hong et al., 2008).
8. Future prospects
Although a growing arsenal of NRTIs has helped improve the treatment of HBV infection, NRTI resistance remains a major problem for the hundreds of millions of people that are still affected by HBV. A key challenge is the discovery of novel drugs that block other functions of HBV or that target novel binding sites. Such drugs may include compounds that affect entry, assembly, or disassembly of the virus, inhibitors of RT that bind at the NNRTI-binding pocket, or that block the RNase H function of RT, or even novel NRTIs with distinct resistance profile. Such compounds would help improve existing therapeutic regimens and provide new opportunities for more efficient combination therapies.
Another future challenge would be to address the effect of genotypic variability on the therapeutic outcome of HBV treatments. Understanding the structural and biochemical interactions between residues that vary in different genotypes and residues that are involved in NRTI resistance may also help understand the molecular basis of resistance differences among different genotypes. Further studies are needed to systematically examine the impact of these residues and their interactions in viral fitness, efficiency of replication, and drug resistance.
Acknowledgments
This work was supported by the Ministry of Knowledge and Economy, Bilateral International Collaborative Research and Development Program, Republic of Korea and National Institutes of Health Grants AI076119, AI094715, AI100890 and AI079801.
Abbreviations
| HBV | Hepatitis B Virus |
| HBsAg | HBV Surface Antigen |
| HBeAg | HBV e Antigen |
| RT | Reverse Transcriptase |
| NRTI | Nucleoside Reverse Transcriptase Inhibitor |
| LVD/3TC | Lamivudine |
| ADV | Adefovir Dipivoxil |
| ETV | Entecavir |
| LdT | Telbivudine |
| TDF | Tenofovir Disoproxil Fumarate |
| FTC | Emtricitabine |
| IFNα | Interferon Alpha |
| peg IFNα | Pegylated Interferon Alpha |
| RFMP | Restriction Fragment Mass Polymorphism |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle SW, Kletzmayr J, Watschinger B, Schmied B, Vetter N, Puchhammer-Stockl E. Comparison of sequence analysis and the INNO-LiPA HBV DR line probe assay for detection of lamivudine-resistant hepatitis B virus strains in patients under various clinical conditions. J Clin Microbiol. 2001;39:1972–1974. [Europe PMC free article] [Abstract] [Google Scholar]
- Ahn SH, Kim do Y, Chang HY, Hong SP, Shin JS, Kim YS, Kim H, Kim JK, Paik YH, Lee KS, Chon CY, Moon YM, Han KH. Association of genetic variations in CCR5 and its ligand, RANTES with clearance of hepatitis B virus in Korea. J Med Virol. 2006;78:1564–1571. [Abstract] [Google Scholar]
- Akuta N, Suzuki F, Kobayashi M, Tsubota A, Suzuki Y, Hosaka T, Someya T, Saitoh S, Arase Y, Ikeda K, Kumada H. The influence of hepatitis B virus genotype on the development of lamivudine resistance during long-term treatment. J Hepatol. 2003;38:315–321. [Abstract] [Google Scholar]
- Amini-Bavil-Olyaee S, Herbers U, Mohebbi SR, Sabahi F, Zali MR, Luedde T, Trautwein C, Tacke F. Prevalence, viral replication efficiency and antiviral drug susceptibility of rtQ215 polymerase mutations within the hepatitis B virus genome. J Hepatol. 2009;51:647–654. [Abstract] [Google Scholar]
- Angus P, Vaughan R, Xiong S, Yang H, Delaney W, Gibbs C, Brosgart C, Colledge D, Edwards R, Ayres A, Bartholomeusz A, Locarnini S. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003;125:292–297. [Abstract] [Google Scholar]
- Bai YJ, Zhao JR, Lv GT, Zhang WH, Wang Y, Yan XJ. Rapid and high throughput detection of HBV YMDD mutants with fluorescence polarization. World J Gastroenterol. 2003;9:2344–2347. [Europe PMC free article] [Abstract] [Google Scholar]
- Baldick CJ, Tenney DJ, Mazzucco CE, Eggers BJ, Rose RE, Pokornowski KA, Yu CF, Colonno RJ. Comprehensive evaluation of hepatitis B virus reverse transcriptase substitutions associated with entecavir resistance. Hepatology. 2008;47:1473–1482. [Abstract] [Google Scholar]
- Bartholomeusz A, Locarnini S. Hepatitis B virus mutations associated with antiviral therapy. J Med Virol. 2006;78(Suppl 1):S52–55. [Abstract] [Google Scholar]
- Bartholomeusz A, Tehan BG, Chalmers DK. Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir Ther. 2004;9:149–160. [Abstract] [Google Scholar]
- Bartholomew MM, Jansen RW, Jeffers LJ, Reddy KR, Johnson LC, Bunzendahl H, Condreay LD, Tzakis AG, Schiff ER, Brown NA. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. [Abstract] [Google Scholar]
- Benhamou Y, Bochet M, Thibault V, Calvez V, Fievet MH, Vig P, Gibbs CS, Brosgart C, Fry J, Namini H, Katlama C, Poynard T. Safety and efficacy of adefovir dipivoxil in patients co-infected with HIV-1 and lamivudine-resistant hepatitis B virus: an open-label pilot study. Lancet. 2001;358:718–723. [Abstract] [Google Scholar]
- Bhattacharya D, Thio CL. Review of hepatitis B therapeutics. Clin Infect Dis. 2010;51:1201–1208. [Europe PMC free article] [Abstract] [Google Scholar]
- Billioud G, Pichoud C, Parent R, Zoulim F. Decreased infectivity of nucleoside analogs-resistant hepatitis B virus mutants. J Hepatol 2012 [Abstract] [Google Scholar]
- Borroto-Esoda K, Miller MD, Arterburn S. Pooled analysis of amino acid changes in the HBV polymerase in patients from four major adefovir dipivoxil clinical trials. J Hepatol. 2007;47:492–498. [Abstract] [Google Scholar]
- Bozdayi AM, Uzunalimoglu O, Turkyilmaz AR, Aslan N, Sezgin O, Sahin T, Bozdayi G, Cinar K, Pai SB, Pai R, Bozkaya H, Karayalcin S, Yurdaydin C, Schinazi RF. YSDD: a novel mutation in HBV DNA polymerase confers clinical resistance to lamivudine. J Viral Hepat. 2003;10:256–265. [Abstract] [Google Scholar]
- Cane PA, Cook P, Ratcliffe D, Mutimer D, Pillay D. Use of real-time PCR and fluorimetry to detect lamivudine resistance-associated mutations in hepatitis B virus. Antimicrob Agents Chemother. 1999;43:1600–1608. [Europe PMC free article] [Abstract] [Google Scholar]
- Chan HL, Wong VW, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. Early virological suppression is associated with good maintained response to adefovir dipivoxil in lamivudine resistant chronic hepatitis B. Aliment Pharmacol Ther. 2007;25:891–898. [Abstract] [Google Scholar]
- Chang TT, Lai CL. Hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;355:322–323. author reply 323. [Abstract] [Google Scholar]
- Chen CH, Lee CM, Hung CH, Wang JH, Hu TH, Changchien CS, Lu SN. Hepatitis B virus genotype B results in better immediate, late and sustained responses to peginterferon-alfa in hepatitis-B-e-antigen-positive patients. J Gastroenterol Hepatol. 2011;26:461–468. [Abstract] [Google Scholar]
- Chien RN, Yeh CT, Tsai SL, Chu CM, Liaw YF. Determinants for sustained HBeAg response to lamivudine therapy. Hepatology. 2003;38:1267–1273. [Abstract] [Google Scholar]
- Chong Y, Stuyver L, Otto MJ, Schinazi RF, Chu CK. Mechanism of antiviral activities of 3′-substituted L-nucleosides against 3TC-resistant HBV polymerase: a molecular modelling approach. Antivir Chem Chemother. 2003;14:309–319. [Abstract] [Google Scholar]
- Clarke B, Bloor S. Molecular genotyping of hepatitis B virus. J Clin Virol. 2002;25(Suppl 3):S41–45. [Abstract] [Google Scholar]
- Damerow H, Yuen L, Wiegand J, Walker C, Bock CT, Locarnini S, Tillmann HL. Mutation pattern of lamivudine resistance in relation to hepatitis B genotypes: hepatitis B genotypes differ in their lamivudine resistance associated mutation pattern. J Med Virol. 2010;82:1850–1858. [Abstract] [Google Scholar]
- Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, Arnold E. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J Virol. 2001;75:4771–4779. [Europe PMC free article] [Abstract] [Google Scholar]
- De Clercq E, Férir G, Kaptein S, Neyts J. Antiviral Treatment of Chronic Hepatitis B Virus (HBV) Infections. Viruses. 2010;2:1279–1305. [Europe PMC free article] [Abstract] [Google Scholar]
- de Vries-Sluijs TE, Reijnders JG, Hansen BE, Zaaijer HL, Prins JM, Pas SD, Schutten M, Hoepelman AI, Richter C, Mulder JW, de Man RA, Janssen HL, van der Ende ME. Long-term therapy with tenofovir is effective for patients co-infected with human immunodeficiency virus and hepatitis B virus. Gastroenterology. 2010;139:1934–1941. [Abstract] [Google Scholar]
- Delaney WEt, Locarnini S, Shaw T. Resistance of hepatitis B virus to antiviral drugs: current aspects and directions for future investigation. Antivir Chem Chemother. 2001;12:1–35. [Abstract] [Google Scholar]
- Delaney WEt, Yang H, Westland CE, Das K, Arnold E, Gibbs CS, Miller MD, Xiong S. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77:11833–11841. [Europe PMC free article] [Abstract] [Google Scholar]
- Delaugerre C, Marcelin AG, Thibault V, Peytavin G, Bombled T, Bochet MV, Katlama C, Benhamou Y, Calvez V. Human immunodeficiency virus (HIV) Type 1 reverse transcriptase resistance mutations in hepatitis B virus (HBV)-HIV-coinfected patients treated for HBV chronic infection once daily with 10 milligrams of adefovir dipivoxil combined with lamivudine. Antimicrob Agents Chemother. 2002;46:1586–1588. [Europe PMC free article] [Abstract] [Google Scholar]
- Devesa M, Rodriguez C, Leon G, Liprandi F, Pujol FH. Clade analysis and surface antigen polymorphism of hepatitis B virus American genotypes. J Med Virol. 2004;72:377–384. [Abstract] [Google Scholar]
- Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486–1500. [Abstract] [Google Scholar]
- Dienstag JL, Perrillo RP, Schiff ER, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. [Abstract] [Google Scholar]
- Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, Crowther L, Condreay LD, Woessner M, Rubin M, Brown NA. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. [Abstract] [Google Scholar]
- Erhardt A, Reineke U, Blondin D, Gerlich WH, Adams O, Heintges T, Niederau C, Haussinger D. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology. 2000;31:716–725. [Abstract] [Google Scholar]
- Foli A, Sogocio KM, Anderson B, Kavlick M, Saville MW, Wainberg MA, Gu Z, Cherrington JM, Mitsuya H, Yarchoan R. In vitro selection and molecular characterization of human immunodeficiency virus type 1 with reduced sensitivity to 9-[2-(phosphonomethoxy)ethyl]adenine (PMEA) Antiviral Res. 1996;32:91–98. [Abstract] [Google Scholar]
- Fung SK, Chae HB, Fontana RJ, Conjeevaram H, Marrero J, Oberhelman K, Hussain M, Lok AS. Virologic response and resistance to adefovir in patients with chronic hepatitis B. J Hepatol. 2006;44:283–290. [Abstract] [Google Scholar]
- Fung SK, Wong F, Hussain M, Lok AS. Sustained response after a 2-year course of lamivudine treatment of hepatitis B e antigen-negative chronic hepatitis B. J Viral Hepat. 2004;11:432–438. [Abstract] [Google Scholar]
- Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132:1574–1585. [Abstract] [Google Scholar]
- Gish R, Jia JD, Locarnini S, Zoulim F. Selection of chronic hepatitis B therapy with high barrier to resistance. Lancet Infect Dis 2012 [Abstract] [Google Scholar]
- Hall JG, Eis PS, Law SM, Reynaldo LP, Prudent JR, Marshall DJ, Allawi HT, Mast AL, Dahlberg JE, Kwiatkowski RW, de Arruda M, Neri BP, Lyamichev VI. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci U S A. 2000;97:8272–8277. [Europe PMC free article] [Abstract] [Google Scholar]
- Han KH, Hong SP, Choi SH, Shin SK, Cho SW, Ahn SH, Hahn JS, Kim SO. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir Ther. 2011;16:77–87. [Abstract] [Google Scholar]
- Hong SP, Kim NK, Hwang SG, Chung HJ, Kim S, Han JH, Kim HT, Rim KS, Kang MS, Yoo W, Kim SO. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J Hepatol. 2004;40:837–844. [Abstract] [Google Scholar]
- Hong SP, Shin SK, Lee EH, Kim EO, Ji SI, Chung HJ, Park SN, Yoo W, Folk WR, Kim SO. High-resolution human papillomavirus genotyping by MALDI-TOF mass spectrometry. Nat Protoc. 2008;3:1476–1484. [Abstract] [Google Scholar]
- Hou J, Schilling R, Janssen HL, Hansen BE, Heijtink R, Sablon E, Williams R, Lau GK, Schalm SW, Naoumov NV. Genetic characteristics of hepatitis B virus genotypes as a factor for interferon-induced HBeAg clearance. J Med Virol. 2007;79:1055–1063. [Abstract] [Google Scholar]
- Hsieh TH, Tseng TC, Liu CJ, Lai MY, Chen PJ, Hsieh HL, Chen DS, Kao JH. Hepatitis B virus genotype B has an earlier emergence of lamivudine resistance than genotype C. Antivir Ther. 2009;14:1157–1163. [Abstract] [Google Scholar]
- Inoue J, Ueno Y, Wakui Y, Niitsuma H, Fukushima K, Yamagiwa Y, Shiina M, Kondo Y, Kakazu E, Tamai K, Obara N, Iwasaki T, Shimosegawa T. Four-year study of lamivudine and adefovir combination therapy in lamivudine-resistant hepatitis B patients: influence of hepatitis B virus genotype and resistance mutation pattern. J Viral Hepat. 2011;18:206–215. [Abstract] [Google Scholar]
- Jain MK, Comanor L, White C, Kipnis P, Elkin C, Leung K, Ocampo A, Attar N, Keiser P, Lee WM. Treatment of hepatitis B with lamivudine and tenofovir in HIV/HBV-coinfected patients: factors associated with response. J Viral Hepat. 2007;14:176–182. [Abstract] [Google Scholar]
- Jain MK, Zoellner CL. Entecavir can select for M184V of HIV-1: a case of an HIV/hepatitis B (HBV) naive patient treated for chronic HBV. AIDS. 2007;21:2365–2366. [Abstract] [Google Scholar]
- Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123–12. [Abstract] [Google Scholar]
- Kamili S, Sozzi V, Thompson G, Campbell K, Walker CM, Locarnini S, Krawczynski K. Efficacy of hepatitis B vaccine against antiviral drug-resistant hepatitis B virus mutants in the chimpanzee model. Hepatology. 2009;49:1483–1491. [Abstract] [Google Scholar]
- Kao JH, Liu CJ, Chen DS. Hepatitis B viral genotypes and lamivudine resistance. J Hepatol. 2002;36:303–304. [Abstract] [Google Scholar]
- Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol. 2000;33:998–1002. [Abstract] [Google Scholar]
- Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. [Abstract] [Google Scholar]
- Kim HS, Han KH, Ahn SH, Kim EO, Chang HY, Moon MS, Chung HJ, Yoo W, Kim SO, Hong SP. Evaluation of methods for monitoring drug resistance in chronic hepatitis B patients during lamivudine therapy based on mass spectrometry and reverse hybridization. Antivir Ther. 2005a;10:441–449. [Abstract] [Google Scholar]
- Kim YJ, Kim SO, Chung HJ, Jee MS, Kim BG, Kim KM, Yoon JH, Lee HS, Kim CY, Kim S, Yoo W, Hong SP. Population genotyping of hepatitis C virus by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of short DNA fragments. Clin Chem. 2005b;51:1123–1131. [Abstract] [Google Scholar]
- Kumada H. Continued lamivudine therapy in patients with chronic hepatitis B. Intervirology. 2003;46:377–387. [Abstract] [Google Scholar]
- Lacombe K, Ollivet A, Gozlan J, Durantel S, Tran N, Girard PM, Zoulim F. A novel hepatitis B virus mutation with resistance to adefovir but not to tenofovir in an HIV-hepatitis B virus-co-infected patient. AIDS. 2006;20:2229–2231. [Abstract] [Google Scholar]
- Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J, Stephenson SL, Gray DF. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. [Abstract] [Google Scholar]
- Langley DR, Walsh AW, Baldick CJ, Eggers BJ, Rose RE, Levine SM, Kapur AJ, Colonno RJ, Tenney DJ. Inhibition of hepatitis B virus polymerase by entecavir. J Virol. 2007;81:3992–4001. [Europe PMC free article] [Abstract] [Google Scholar]
- Le Bouvier GL, McCollum RW, Hierholzer WJ, Jr, Irwin GR, Krugman S, Giles JP. Subtypes of Australia antigen and hepatitis-B virus. JAMA. 1972;222:928–930. [Abstract] [Google Scholar]
- Lee CH, Kim SO, Byun KS, Moon MS, Kim EO, Yeon JE, Yoo W, Hong SP. Predominance of hepatitis B virus YMDD mutants is prognostic of viral DNA breakthrough. Gastroenterology. 2006a;130:1144–1152. [Abstract] [Google Scholar]
- Lee YS, Chung YH, Kim JA, Kim SE, Shin JW, Kim KM, Lim YS, Park NH, Lee HC, Lee YS, Suh DJ. Hepatitis B virus with the rtL80V/I mutation is associated with a poor response to adefovir dipivoxil therapy. Liver Int. 2009;29:552–556. [Abstract] [Google Scholar]
- Lee YS, Suh DJ, Lim YS, Jung SW, Kim KM, Lee HC, Chung YH, Yoo W, Kim SO. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology. 2006b;43:1385–1391. [Abstract] [Google Scholar]
- Liaw YF, Gane E, Leung N, Zeuzem S, Wang Y, Lai CL, Heathcote EJ, Manns M, Bzowej N, Niu J, Han SH, Hwang SG, Cakaloglu Y, Tong MJ, Papatheodoridis G, Chen Y, Brown NA, Albanis E, Galil K, Naoumov NV. 2-Year GLOBE trial results: telbivudine Is superior to lamivudine in patients with chronic hepatitis B. Gastroenterology. 2009;136:486–495. [Abstract] [Google Scholar]
- Liaw YF, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Chien RN, Dent J, Roman L, Edmundson S, Lai CL. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology. 2000;119:172–180. [Abstract] [Google Scholar]
- Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26(Suppl 1):123–130. [Abstract] [Google Scholar]
- Ling R, Mutimer D, Ahmed M, Boxall EH, Elias E, Dusheiko GM, Harrison TJ. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. [Abstract] [Google Scholar]
- Litwin S, Toll E, Jilbert AR, Mason WS. The competing roles of virus replication and hepatocyte death rates in the emergence of drug-resistant mutants: theoretical considerations. J Clin Virol. 2005;34(Suppl 1):S96–S107. [Abstract] [Google Scholar]
- Liu BM, Li T, Xu J, Li XG, Dong JP, Yan P, Yang JX, Yan L, Gao ZY, Li WP, Sun XW, Wang YH, Jiao XJ, Hou CS, Zhuang H. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naive Chinese patients. Antiviral Res. 2010;85:512–519. [Abstract] [Google Scholar]
- Liu CJ, Kao JH, Chen PJ, Chen TC, Lin FY, Lai MY, Chen DS. Overlap lamivudine treatment in patients with chronic hepatitis B receiving adefovir for lamivudine-resistant viral mutants. J Viral Hepat. 2006;13:387–395. [Abstract] [Google Scholar]
- Liu CJ, Kao JH, Chen PJ, Lai MY, Chen DS. Molecular epidemiology of hepatitis B viral serotypes and genotypes in taiwan. J Biomed Sci. 2002;9:166–170. [Abstract] [Google Scholar]
- Liu H, Mao R, Fan L, Xia J, Li Y, Yin Y, Li X, Zhao X, Guo H, Zhu H, Zhang Y, Kang Y, Zhang J. Detection of lamivudine- or adefovir-resistant hepatitis B virus mutations by a liquid array. J Virol Methods. 2011;175:1–6. [Abstract] [Google Scholar]
- Liu X, Schinazi RF. Hepatitis B virus resistance to lamivudine and its clinical implications. Antivir Chem Chemother. 2002;13:143–155. [Abstract] [Google Scholar]
- Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006;44:422–431. [Abstract] [Google Scholar]
- Locarnini SA, Yuen L. Molecular genesis of drug-resistant and vaccine-escape HBV mutants. Antivir Ther. 2010;15:451–461. [Abstract] [Google Scholar]
- Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, Gardner SD, Castiglia M. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714–1722. [Abstract] [Google Scholar]
- Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. [Abstract] [Google Scholar]
- McMahon MA, Jilek BL, Brennan TP, Shen L, Zhou Y, Wind-Rotolo M, Xing S, Bhat S, Hale B, Hegarty R, Chong CR, Liu JO, Siliciano RF, Thio CL. The HBV drug entecavir - effects on HIV-1 replication and resistance. N Engl J Med. 2007;356:2614–2621. [Europe PMC free article] [Abstract] [Google Scholar]
- Michailidis E, Singh K, Kirby KA, Hachiya A, Yoo W, Hong SP, Kim SO, Folk WR, Sarafianos SG. Hepatitis B Virus genotypic differences map structurally close to NRTI resistance hot spots. International Journal of Current Chemistry. 2011;2:253–260. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller RH, Kaneko S, Chung CT, Girones R, Purcell RH. Compact organization of the hepatitis B virus genome. Hepatology. 1989;9:322–327. [Abstract] [Google Scholar]
- Mitsui F, Tsuge M, Kimura T, Kitamura S, Abe H, Saneto H, Kawaoka T, Miki D, Hatakeyama T, Hiraga N, Imamura M, Kawakami Y, Aikata H, Takahashi S, Hayes CN, Igarashi H, Morimoto K, Shimizu M, Chayama K. Importance of serum concentration of adefovir for Lamivudine-adefovir combination therapy in patients with lamivudine-resistant chronic hepatitis B. Antimicrob Agents Chemother. 2010;54:3205–3211. [Europe PMC free article] [Abstract] [Google Scholar]
- Mitsuya H, Jarrett RF, Matsukura M, Di Marzo Veronese F, DeVico AL, Sarngadharan MG, Johns DG, Reitz MS, Broder S. Long-term inhibition of human T-lymphotropic virus type III/lymphadenopathy-associated virus (human immunodeficiency virus) DNA synthesis and RNA expression in T cells protected by 2′,3′-dideoxynucleosides in vitro. Proc Natl Acad Sci U S A. 1987;84:2033–2037. [Europe PMC free article] [Abstract] [Google Scholar]
- Mizokami M, Orito E, Ohba K, Ikeo K, Lau JY, Gojobori T. Constrained evolution with respect to gene overlap of hepatitis B virus. J Mol Evol. 1997;44(Suppl 1):S83–90. [Abstract] [Google Scholar]
- Mukaide M, Tanaka Y, Shin IT, Yuen MF, Kurbanov F, Yokosuka O, Sata M, Karino Y, Yamada G, Sakaguchi K, Orito E, Inoue M, Baqai S, Lai CL, Mizokami M. Mechanism of entecavir resistance of hepatitis B virus with viral breakthrough as determined by long-term clinical assessment and molecular docking simulation. Antimicrob Agents Chemother. 2010;54:882–889. [Europe PMC free article] [Abstract] [Google Scholar]
- Murray KK. DNA sequencing by mass spectrometry. J Mass Spectrom. 1996;31:1203–1215. [Abstract] [Google Scholar]
- Nelson M, Portsmouth S, Stebbing J, Atkins M, Barr A, Matthews G, Pillay D, Fisher M, Bower M, Gazzard B. An open-label study of tenofovir in HIV-1 and Hepatitis B virus co-infected individuals. AIDS. 2003;17:F7–10. [Abstract] [Google Scholar]
- Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. [Abstract] [Google Scholar]
- Norder H, Courouce AM, Magnius LO. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J Gen Virol. 1992a;73(Pt 12):3141–3145. [Abstract] [Google Scholar]
- Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. [Abstract] [Google Scholar]
- Norder H, Hammas B, Lofdahl S, Courouce AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992b;73(Pt 5):1201–1208. [Abstract] [Google Scholar]
- Ogata N, Fujii K, Takigawa S, Nomoto M, Ichida T, Asakura H. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in japanese patients with chronic hepatitis B. J Med Virol. 1999;59:270–276. [Abstract] [Google Scholar]
- Ohba K, Mizokami M, Ohno T, Suzuki K, Orito E, Lau JY, Ina Y, Ikeo K, Gojobori T. Relationships between serotypes and genotypes of hepatitis B virus: genetic classification of HBV by use of surface genes. Virus Res. 1995;39:25–34. [Abstract] [Google Scholar]
- Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69(Pt 10):2575–2583. [Abstract] [Google Scholar]
- Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF, Carrilho FJ, Omata M. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest. 2001;107:449–455. [Europe PMC free article] [Abstract] [Google Scholar]
- Osiowy C, Villeneuve JP, Heathcote EJ, Giles E, Borlang J. Detection of rtN236T and rtA181V/T mutations associated with resistance to adefovir dipivoxil in samples from patients with chronic hepatitis B virus infection by the INNO-LiPA HBV DR line probe assay (version 2) J Clin Microbiol. 2006;44:1994–1997. [Europe PMC free article] [Abstract] [Google Scholar]
- Paik YH, Han KH, Hong SP, Lee HW, Lee KS, Kim SO, Shin JE, Ahn SH, Chon CY, Moon YM. The clinical impact of early detection of the YMDD mutant on the outcomes of long-term lamivudine therapy in patients with chronic hepatitis B. Antivir Ther. 2006;11:447–455. [Abstract] [Google Scholar]
- Pawlotsky JM, Dusheiko G, Hatzakis A, Lau D, Lau G, Liang TJ, Locarnini S, Martin P, Richman DD, Zoulim F. Virologic monitoring of hepatitis B virus therapy in clinical trials and practice: recommendations for a standardized approach. Gastroenterology. 2008;134:405–415. [Europe PMC free article] [Abstract] [Google Scholar]
- Perrillo R. Benefits and risks of interferon therapy for hepatitis B. Hepatology. 2009;49:S103–111. [Abstract] [Google Scholar]
- Rhee SY, Margeridon-Thermet S, Nguyen MH, Liu TF, Kagan RM, Beggel B, Verheyen J, Kaiser R, Shafer RW. Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery. Antiviral Res. 2010;88:269–275. [Europe PMC free article] [Abstract] [Google Scholar]
- Sablon E, Shapiro F. Advances in Molecular Diagnosis of HBV Infection and Drug Resistance. Int J Med Sci. 2005;2:8–16. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc Natl Acad Sci U S A. 1999;96:10027–10032. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. [Europe PMC free article] [Abstract] [Google Scholar]
- Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. [Europe PMC free article] [Abstract] [Google Scholar]
- Schildgen O, Sirma H, Funk A, Olotu C, Wend UC, Hartmann H, Helm M, Rockstroh JK, Willems WR, Will H, Gerlich WH. Variant of hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;354:1807–1812. [Abstract] [Google Scholar]
- Seifer M, Hamatake RK, Colonno RJ, Standring DN. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob Agents Chemother. 1998;42:3200–3208. [Europe PMC free article] [Abstract] [Google Scholar]
- Seifer M, Patty A, Serra I, Li B, Standring DN. Telbivudine, a nucleoside analog inhibitor of HBV polymerase, has a different in vitro cross-resistance profile than the nucleotide analog inhibitors adefovir and tenofovir. Antiviral Res. 2009;81:147–155. [Abstract] [Google Scholar]
- Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol. 2006;44:593–606. [Abstract] [Google Scholar]
- Sheldon J, Ramos B, Garcia-Samaniego J, Rios P, Bartholomeusz A, Romero M, Locarnini S, Zoulim F, Soriano V. Selection of hepatitis B virus (HBV) vaccine escape mutants in HBV-infected and HBV/HIV-coinfected patients failing antiretroviral drugs with anti-HBV activity. J Acquir Immune Defic Syndr. 2007;46:279–282. [Abstract] [Google Scholar]
- Shiina S, Fujino H, Uta Y, Tagawa K, Unuma T, Yoneyama M, Ohmori T, Suzuki S, Kurita M, Ohashi Y. Relationship of HBsAg subtypes with HBeAg/anti-HBe status and chronic liver disease. Part I: Analysis of 1744 HBsAg carriers. Am J Gastroenterol. 1991;86:866–871. [Abstract] [Google Scholar]
- Song Y, Dai E, Wang J, Liu H, Zhai J, Chen C, Du Z, Guo Z, Yang R. Genotyping of hepatitis B virus (HBV) by oligonucleotides microarray. Mol Cell Probes. 2006;20:121–127. [Abstract] [Google Scholar]
- Soriano V, Barreiro P, Nunez M. Management of chronic hepatitis B and C in HIV-coinfected patients. J Antimicrob Chemother. 2006;57:815–818. [Abstract] [Google Scholar]
- Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000a;81:67–74. [Abstract] [Google Scholar]
- Stuyver L, Van Geyt C, De Gendt S, Van Reybroeck G, Zoulim F, Leroux-Roels G, Rossau R. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000b;38:702–707. [Europe PMC free article] [Abstract] [Google Scholar]
- Stuyver LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, Schinazi RF. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33:751–757. [Abstract] [Google Scholar]
- Sugauchi F, Kumada H, Acharya SA, Shrestha SM, Gamutan MT, Khan M, Gish RG, Tanaka Y, Kato T, Orito E, Ueda R, Miyakawa Y, Mizokami M. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol. 2004;85:811–820. [Abstract] [Google Scholar]
- Svicher V, Cento V, Salpini R, Mercurio F, Fraune M, Beggel B, Han Y, Gori C, Wittkop L, Bertoli A, Micheli V, Gubertini G, Longo R, Romano S, Visca M, Gallinaro V, Marino N, Mazzotta F, De Sanctis GM, Fleury H, Trimoulet P, Angelico M, Cappiello G, Zhang XX, Verheyen J, Ceccherini-Silberstein F, Perno CF. Role of hepatitis B virus genetic barrier in drug-resistance and immune-escape development. Dig Liver Dis. 2011;43:975–983. [Abstract] [Google Scholar]
- Tadokoro K, Kobayashi M, Yamaguchi T, Suzuki F, Miyauchi S, Egashira T, Kumada H. Classification of hepatitis B virus genotypes by the PCR-Invader method with genotype-specific probes. J Virol Methods. 2006;138:30–39. [Abstract] [Google Scholar]
- Tchesnokov EP, Obikhod A, Schinazi RF, Gotte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J Biol Chem. 2008;283:34218–34228. [Europe PMC free article] [Abstract] [Google Scholar]
- Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P, Ayres A, Bartholomeusz A, Sievert W, Thompson G, Warner N, Locarnini S, Colonno RJ. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–3507. [Europe PMC free article] [Abstract] [Google Scholar]
- Tenney DJ, Rose RE, Baldick CJ, Levine SM, Pokornowski KA, Walsh AW, Fang J, Yu CF, Zhang S, Mazzucco CE, Eggers B, Hsu M, Plym MJ, Poundstone P, Yang J, Colonno RJ. Two-year assessment of entecavir resistance in Lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present. Antimicrob Agents Chemother. 2007;51:902–911. [Europe PMC free article] [Abstract] [Google Scholar]
- Tenney DJ, Rose RE, Baldick CJ, Pokornowski KA, Eggers BJ, Fang J, Wichroski MJ, Xu D, Yang J, Wilber RB, Colonno RJ. Long-term monitoring shows hepatitis B virus resistance to entecavir in nucleoside-naive patients is rare through 5 years of therapy. Hepatology. 2009;49:1503–1514. [Abstract] [Google Scholar]
- Teo CG, Locarnini SA. Potential threat of drug-resistant and vaccine-escape HBV mutants to public health. Antivir Ther. 2010;15:445–449. [Abstract] [Google Scholar]
- Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25:97–106. [Abstract] [Google Scholar]
- Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, Fyfe J, Sozzi T, Jackson DC. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293:305–313. [Abstract] [Google Scholar]
- Villeneuve JP, Durantel D, Durantel S, Westland C, Xiong S, Brosgart CL, Gibbs CS, Parvaz P, Werle B, Trepo C, Zoulim F. Selection of a hepatitis B virus strain resistant to adefovir in a liver transplantation patient. J Hepatol. 2003;39:1085–108. [Abstract] [Google Scholar]
- Villet S, Billioud G, Pichoud C, Lucifora J, Hantz O, Sureau C, Deny P, Zoulim F. In vitro characterization of viral fitness of therapy-resistant hepatitis B variants. Gastroenterology. 2009;136:168–176. e162. [Abstract] [Google Scholar]
- Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425–1430. [Abstract] [Google Scholar]
- Westland C, Delaney Wt, Yang H, Chen SS, Marcellin P, Hadziyannis S, Gish R, Fry J, Brosgart C, Gibbs C, Miller M, Xiong S. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil1. Gastroenterology. 2003a;125:107–116. [Abstract] [Google Scholar]
- Westland CE, Yang H, Delaney WEt, Gibbs CS, Miller MD, Wulfsohn M, Fry J, Brosgart CL, Xiong S. Week 48 resistance surveillance in two phase 3 clinical studies of adefovir dipivoxil for chronic hepatitis B. Hepatology. 2003b;38:96–103. [Abstract] [Google Scholar]
- Westland CE, Yang H, Delaney WEt, Wulfsohn M, Lama N, Gibbs CS, Miller MD, Fry J, Brosgart CL, Schiff ER, Xiong S. Activity of adefovir dipivoxil against all patterns of lamivudine-resistant hepatitis B viruses in patients. J Viral Hepat. 2005;12:67–73. [Abstract] [Google Scholar]
- Whalley SA, Brown D, Teo CG, Dusheiko GM, Saunders NA. Monitoring the emergence of hepatitis B virus polymerase gene variants during lamivudine therapy using the LightCycler. J Clin Microbiol. 2001;39:1456–1459. [Europe PMC free article] [Abstract] [Google Scholar]
- Yeh CT, Chien RN, Chu CM, Liaw YF. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology. 2000;31:1318–1326. [Abstract] [Google Scholar]
- Yeon JE, Yoo W, Hong SP, Chang YJ, Yu SK, Kim JH, Seo YS, Chung HJ, Moon MS, Kim SO, Byun KS, Lee CH. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006;55:1488–1495. [Europe PMC free article] [Abstract] [Google Scholar]
- Yim HJ, Hussain M, Liu Y, Wong SN, Fung SK, Lok AS. Evolution of multi-drug resistant hepatitis B virus during sequential therapy. Hepatology. 2006;44:703–712. [Abstract] [Google Scholar]
- Zhang R, Deng Y, Muller CP, Ou ZY, Ma L, Wang M, Li PQ, He YS. Determination of hepatitis B virus genotype by flow-through reverse dot blot. J Clin Virol. 2007;39:94–100. [Abstract] [Google Scholar]
- Zhang X, Zoulim F, Habersetzer F, Xiong S, Trepo C. Analysis of hepatitis B virus genotypes and pre-core region variability during interferon treatment of HBe antigen negative chronic hepatitis B. J Med Virol. 1996;48:8–16. [Abstract] [Google Scholar]
- Zhong Y, Lv J, Li J, Xing X, Zhu H, Su H, Chen L, Zhou X. Prevalence, virology and antiviral drugs susceptibility of hepatitis B virus rtN238H polymerase mutation from 1865 Chinese patients with chronic hepatitis B. Antiviral Res. 2012;93:185–190. [Abstract] [Google Scholar]
- Zhu Y, Curtis M, Qi X, Miller MD, Borroto-Esoda K. Anti-hepatitis B virus activity in vitro of combinations of tenofovir with nucleoside/nucleotide analogues. Antivir Chem Chemother. 2009;19:165–176. [Abstract] [Google Scholar]
- Zollner B, Petersen J, Puchhammer-Stockl E, Kletzmayr J, Sterneck M, Fischer L, Schroter M, Laufs R, Feucht HH. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology. 2004;39:42–50. [Abstract] [Google Scholar]
- Zoulim F. Assessing hepatitis B virus resistance in vitro and molecular mechanisms of nucleoside resistance. Semin Liver Dis. 2002;22(Suppl 1):23–31. [Abstract] [Google Scholar]
- Zoulim F, Locarnini S. Management of treatment failure in chronic hepatitis B. J Hepatol. 2012;56(Suppl 1):S112–122. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.biocel.2012.04.006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3522522?pdf=render
Citations & impact
Impact metrics
Article citations
Discovery of dual S-RBD/NRP1-targeting peptides: structure-based virtual screening, synthesis, biological evaluation, and molecular dynamics simulation studies.
J Enzyme Inhib Med Chem, 38(1):2212327, 01 Dec 2023
Cited by: 5 articles | PMID: 37194732 | PMCID: PMC10193894
In vitro evaluation of tropolone absorption, metabolism, and clearance.
Antiviral Res, 220:105762, 22 Nov 2023
Cited by: 0 articles | PMID: 37996012
Biology of the hepatitis B virus (HBV) core and capsid assembly modulators (CAMs) for chronic hepatitis B (CHB) cure.
Glob Health Med, 5(4):199-207, 01 Aug 2023
Cited by: 2 articles | PMID: 37655181 | PMCID: PMC10461335
Review Free full text in Europe PMC
Improved pharmacokinetics of tenofovir ester prodrugs strengthened the inhibition of HBV replication and the rebalance of hepatocellular metabolism in preclinical models.
Front Pharmacol, 13:932934, 29 Aug 2022
Cited by: 7 articles | PMID: 36105197 | PMCID: PMC9465247
Design, Synthesis, and Evaluation of a Set of Carboxylic Acid and Phosphate Prodrugs Derived from HBV Capsid Protein Allosteric Modulator NVR 3-778.
Molecules, 27(18):5987, 14 Sep 2022
Cited by: 1 article | PMID: 36144715 | PMCID: PMC9505734
Go to all (28) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Hepatitis B virus reverse transcriptase - Target of current antiviral therapy and future drug development.
Antiviral Res, 123:132-137, 25 Sep 2015
Cited by: 42 articles | PMID: 26408354 | PMCID: PMC4639421
Review Free full text in Europe PMC
Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery.
Antiviral Res, 88(3):269-275, 25 Sep 2010
Cited by: 36 articles | PMID: 20875460 | PMCID: PMC4374605
Hepatitis B Virus genotypic differences map structurally close to NRTI resistance hot spots.
Int J Curr Chem, 2(4):253-260, 01 Oct 2011
Cited by: 3 articles | PMID: 22505793 | PMCID: PMC3325108
[Dissection of mechanism for the adefovir dipivoxil resistance in chronic hepatitis B patients].
Zhonghua Gan Zang Bing Za Zhi, 17(10):730-734, 01 Oct 2009
Cited by: 1 article | PMID: 19874686
Funding
Funders who supported this work.
Ministry of Knowledge and Economy, Bilateral International Collaborative Research and Development Program
NIAID NIH HHS (9)
Grant ID: R01 AI100890
Grant ID: R37 AI076119
Grant ID: R01 AI076119
Grant ID: R21 AI094715
Grant ID: R33 AI079801
Grant ID: AI076119
Grant ID: AI094715
Grant ID: AI079801
Grant ID: R21 AI079801
National Institutes of Health (3)
Grant ID: AI079801
Grant ID: AI076119
Grant ID: AI094715





