Abstract
Free full text

The XXmotif web server for eXhaustive, weight matriX-based motif discovery in nucleotide sequences
Abstract
The discovery of regulatory motifs enriched in sets of DNA or RNA sequences is fundamental to the analysis of a great variety of functional genomics experiments. These motifs usually represent binding sites of proteins or non-coding RNAs, which are best described by position weight matrices (PWMs). We have recently developed XXmotif, a de novo motif discovery method that is able to directly optimize the statistical significance of PWMs. XXmotif can also score conservation and positional clustering of motifs. The XXmotif server provides (i) a list of significantly overrepresented motif PWMs with web logos and E-values; (ii) a graph with color-coded boxes indicating the positions of selected motifs in the input sequences; (iii) a histogram of the overall positional distribution for selected motifs and (iv) a page for each motif with all significant motif occurrences, their P-values for enrichment, conservation and localization, their sequence contexts and coordinates. Free access: http://xxmotif.genzentrum.lmu.de.
INTRODUCTION
To understand how cells read off information from the genome at the right time at the right position, we have to learn the sequence motifs that the regulatory factors recognize and bind to. A large variety of experimental methods yield sequences that are enriched in binding sites of regulatory factors. Methods that can discover these enriched motifs have therefore proven to be of great practical importance for modern biological research and a multitude of motif discovery methods have been developed (1–4). Most of the tools can only be run on the command line, making them inaccessible to the majority of biologists. However, a few web services for de novo motif discovery exist.
The most popular one is the MEME Suite server (5), within which the position weight matrix (PWM)-based MEME and GLAM2 motif discovery programs can be run (6,7), alongside several related tools to compare the discovered motifs with libraries of literature motifs and to search for matches to the discovered motifs in sequence databases. With a higher order background model to describe sequences that should not carry the sought motifs, MEME has shown state-of-the-art performance (8). To use higher order models, users have to upload their own model file generated using a MEME command line tool, which will limit most users to the zero-order model with lower sensitivity. The SCOPE web server combines three pattern-based motif discovery tools, which are specialized to find non-degenerate, degenerate and gapped motifs, into a single prediction using a ‘winner takes all’ learning rule (9). The RegAnalyst server runs a motif discovery method that searches for the most enriched patterns using fixed thresholds for the maximum number of allowed mismatches. It was originally developed for mycobacterial and yeast sequences, on which it was reported to have higher sensitivity than SCOPE (10). The WebMOTIFS server takes gene names from human, mouse or Saccharomyces cerevisiae as input, extracts promoter sequences, launches four motif discovery programs and displays the results in a uniform format (11). RSAT is a web toolbox for regulatory sequence analysis that also offers several simple tools and Gibbs sampling for motif discovery (12). Finally, AMADEUS (13) is a software tool with a nicely designed graphical user interface that presents an alternative to these web services.
Although various published tools can score conservation in multiple sequence alignments of related species and a few can exploit the positional clustering of motifs, to our knowledge, none of the web services offers this useful functionality. In contrast, the XXmotif web server can combine enrichment P-values for PWMs with P-values for sequence conservation and for positional clustering of motif occurrences.
METHOD SUMMARY
The binding site motifs of regulatory factors are described either with PWMs or with patterns, such as consensus sequences with degenerate IUPAC characters, sometimes allowing for mismatches (14). A PWM is a statistical model – represented by 4 ×
× l matrix – that has weights for the four bases at each of the l binding sites positions. In contrast to patterns that either do or do not match, a PWM gives a more nuanced description of the binding affinity landscape. From a thermodynamic point of view, a PWM approximates the binding energy under the assumption that each position contributes independently of the others. Although it is straightforward to calculate enrichment P-values for patterns, this is more challenging to do for PMWs and usually involves time-consuming random sampling. Therefore, all PWM-based methods to date have taken a likelihood-based approach for finding enriched PWMs. XXmotif is the first PWM-based method to directly optimize the motif enrichment P-value in its PWM stage.
l matrix – that has weights for the four bases at each of the l binding sites positions. In contrast to patterns that either do or do not match, a PWM gives a more nuanced description of the binding affinity landscape. From a thermodynamic point of view, a PWM approximates the binding energy under the assumption that each position contributes independently of the others. Although it is straightforward to calculate enrichment P-values for patterns, this is more challenging to do for PMWs and usually involves time-consuming random sampling. Therefore, all PWM-based methods to date have taken a likelihood-based approach for finding enriched PWMs. XXmotif is the first PWM-based method to directly optimize the motif enrichment P-value in its PWM stage.
XXmotif consists of three stages: a masking stage, a pattern stage, and a PWM stage. In the masking stage, repeat regions, compositionally biased segments and homologous segment pairs are masked out.
When parts of sequences in the input set are identical or similar over longer regions, this region can give rise to a significant motif even if just these two occurrences are observed. The reason is that the motif is so long and informative that it would be very unlikely to observe even two such motifs by chance. Hence, to avoid reporting false motifs stemming from regions of local homology, XXmotif masks regions of local homology found by an all-against-all sequence comparison using BLAST. For similar reasons, XXmasker also masks perfect repeats of length 50 or more base pairs.
In the pattern stage, XXmotif calculates enrichment P-values for seed patterns, consisting of all 5-mers with up to two degenerate IUPAC characters, and all palindromic and tandemic 6-mer seeds with gaps up to size 11. For each seed pattern, an enrichment P-value is calculated using a binomial distribution and a length- and gap-dependent Bonferroni correction factor. For each non-degenerate seed (i.e. without IUPAC characters), the five most significant matching IUPAC seed patterns are extended, allowing gaps of up to 3, until the P-value cannot be improved anymore. IUPAC strings are then converted to PWMs by counting the nucleotides at each position in the matching sequence segments. In the PWM stage, thousands of candidate PWMs are iteratively optimized: similar PWMs are merged, and PWMs are extended (allowing gaps up to 2) or shortened, until their enrichment P-value cannot be improved anymore. Enrichment P-values give the statistical significance of the enrichment of a PWM in the positive sequence set compared with the expectation derived from the background model. Enrichment P-values are calculated from the single-site P-values for each possible motif position. A single-site P-value quantifies the significance of the match of a single site to the PWM. It is the probability that a random site (generated from the background model) will obtain at least the same score. Hence, the better the PWM score of the single site, the more significant and the nearer to zero is its single-site P-value. We developed an efficient branch-and-bound algorithm to compute the single-site P-values for all sites in the positive sequence set. The enrichment P-value is calculated from all single-site P-values in the input sequences using order statistics: the enrichment P-value is the probability to obtain by chance on a same-size set of background sequences at least K out of N possible motif positions with better single-site P-values than the Kth single-site P-value actually observed. We choose K that optimizes this enrichment P-value. Finally, the enrichment P-value can be combined with the P-values for conservation and localization into a total P-value. E-values are obtained by multiplying the total motif P-values with a Bonferroni-like correction factor, which penalizes model complexity similar to the Akaike information criterion. For a detailed description, see (H. Hartmann, E.W. Guthoehrlein, M. Siebert, S. Luehr and J. Söding, submitted for publication).
XXmotif has been compared with various versions of five state-of-the-art methods for motif discovery (MEME (7), Weeder (15), PRIORITY (16), AMADEUS (5) and ERMIT (17)) on a standard benchmark set containing 352 datasets of ChIP-enriched sequences from S. cerevisiae (18), and the other containing 34 sets of metazoan sequences obtained with a wide range of experimental approaches (5). XXmotif showed 20–50% higher sensitivity (number of correctly identified motifs) than the other tools on the Harbison datasets (18) and 15–300% on the metazoan datasets. The quality of the reported PWMs was measured in a partial area under receiver operating characteristic curve (pAUC) analysis and showed between 30 and 75% higher values than the other tools (H. Hartmann, E.W. Guthoehrlein, M. Siebert, S. Luehr and J. Söding, submitted for publication).
INPUT
On the ‘Data upload page’ (Figure 1A), users can enter the input sequence set and an optional background sequence set (up to 25 MB per file). The background sequences are used to learn the statistical background model, which describes how ‘normal’ sequences look like. XXmotif will then try to find motifs that are enriched in the input set in comparison to the expectation derived from the background model. When no background sequences are supplied, a second-order background model is trained from the input sequences.
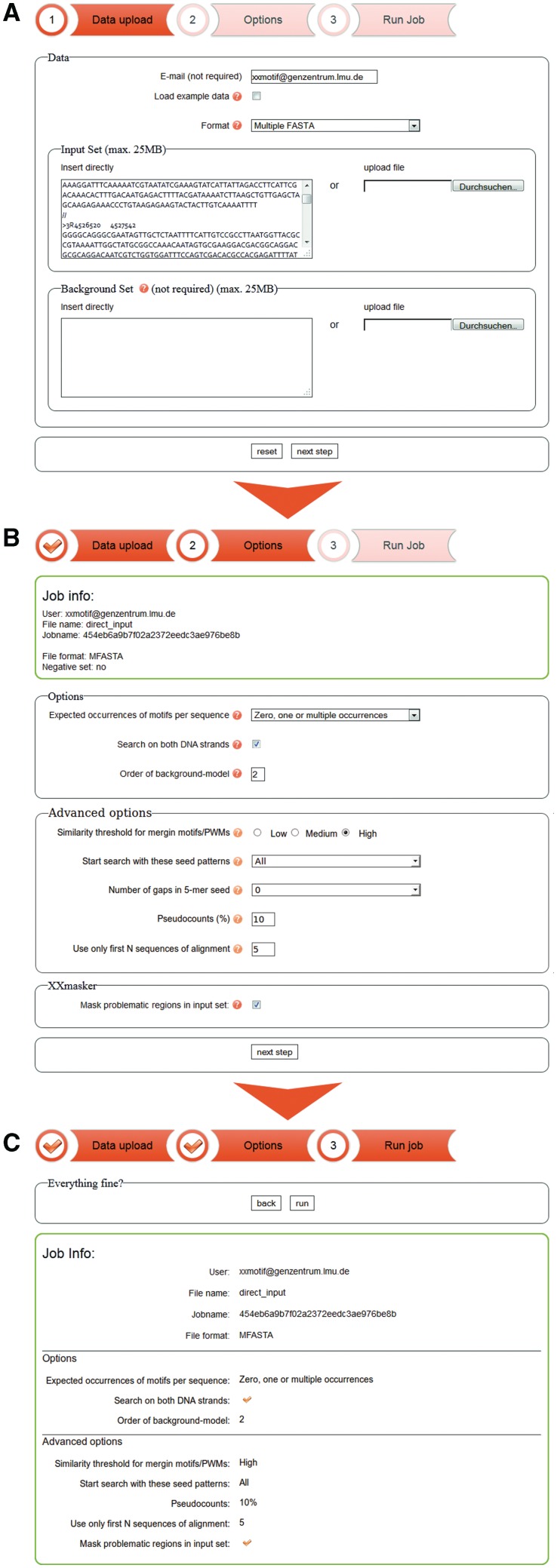
Pages for submitting a job to the XXmotif web server: (A) upload input and background sequence sets, (B) set options for the motif search and (C) verify and submit.
It is not trivial to supply a suitable background set. It should have a trinucleotide distribution similar to the positive sequences while not being enriched for the motifs we seek. More concretely, the background set should have a similar mono-, di- and trinucleotide composition as the positive set. If this is not the case, XXmotif may run very slowly – because it tries to extend sequences too much – and it may produce falsely significant motifs. If in doubt, it is better to omit the background set altogether and to let XXmotif learn the background model from the positive set. We are about to add an automatic quality test that will warn the user if the background set is not well chosen.
To increase the sensitivity of the motif search, XXmotif can calculate motif conservation P-values during the search, which are combined with the enrichment P-values. In this case, the user can upload a set of input and background multiple sequence alignments, using the ‘multiple FASTA’ format.
On the ‘Options’ page, the suggested default options can be modified (Figure 1B). First, the user can specify how many motif occurrences per input sequence are expected. For most transcription factor and microRNA binding sites, we would expect multiple occurrences, for example. For core promoter motifs or splice sites, we would expect zero or one occurrence per sequence. When selecting the latter option, only the best occurrence per sequence is scored, whereas with the former option, all occurrences above a certain single-site significance P-value are scored. Searching on both strands is recommended for all motifs that should occur with similar probabilities on both strands (i.e. as reverse complements of each other). This is true for most transcription factor and microRNA binding sites, for example, but not for core promoter or splice site motifs. The order of the background model specifies how long the patterns are that XXmotif learns from the background sequence set. An eighth-order model learns the frequencies of 9-mer nucleotides to model the correlations between nearby nucleotides. This is the default option selected when a background set is supplied by the user. When the background model is learned from the positive set, the default order is set to 2. If we were to train a model of order 8 from the positive set, no motif shorter than 10 nucleotides could become significant.
Under ‘Advanced options’, the user can first specify one of three similarity thresholds for merging motifs (low, medium and high). Setting this threshold to ‘high’ will produce longer lists of motifs consisting of groups of similar, partially redundant motifs, which were not similar enough to be merged with each other. Setting the threshold to ‘low’ will produce shorter, non-redundant lists of motifs, as similar motifs are merged into a single PWM. However, to be able to discern PWMs of factors with similar binding affinities, the ‘high’ threshold is preferable, as it prevents XXmotif from merging the similar but distinct motifs.
The user can further specify which 5-mer and 6-mer patterns are evaluated as seed patterns to initiate the search. The number of uninformative (gap) positions in the 5-mer seeds can also be set. When setting this parameter to 1, all seeds of the types XXXXX, XNXXXX, XXNXXX, XXXNXX and XXXXNX will be assessed, for example, where X stands for an informative position and N stands for ‘any nucleotide’. Usually, it is sufficient to choose zero here. XXmotif also allows changing the amount of pseudocounts added to the nucleotide counts in motif occurrences. The addition of pseudocounts ensures that the PWM constructed from the motif occurrences in the positive set can predict motif occurrences in new datasets better than without pseudocounts. This parameter does not normally need to be changed. With a check box, the XXmasker tool can be switched off, which masks repeat regions and regions of local homology (see Method Summary section).
Upon pressing ‘next step’, a summary of all selected options is presented (Figure 1C), and corrections can be made using the ‘back’ button. After job submission, the user is directed to a status page, which can be bookmarked and automatically redirects to the results page when the job is finished. If the user has provided an email address, a notification with the result page URL is sent. XXmotif runs around 5 min on 100 sequences of length 1000. Run time scales approximately linearly with the average sequence length and the number of sequences in the positive sequence set.
min on 100 sequences of length 1000. Run time scales approximately linearly with the average sequence length and the number of sequences in the positive sequence set.
OUTPUT
The results page lists the web logos, E-values and number of sites of matched motifs found up to an E-value of 100 (Figure 2A). When both strands were searched, the reverse complement versions of the motifs are also plotted. More detailed results are hidden behind expandable boxes.
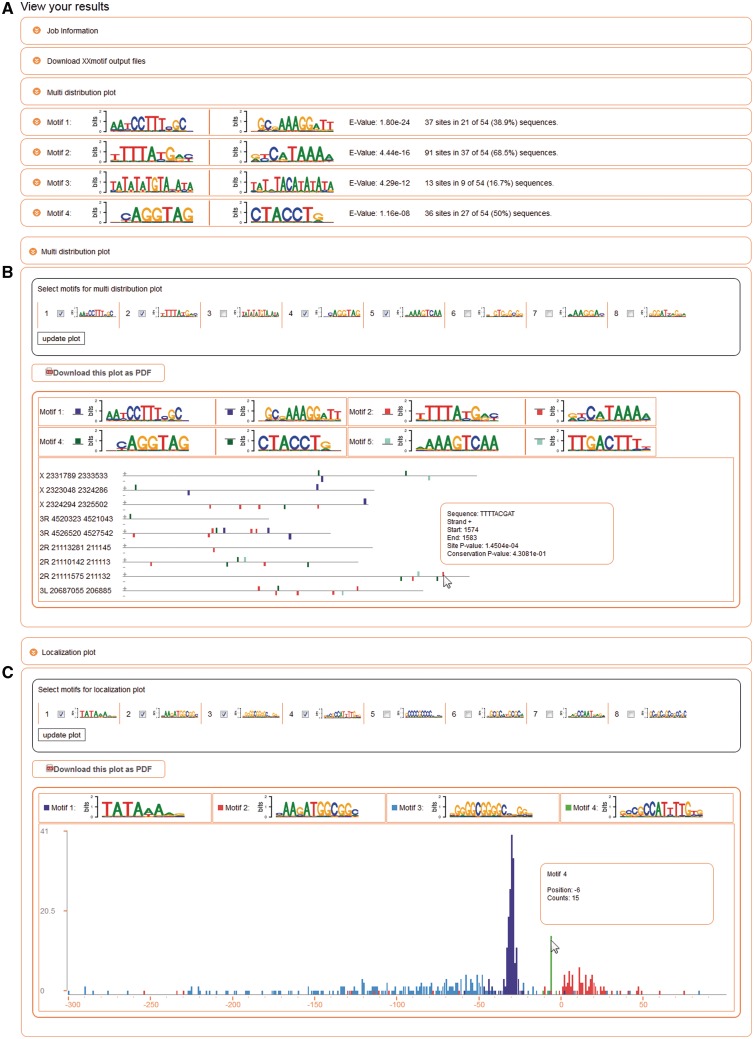
Sample results with boxes that can be expanded with the orange buttons on the left. (A) Summary list of discovered motifs sorted by significance (E-value). (B) The ‘multi distribution plot’ depicts positions and strand of motif occurrences on the input sequences. Motifs can be selected in the upper part. The single-site P-values are represented by the height of the box, their length corresponds to the motif length. (C) The ‘localization plot’ is a histogram view of the positional distribution of selected motifs relative to an anchor point. All plots can be downloaded in PDF format.
The ‘multi distribution plot’ (Figure 2B) depicts with colored boxes the position and strand of significant motif occurrences within the input sequences. The motifs to display in this plot can be selected by the user in the upper part of the plot. This allows plotting clustered binding sites marking, for example, cis-regulatory elements, co-occurring pairs of motifs and other positional biases. Setting the mouse over a particular motif site will show the site’s sequence, strand, start and end position, the single-site P-value measuring the match quality with the PWM and a conservation P-value (if multiple sequence alignments had been supplied). Only sequences with at least one motif site are shown. Most significant motifs are drawn last and may hide less significant ones.
When the input sequences are all of the same length, a ‘localization plot’ can be displayed (Figure 2C). This graph is useful to analyze positional preferences with respect to the fixed-length sequence window of the input sequences. It shows in a histogram view the positional distributions of all user-selected motif occurrences with each motif in a different color. For instance, Motif 1 (TATA-Box) in Figure 2B is exactly positioned between −33 to −27 bp with respect to the transcription start site (TSS) at Position 0, whereas Motif 2 (YY1) is located mainly downstream of the TSS. Mouse-over in the histogram provides the position with respect to the anchor point and the number of counts of the motif. For instance, Motif 4 in Figure 2C has 15 counts sharply peaked at Position −6 with respect to the TSS and a ‘CA’ dinucleotide at Position −1, indicating an initiator like function.
bp with respect to the transcription start site (TSS) at Position 0, whereas Motif 2 (YY1) is located mainly downstream of the TSS. Mouse-over in the histogram provides the position with respect to the anchor point and the number of counts of the motif. For instance, Motif 4 in Figure 2C has 15 counts sharply peaked at Position −6 with respect to the TSS and a ‘CA’ dinucleotide at Position −1, indicating an initiator like function.
Detailed information about each motif can be obtained by clicking the expand buttons in the motif summary list. Two single motif graphs can then be viewed (Figure 3). The ‘motif distribution plot’ is similar to the ‘multi distribution plot’ and indicates the positions of significant matches of the selected motif on the input sequences. The ‘motif site table’ lists all significant matches with their sequence identifiers, strands, positions, the single-site P-values and the sequence contexts of the motif.
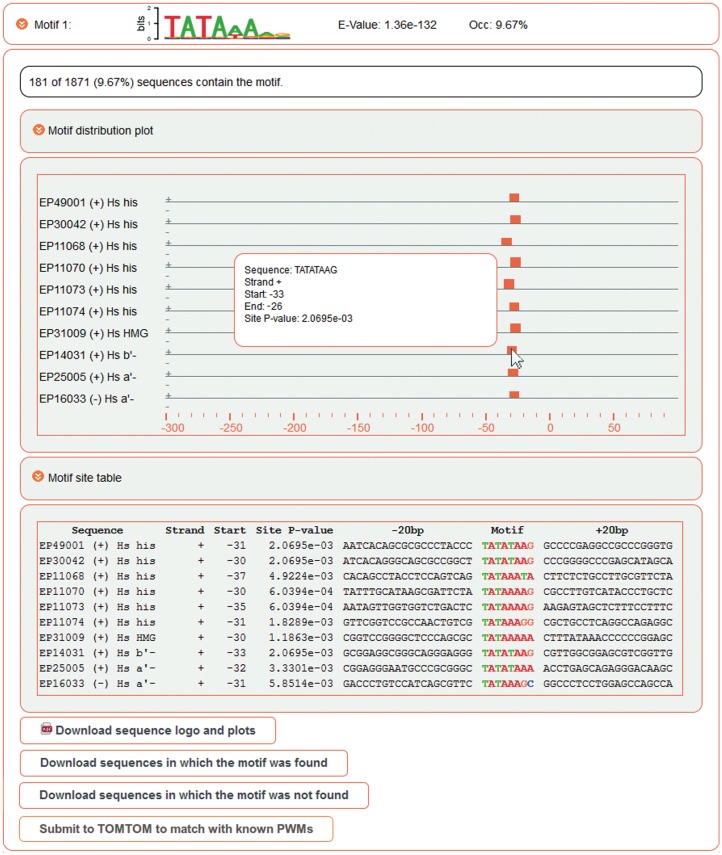
Detailed motif view. The first box (motif distribution plot) plots the position of significant motif matches within the input sequences. The second box (motif site table) gives detailed information on all significant motif matches.
All plots can be downloaded with the buttons below them. All data files generated by the XXmotif program, such as lists of motifs with their occurrence positions, P-values and site sequences, PWM weight coefficients and images of motif logos can be downloaded by expanding the box ‘Download XXmotif output files’.
DOCUMENTATION
Two sample input sets and pre-computed results allow the user to get a quick overview of the server’s usage and results. Help buttons and mouse-over explanations are available for all input options. More general help is listed on the FAQ page.
IMPLEMENTATION
The XXmotif web server runs on an Apache server and is implemented using PHP, PERL and R scripts. The user interface is dynamically generated HTML content with JavaScripts from the jQuery library. Submitted jobs are processed on a Scientific Linux computer cluster.
CONCLUSION
With the XXmotif web server, we aim to make a very sensitive and reliable motif discovery method easily accessible to non-expert users. The server has clearly structured input and results pages and offers various useful interactive analyses. It is unique in being able to include evidence from motif conservation and positional clustering in the motif search.
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG) [SFB646]. We acknowledge financial support by the DFG, the Center for Protein Science Munich (CIPSM), and a research professorship from the Ludwig-Maximilians-Universität München, financed through the Excellence Initiative of the German Bundesministerium für Bildung und Forschung.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gks602
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/40/W1/W104/4936010/gks602.pdf
Citations & impact
Impact metrics
Article citations
In-silico identification and comparison of transcription factor binding sites cluster in anterior-posterior patterning genes in Drosophila melanogaster and Tribolium castaneum.
PLoS One, 18(8):e0290035, 17 Aug 2023
Cited by: 0 articles | PMID: 37590227 | PMCID: PMC10434971
Gene expression patterns associated with caudal fin shape in the cichlid Lamprologus tigripictilis.
Hydrobiologia, 850(10-11):2257-2273, 18 Nov 2022
Cited by: 0 articles | PMID: 37325486 | PMCID: PMC10261199
Large-scale analysis of Drosophila core promoter function using synthetic promoters.
Mol Syst Biol, 18(2):e9816, 01 Feb 2022
Cited by: 9 articles | PMID: 35156763 | PMCID: PMC8842121
Decoding myofibroblast origins in human kidney fibrosis.
Nature, 589(7841):281-286, 11 Nov 2020
Cited by: 304 articles | PMID: 33176333 | PMCID: PMC7611626
FisherMP: fully parallel algorithm for detecting combinatorial motifs from large ChIP-seq datasets.
DNA Res, 26(3):231-242, 01 Jun 2019
Cited by: 3 articles | PMID: 30957858 | PMCID: PMC6589551
Go to all (31) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
P-value-based regulatory motif discovery using positional weight matrices.
Genome Res, 23(1):181-194, 18 Sep 2012
Cited by: 40 articles | PMID: 22990209 | PMCID: PMC3530678
The BaMM web server for de-novo motif discovery and regulatory sequence analysis.
Nucleic Acids Res, 46(w1):W215-W220, 01 Jul 2018
Cited by: 20 articles | PMID: 29846656 | PMCID: PMC6030882
abc4pwm: affinity based clustering for position weight matrices in applications of DNA sequence analysis.
BMC Bioinformatics, 23(1):83, 03 Mar 2022
Cited by: 1 article | PMID: 35240993 | PMCID: PMC8896320
DNA Motif Databases and Their Uses.
Curr Protoc Bioinformatics, 51:2.15.1-2.15.6, 03 Sep 2015
Cited by: 11 articles | PMID: 26334922
Review





