Abstract
Free full text

piRNAs initiate an epigenetic memory of non-self RNA in the C. elegans germline
SUMMARY
Organisms exhibit a fascinating array of gene-silencing pathways, which have evolved in part, to confront invasive nucleic acids such as transposons and viruses. A key question raised by the existence of these pathways is how do they distinguish “self” from “non-self” nucleic acids? Evidence exists for a number of mechanisms that might facilitate detection of foreign sequences including mechanisms that sense copy-number, unpaired DNA, or aberrant RNA (e.g. dsRNA). Here we describe an RNA-induced epigenetic silencing pathway, RNAe, that permanently silences single-copy transgenes. We show that the Piwi Argonaute PRG-1 and its genomically encoded piRNA cofactors initiate RNAe, while maintenance depends on chromatin factors and the WAGO Argonaute pathway. Our findings support a model in which PRG-1 scans for foreign sequences, while two other Argonaute pathways serve as epigenetic memories of “self” and “non-self” RNAs. These findings suggest how organisms may utilize RNAi-related mechanisms not only to recognize and silence foreign genes, but also to keep inventory of all genes expressed in the germ-line.
INTRODUCTION
All organisms balance the need to maintain genetic variation against the danger of accumulating potentially deleterious genes or pathogenic sequences (Antonovics et al., 2011). The experimental introduction of DNA (transgenes) into the germ line provides an opportunity to probe an organism’s response to foreign DNA (Rulicke and Hubscher, 2000), and has revealed that organisms use a variety of mechanisms to silence transgenes in the germ line (Birchler et al., 2003; Brodersen and Voinnet, 2006). Interestingly, some mutants that disrupt transgene silencing also de-silence endogenous genes, including self-replicating elements called transposons (Ketting et al., 1999; Tabara et al., 1999). Thus, the mechanisms involved in transgene silencing protect the genome from invasive DNA elements.
In many organisms transgene silencing has been linked to factors that are also required for the RNAi pathway (Bosher and Labouesse, 2000). RNAi was first identified as a sequence-specific response triggered by double-stranded (ds) RNA (Fire et al., 1998). During RNAi, dsRNA is processed by the RNase III-related protein, Dicer, into ~21 nucleotide (nt) short-interfering (si) RNAs (Bernstein et al., 2001; Carmell and Hannon, 2004; Zamore et al., 2000), which are loaded onto Argonaute (AGO) proteins to form the key effectors of RNA-induced silencing complexes (Hammond et al., 2001; Liu et al., 2004; Meister et al., 2004). AGOs are RNase H-related proteins that use the base-pairing potential of small RNA cofactors to guide sequence-specific binding to target sequences (Song et al., 2004). In some cases, AGOs directly cleave their targets; in other cases, AGOs recruit co-factors that direct mRNA destruction or other modes of regulation.
Despite a clear overlap between the mechanisms that mediate RNAi and the silencing of transposons and transgenes, several findings point to distinct triggering mechanisms. For example, the AGO protein RDE-1 is essential for the dsRNA response in C. elegans, but is not required for transposon or transgene silencing (Ketting et al., 1999; Tabara et al., 1999). RDE-1 engages siRNAs produced by Dicer and mediates the initial search for target RNAs in the cell (Parrish and Fire, 2001; Yigit et al., 2006). RDE-1 is thought to recruit a cellular RNA-dependent RNA polymerase (RdRP), which then utilizes the target mRNA as a template for the production of secondary siRNAs, termed 22G-RNAs (Gu et al., 2009; Pak and Fire, 2007; Sijen et al., 2001; Sijen et al., 2007; Yigit et al., 2006). The 22G-RNAs are loaded onto members of an expanded, partially redundant, group of Worm-specific AGOs (WAGOs). WAGOs that localize to the cytoplasm are thought to mediate mRNA turnover, whereas WAGOs that localize to the nucleus mediate transcriptional silencing (Gu et al., 2009; Guang et al., 2008). Many components of the RNAi pathway that function downstream of RDE-1 are required for transposon and transgene silencing, including the RdRP system (Gu et al., 2009; Smardon et al., 2000), the polynucleotide polymerase RDE-3 (Chen et al., 2005), the nuclease MUT-7 (Ketting et al., 1999), and the WAGO proteins (Yigit et al., 2006), among others (Robert et al., 2004). The fact that RDE-1 is not required for transposon and transgene silencing suggests that features unique to transposons and transgenes underlie the initial recruitment of RdRP to these targets and that dsRNA is unlikely to be the trigger.
In the germ line, RdRPs not only produces 22G-RNAs that interact with WAGOs, but also produce 22G-RNAs that interact with a distinct AGO, CSR-1, required for fertility and chromosome segregation (Claycomb et al., 2009; Yigit et al., 2006). However some factors, including RDE-3 and MUT-7, are only required for WAGO 22G-RNA accumulation (Gu et al., 2009), indicating that the CSR-1 and WAGO 22G pathways also involve distinct mechanisms. Indeed, the WAGO and CSR-1 22G pathways together target virtually all germ-line-expressed mRNAs, however their targets are largely non-overlapping (Gu et al., 2009). Furthermore, unlike the WAGO pathway, the CSR-1 22G pathway does not appear to silence its targets (Claycomb et al., 2009). Instead, the CSR-1 pathway may help to define and maintain euchromatic regions along the holocentric chromosomes in order to support the proper assembly of kinetochores.
In most animals, the Piwi-family AGOs are required for fertility and transposon silencing (Cox et al., 1998; Juliano et al., 2011). In C. elegans, however, the Piwi-related gene product PRG-1 has only been linked to the silencing of one transposon family, Tc3 (Batista et al., 2008; Das et al., 2008). Interestingly, PRG-1 appears to recruit RdRP and the WAGO 22G pathway to maintain Tc3 silencing. Piwi-interacting (pi) RNAs (21U-RNAs in C. elegans) are genomically encoded and appear to be expressed as Pol II transcripts whose single-stranded products are processed and loaded onto Piwi (Aravin and Hannon, 2008; Kim et al., 2009). More than 15,000 distinct piRNA species exist in C. elegans, while millions of species are expressed in the testes of mammals (Aravin et al., 2006; Batista et al., 2008; Das et al., 2008; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006). The majority of these piRNAs map uniquely to the genome and lack obvious targets. As such, their function remains entirely unknown.
Here we use a homologous gene-targeting method, called “Mos1-mediated single copy insertion” (MosSCI), to show that strains bearing identical single-copy transgenes inserted at the same chromosomal site can exhibit opposite and remarkably stable epigenetic fates, either expressed or silenced. Transgenes consisting of an endogenous germ-line-expressed gene fused to a relatively long foreign sequence (e.g. gfp) were prone to silencing. By contrast, otherwise identical transgenes fused to a short foreign sequence (e.g. flag) always expressed. Our genetic and molecular analyses reveal that silencing is dependent on nuclear and cytoplasmic WAGOs and is correlated with the accumulation of 22G-RNAs targeting the foreign portion of the transgene. Importantly, PRG-1 is required to initiate, but not to maintain silencing. We propose that PRG-1 and its 21U-RNA co-factors scan for foreign RNA sequences and initiate WAGO-maintained gene silencing, while endogenous mRNAs are protected from silencing, perhaps by the CSR-1/22G-RNA pathway.
RESULTS
Heritable and dominant silencing of single-copy transgenes
Single-copy insertions can overcome barriers to transgene expression in the germ line (Rieckher et al., 2009). Indeed, the single-copy insertion of transgenes at a defined chromosomal locus via the recently developed MosSCI approach reproducibly achieves germ-line expression (Frokjaer-Jensen et al., 2008). However, while using MosSCI, we were surprised to find that not all single-copy transgenes were expressed in the germ line (Figure 1A and B and Table 1). The failure to express was only common for transgene fusions to lengthy foreign sequences, such as gfp (Table 1); transgenes with the flag epitope sequences were nearly always fully expressed (Table 1). Furthermore, we observed that transgenes where gfp was inserted at the 5′ (rather than 3′) end of the construct were much less likely to be expressed (Table 1). PCR and sequence analyses indicated that non-expressed transgenes are structurally identical to expressed transgenes, suggesting that the former are actively silenced.
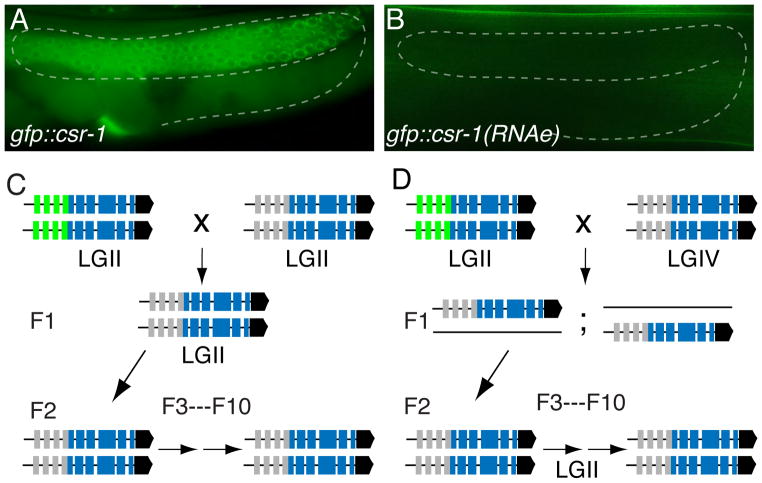
(A, B) Fluorescence micrographs of adult hermaphrodite germ lines from (A) neSi9 gfp::csr-1, GFP positive and, (B) neSi8 gfp::csr-1(RNAe), GFP negative transgenic lines. GFP::CSR-1 is expressed prominently in the peri-nuclear P-granules in the syncytial germ line (dashed outline) and is also visible in the cytoplasm of maturing oocytes.
(C, D) Schematic diagrams illustrating the results of genetic crosses between expressed (green) and silenced (gray) gfp::csr-1 transgenic lines (>100 animals scored per generation after F2). In (C) neSi8 gfp::csr-1(RNAe) hermaphrodites were mated with neSi9 gfp::csr-1 males. In (D) neSi10 gfp::csr-1(RNAe) hermaphrodites, integrated on chromosome IV (LGIV), were mated to neSi9 gfp::csr-1 males, integrated on LGII (LGII). In the F2 generation, the neSi9 gfp::csr-1 allele was segregated away from neSi10 and propagated for 8 more generations.
Table 1
Transgene silencing in MosSCI lines
| Transgene | Positive (%) | n |
|---|---|---|
| gfp::csr-1 | 6.3 | 16 |
| gfp::csr-1a | 75 | 12 |
| gfp::csr-1b | 0 | >50 |
| flag::csr-1 | 83 | 6 |
| gfp::rde-3 | 17 | 18 |
| flag::rde-3 | 100 | 7 |
| rde-3::gfp | 93 | 14 |
| rde-3::flag | 100 | 6 |
| gfp::cdk-1 | 0 | 5 |
| gfp::cdk-1a | 0 | 17 |
| gfp::cdk-1b | 0 | 21 |
| gfp::cdk-1c | 0 | 3 |
| gfp::cdk-1d | 100 | 5 |
| cdk-1::gfp | 100 | 6 |
| gfp::pie-1a | 96 | 23 |
| gfp::pie-1b | 76 | 21 |
We next crossed a silent line to an expressing line to see which phenotype dominates. Strikingly, we found that 100 % of the F1 cross-progeny (n=12) and F2 self-progeny (n=24) failed to express gfp in the germ line (Figure 1C). Identical results were obtained even when the silent and active alleles were inserted on separate chromosomes (Figure 1D), suggesting that chromosomal pairing is not required for transfer of the silent state. Although transgenes with 3′ gfp insertions were less prone to silencing during transgene formation, they were fully silenced when crossed to a silent line (Figure 3A and data not shown).
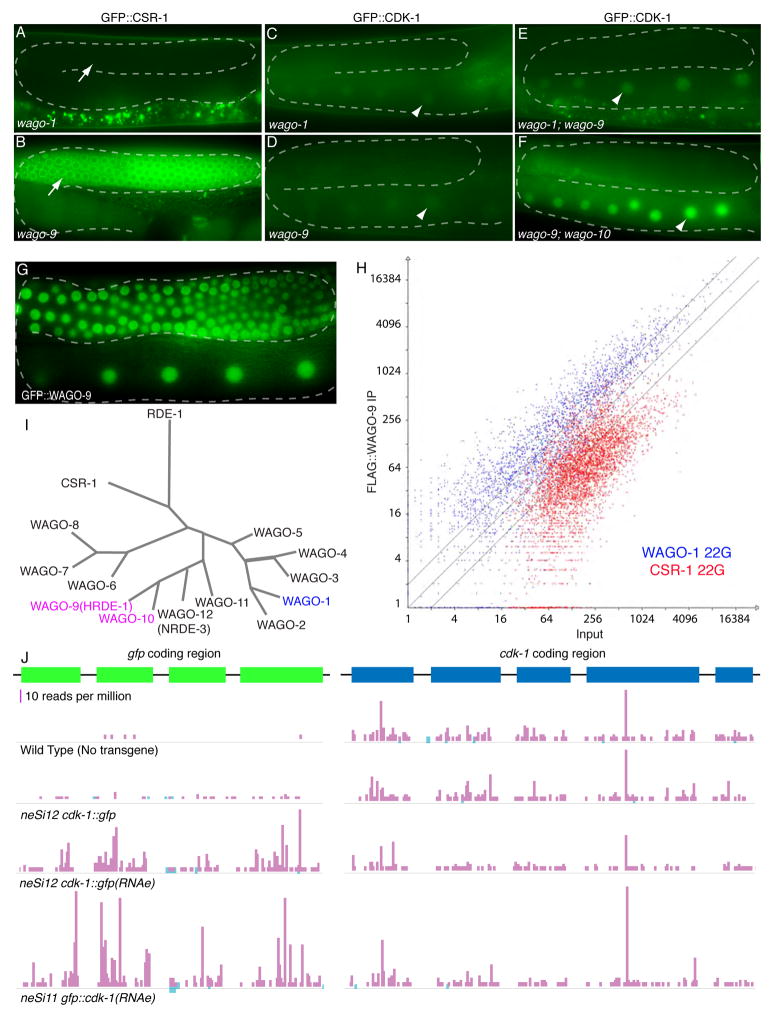
(A–F) Fluorescence microscopy of transgene desilencing in wago mutant backgrounds. The transgenes used were neSi8 gfp::csr-1(RNAe), which localizes to P-granules when expressed (indicated by arrow in A and B), and neSi11 gfp::cdk-1(RNAe), which is most prominent in oocyte nuclei (indicated by arrowheads in C–F).
(G) WAGO-9 is a germ-line expressed nuclear Argonaute. Fluorescence micrograph of GFP::WAGO-9 in the adult hermaphrodite germ line. The dashed lines in the micrograph indicate the position of the syncytial germ line.
(H) WAGO-9-associated small RNAs overlap extensively with WAGO-1 small RNAs. The plot shows the enrichment of 22G-RNAs in FLAG::WAGO-9 IP relative to input. Each point in the graph corresponds to previously identified WAGO-1 (blue) and CSR-1 (red) target genes. The x- and y-axes represent the number of 22Gs (log2 scale) targeting each gene in the Input and WAGO-9 IP samples, respectively. The diagonal lines signify 2-fold enrichment (upper), identity (middle), and 2-fold depletion (lower) of 22G-RNAs in the WAGO-9 IP.
(I) Phylogenetic tree of WAGOs, CSR-1 and RDE-1. Adapted from (Yigit et al., 2006).
(J) Small RNA density along the gfp and cdk-1 coding regions of wild-type and indicated transgenic lines. Vertical bars represent the 5′ nt of a small RNA, and the height of each bar indicates the number of reads that start at that position. The strand is represented by color; sense (light blue) and antisense (pink). Scale bar indicates 10 reads per million. Strain neSi12 cdk-1::gfp(RNAe) was generated by crossing neSi12 cdk-1::gfp to neSi11 gfp::cdk-1(RNAe).
Either parent could contribute the dominant silencing signal. However, when the silent allele was male-derived, it took more than one generation to completely silence the active allele (data not shown). Nevertheless, in the F3 and subsequent generations, 100 % of the descendants were GFP negative (n>100). The silent phenotype was fully penetrant with no evidence of expression or reversion even after the formerly active allele was re-segregated as a homozygote (Figure 1D). These results clearly indicate that the failure to express these single-copy transgenes represents an active silencing process that involves a dominant trans-acting silencing signal. Because the silent state is stable indefinitely over many generations and (as shown below) involves a small-RNA silencing signal, we refer to this phenomenon as RNA-induced epigenetic silencing (RNAe). We identify transgenes exhibiting this type of silencing by including the term “(RNAe)” after the transgene name [e.g. neSi11 gfp-1::cdk-1-(RNAe)].
High-copy transgenes in C. elegans can induce co-suppression of endogenous homologous genes (Dernburg et al., 2000; Ketting and Plasterk, 2000). Several of the transgenes we analyzed are fusion constructs with essential genes (e.g. gfp::cdk-1) and should result in obvious visible phenotypes if the corresponding endogenous locus was co-suppressed. However, no phenotypic evidence of co-suppression was observed in the silent lines analyzed (data not shown), suggesting that despite the dominant nature of the silencing signal, silencing does not spread to the endogenous locus. To ask if there is a partial suppression of the endogenous locus, we performed Western blot analysis to determine the relative expression of the transgene and endogenous protein products in both active and silent lines. Consistent with the lack of phenotypic evidence for co-suppression, we observed identical levels of endogenous protein expression in both the active and silent transgenic lines (Figure 2A).
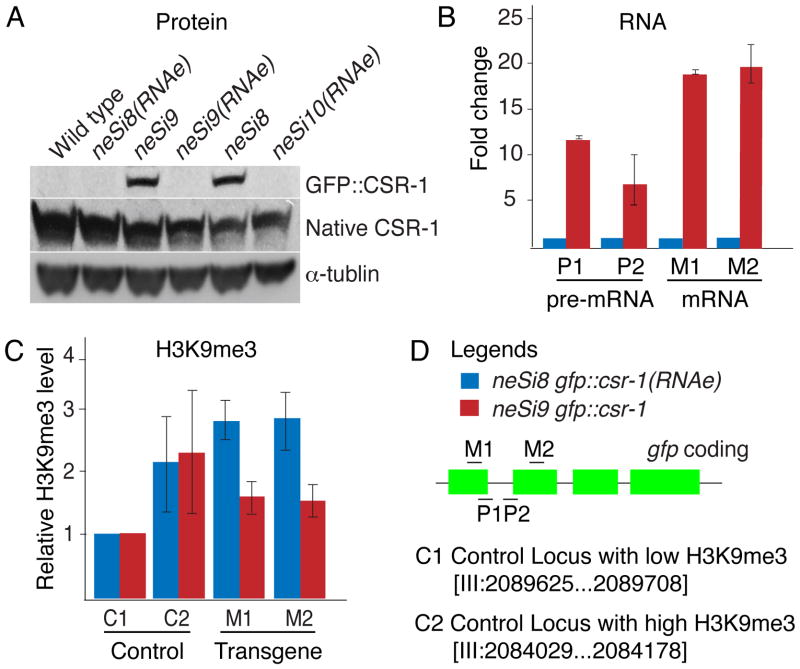
(A) Analysis of protein expression in wild-type and transgenic strains (as indicated). The blot was probed with anti-GFP (GFP::CSR-1), anti-CSR-1 (Native CSR-1) and anti-α-tubulin (α-tubulin) antibodies (as indicated). The neSi9 gfp::csr-1(RNAe) strain was generated by crossing neSi9 gfp::csr-1 to neSi10 gfp::csr-1(RNAe). The neSi8 gfp::csr-1 strain was generated by crossing neSi8 gfp::csr-1(RNAe) to rde-3.
(B, C) qPCR analysis of gfp::csr-1 mRNA, pre-mRNA, and H3K9me3 levels in silent (blue) and expressed (red) transgenic lines. The strains and probes used are indicated in (D). In (B) gfp::csr-1 expression was normalized to the clp-3 mRNA. The data is shown as fold-change between the expressed and silent gfp::csr-1 alleles. Error bars represent the standard deviation for two experimental replicates. In (C), error bars indicate the standard deviation for three experimental replicates.
RNAe requires chromatin factors and correlates with H3K9me3
To ask if silencing is regulated transcriptionally or post-transcriptionally, we isolated total RNA from otherwise identical silent and active gfp::csr-1 strains and measured the abundance of pre-mRNAs and mRNAs by real-time quantitative PCR (qPCR). We found that both the pre-mRNA and mRNA levels were significantly reduced in the silent line compared to the active line (Figure 2B and D). Moreover, although a reduction at the pre-mRNA level appeared to account for the majority of silencing, a further reduction was evident at the mRNA level, suggesting that silencing is achieved at both transcriptional and post-transcriptional levels (Figure 2B and 2D).
Previous work has shown that the methylation of lysine 9 on histone H3 (H3K9me), a histone modification associated with silent chromatin, is enriched on high-copy number transgenes in the germ line (Bessler et al., 2010; Kelly et al., 2002). Furthermore, germ-line silencing of high-copy transgenes is dependent on a number of chromatin-associated factors, including the Polycomb-Group complex (MES-2/-3/-6), Trithorax-related (MES-4) and the heterochromatin proteins (HPL-1 and -2) (Couteau et al., 2002; Grishok et al., 2005; Kelly and Fire, 1998; Kelly et al., 1997). Consistent with these previous findings, we found that transgene sequences from a silent MosSCI allele, but not an active MosSCI allele, were enriched in Chromatin Immunoprecipitation (ChIP) experiments using antibodies specific for H3K9me3 (Figure 2C and D). Furthermore, we found that mes-3, mes-4, and hpl-2 mutants all de-silenced the gfp::csr-1 and gfp::cdk-1 transgenes (Table 2). These findings suggest that the maintenance of single-copy transgene silencing involves a chromatin component similar to that described for maintenance of high-copy transgene silencing.
Table 2
Genetic test for maintenance of gene silencing
| Gene(allele) | Gene function | Transgene expression
| |
|---|---|---|---|
| gfp::csr-1 | gfp::cdk-1 | ||
|
| |||
| rde-1(ne300) | Argonaute in RNAi | − | − |
|
| |||
| prg-1(tm872) | Piwi homolog | − | − |
|
| |||
| rde-3(ne3370) | Poly(A) polymerase | + | + |
|
| |||
| mut-7(ne4255) | 3′ to 5′ exonuclease | + | + |
|
| |||
| hpl-1(tm1624) | HP1 homolog | − | − |
|
| |||
| hpl-2(tm1489) | HP1 homolog | + c | + b |
|
| |||
| hpl-1(tm1624) | HP1 homolog | + | + |
| hpl-2(tm1489) | HP1 homolog | ||
|
| |||
| met-1(n4337) | Methyltransferase | − | NA |
| met-2(n4256) | Methyltransferase | ||
|
| |||
| mes-3(bn35) a | Polycomb complex | + b | + b |
|
| |||
| mes-4(bn23) a | Trithorax complex | + b | + b |
|
| |||
| wago-1(tm1414) | Cytoplasmic WAGO | − | + b |
|
| |||
| nrde-3(tm1116) | Nuclear WAGO | − | NA |
|
| |||
| wago-9(tm1200) | Nuclear WAGO | + | + b |
|
| |||
| wago-1(tm1414) | Cytoplasmic WAGO | NA | + |
| wago-9(tm1200) | Nuclear WAGO | ||
|
| |||
| wago-9(tm1414) | Nuclear WAGO | NA | + |
| wago-10(tm1186) | Nuclear WAGO | ||
|
| |||
| wago-9(tm1414) | Nuclear WAGO | NA | + |
| wago-10(tm1186) | Nuclear WAGO | ||
| nrde-3(tm1116) | Nuclear WAGO | ||
|
| |||
| wago-9(tm1414) | Nuclear WAGO | NA | + |
| wago-10(tm1186) | Nuclear WAGO | ||
| wago-11(tm1127) | Nuclear WAGO | ||
| nrde-3(tm1116) | Nuclear WAGO | ||
Maintenance of silencing requires RNAi-related factors
The trans-acting nature of the silencing phenomenon suggested the possible involvement of an RNAi-related small RNA pathway. To explore this possibility we crossed a silent transgenic strain into strains bearing mutations in RNAi components. Two downstream factors in the exo-RNAi pathway, rde-3 and mut-7, which encode a beta-nucleotidyl transferase and a 3′–5′ exonuclease respectively (Chen et al., 2005; Ketting et al., 1999), are known to be required for the maintenance of transposon silencing and have been implicated in co-suppression (Dernburg et al., 2000; Ketting and Plasterk, 2000) and high-copy number transgene silencing (Tabara et al., 1999). Consistent with the involvement of these factors in the maintenance of RNAe we found that crossing a silent transgene into these mutant strains resulted in fully restored transgene expression (Table 2).
We also examined the consequences of crossing strains de-silenced in the rde-3 mutant background back into a wild-type rde-3(+) background. We found that for a gfp::csr-1 transgene de-silenced by rde-3, 27 % of rde-3(+) segregants (n=15) retained expression after outcross (Figure 2A and Supplementary figure 1). However, in contrast, strains bearing the gfp::cdk-1 transgene, also desilenced by rde-3, were always rapidly and fully re-silenced by reintroducing rde-3(+) (n>20).
Nuclear and Cytoplasmic WAGOs are required for silencing maintenance
Because RDE-3 and MUT-7 are required for the accumulation of RdRP-derived 22G-RNAs that engage WAGOs (Gu et al., 2009; Yigit et al., 2006), we asked whether WAGOs are required for the maintenance of single-copy transgene silencing by crossing silent lines with several different wago mutant strains. We found that a mutation in the predominantly cytoplasmic germ-line WAGO, wago-1(tm1414) (Gu et al., 2009), partially de-silenced a gfp::cdk-1 transgene but did not de-silence a gfp::csr-1 transgene (Table 2 and Figure 3A and C).
The finding that wago-1 mutants failed to de-silence gfp::csr-1 and only partially de-silenced gfp::cdk-1 suggested that additional WAGOs contribute to RNAe (Figure 3I). Furthermore, because RNAe involves a chromatin component, we suspected that nuclear WAGOs might be important for RNAe. The nuclear WAGO, NRDE-3/WAGO-12, is required for nuclear RNAi and transcriptional silencing in somatic tissues (Burton et al., 2011; Guang et al., 2008), and nrde-3 mutants failed to de-silence a gfp::csr-1 transgene in the germ line (Table 2). However, within the WAGO sub-clade that includes NRDE-3 (Figure 3I), we identified WAGO-9 (HRDE-1/C16C10.3) as a nuclear WAGO that is restricted to the germ line (Figure 3G). Furthermore, we found that wago-9(tm1200) mutants fully de-silenced a gfp::csr-1 transgene and partially de-silenced a gfp::cdk-1 transgene (Figure 3B and D), the converse of the relationship between wago-1(tm1414) and these RNAe lines. The de-silencing of gfp::cdk-1 was increased in a wago-1; wago-9 double mutant (Figure 3E).
Because gfp::cdk-1 was not completely desilenced by these wago mutant combinations, we asked if additional members of the nuclear WAGO sub-clade play a role in gfp::cdk-1 silencing. Indeed, gfp::cdk-1 was strongly de-silenced in a wago-9; wago-10 (t22h9.3); wago-11(f49f6a.1); nrde-3 quadruple mutant, as well as in a wago-9; wago-10 double mutants (Table 2 and Figure 3F). Taken together, these findings indicate that cytoplasmic and nuclear WAGOs contribute to RNAe in parallel and that the input from cytoplasmic and nuclear WAGOs varies between individual RNAe lines.
The small RNAs that associate with WAGO-1 were previously identified by immunoprecipitation (IP) of FLAG::WAGO-1 followed by deep sequencing of associated small RNAs (Gu et al., 2009). We performed similar studies using a flag::wago-9 transgene. We found that the targets of WAGO-9 are largely overlapping with those of WAGO-1 (Figure 3H). These observations suggest that nuclear and cytoplasmic WAGOs share targets and are likely to share a common 22G biogenesis pathway.
Silencing correlates with accumulation of 22Gs targeting GFP
To examine the small RNA profile associated with germ-line silencing, we dissected gonads from different transgenic lines, including active, silent, and converted lines (e.g. active to silent and silent to active lines), and prepared small RNA libraries for deep sequencing (Figure 3J and Supplementary Figure 1). Strikingly, each silenced line exhibited a marked accumulation of 22G-RNAs that were restricted to the gfp portion of the transgene sequence (Figure 3J and Supplementary Figure 1). Consistent with the idea that these 22Gs are WAGO-pathway dependent, we found that 22G-RNA levels targeting gfp were significantly reduced in lines converted from silent to active by crossing through an rde-3 mutant background (Supplementary Figure 1).
Native germ-line-expressed genes are recognized by low levels of 22G-RNAs that engage CSR-1 (CSR-1-22Gs) (Claycomb et al., 2009). We found that the transgene sequences corresponding to endogenous germ-line expressed mRNA sequences, always exhibited low 22G-RNA levels similar to those observed for the endogenous sequences in wild-type non-transgenic animals (Figure 3J and Supplementary Figure 1). These findings suggest that the WAGO-mediated silencing signal only targets the foreign sequences of the transgene.
Initiation of silencing requires the Piwi Argonaute PRG-1
Despite interacting with distinct small RNA species, both PRG-1 and RDE-1 function as primary AGOs upstream of WAGO-22G mediated silencing (Batista et al., 2008; Das et al., 2008; Pak and Fire, 2007; Sijen et al., 2007; Yigit et al., 2006). However, we found that neither prg-1 nor rde-1 mutants could activate an already established silent transgene (Table 2). To explore the possibility that either PRG-1 or RDE-1 is involved in the initiation of RNAe, we generated new transgenic lines by directly injecting into prg-1 and rde-1 mutants. We chose to inject the gfp::cdk-1 construct, because 100 % of MosSCI lines were silent when established in the wild-type background (n=21) (Table 1). In an rde-1(ne300) mutant strain, we found that the cdk-1::gfp transgene was silenced in all three newly isolated lines (Table 1). Strikingly, however, when we repeated the same experiments with prg-1(tm872) mutants, the cdk-1::gfp transgene was fully active in all five independently generated transgenic lines (Figure 4 and Table 1). Taken together, these findings suggest that PRG-1 and piRNAs are involved in the initiation of transgene silencing, whereas dsRNA (e.g. from bi-directional transcription of the transgene) is not involved.
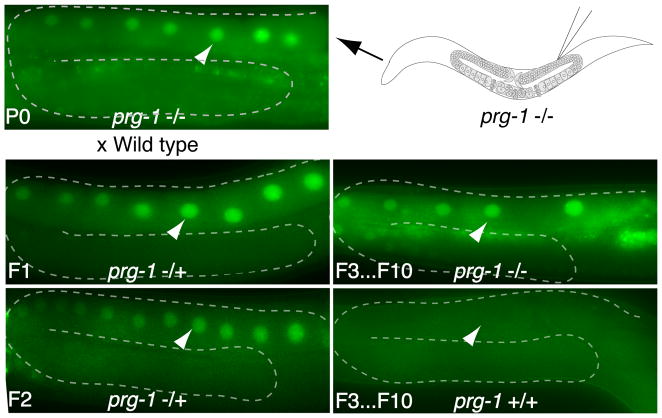
prg-1(tm872) mutant worms injected with the gfp::cdk-1 construct (top right) give rise to MosSCI lines that express GFP::CDK-1 (P0, top left). The micrographs show the expression status of GFP::CDK-1 in oocyte nuclei (arrowheads) before (P0) and after outcrossing to wild type (F1 and F2 panels), and after segregating homozygous prg-1(+) and prg-1(−) strains for several generations (F3–F10 panels). More than 10 worms were examined per generation. Results are detailed in the text.
When established in the wild-type background, the epigenetic state of a transgene, whether active and silent, is stably maintained over many generations. If PRG-1 is only required for the initiation of silencing, then we expected that active transgenes established in the prg-1 mutant background would remain stably active even after outcrossing to a wild-type strain. However, we found that the active transgenes established in the prg-1 mutant rapidly became silent after outcrossing. We found that 96 % of the heterozygous F1 progeny (n=24) exhibited active expression of gfp::cdk-1. However, by the F3 generation, only 9 % of heterozygous or homozygous wild type exhibited active expression (n=66). Conversely, among the F3 that were once again homozygous for prg-1, 77 % (n=30) maintained the active state. These findings raise the possibility that PRG-1 may not be the most upstream factor in the initiation of silencing. Alternatively, PRG-1 may be upstream of competing pathways: one that initiates silencing and one that initiates anti-silencing (see DISCUSSION).
A trans-acting anti-silencing signal
The findings described above indicate that extremely stable silencing associated with single-copy transgenes is initiated by piRNAs and requires the same downstream factors that are required for RDE-1-dependent dsRNA-induced silencing. However, to our knowledge, dsRNA-induced silencing always results in reversible silencing (Alcazar et al., 2008; Vastenhouw et al., 2006). We therefore wondered if PRG-1 somehow initiates a more stable mode of silencing than that initiated through RDE-1. To test this idea, we used gfp dsRNA to initiate silencing of active GFP(+) transgenes and monitored expression for multiple generations after removal of the dsRNA trigger. In each generation, we scored 10 animals from each of 10 independent lines for a total of 100 worms per generation. For the gfp::csr-1 transgene, we found that, as expected, 100% of the animals were silenced in the F1 generation. Remarkably, however, 100% of gfp::csr-1 worms remained silent in all ten lines for greater than 10 generations with no evidence of reversion. Similar results were obtained for the cdk-1::gfp transgene. This transgene, which was less prone to silencing during initial transgenesis, remained completely silent in 6 of 10 lines, whereas 4 lines recovered expression. Thus, the susceptibility of these active transgene lines to piRNA induced silencing mirrors their susceptibility to dsRNA-induced permanent silencing.
The above data suggest that the MosSCI transgenes studied here are more sensitive than endogenous genes to permanent silencing by RNAi. To ask if this is generally true of transgenes, we asked whether exposure to gfp(RNAi) could permanently silence low-copy transgenes generated several years ago by different methods. For this analysis we chose two different transgenes generated by different approaches, gfp::wrm-1 (Nakamura et al., 2005), which was produced by injecting an engineered yeast artificial chromosome, and oma-1::gfp (Lin, 2003), which was generated by biolistic gold-particle mediated transformation (Praitis, 2006). We found that although both of these transgenes were efficiently silenced by RNAi in F1 (100 %, n=100), they always fully recovered expression after removal of the dsRNA trigger (100 % GFP+ by the F3 generation).
Considering the resistance of gfp::wrm-1 and oma-1::gfp to permanent silencing by dsRNA, we wondered if they might also be resistant to trans-silencing in crosses with silent transgenes. Surprisingly, not only were both gfp::wrm-1 and oma-1::gfp resistant to trans-silencing, we found that both transgenes could dominantly activate the expression of a silent transgene in the F1 cross progeny (Figure 5A–C). Expression was initially low in the F1 and F2, but, when propagated along with gfp::wrm-1 or oma-1::gfp transgenes, the trans-activated transgene alleles became fully expressed by the third generation (Figure 5A–C). Finally, after propagating the activated transgene lines in the presence of gfp::wrm-1 or oma-1::gfp for a few generations, we segregated the transgenes away from each other. We found that gfp::cdk-1 returned to its silent state (Figure 5B), while cdk-1::gfp remained stably expressed after exposure to the active transgene (Figure 5C). These findings indicate that a trans-acting dominant mechanism can activate a silent transgene and suggests that activating and silencing signals compete with each other for dominance when transgene alleles interact.
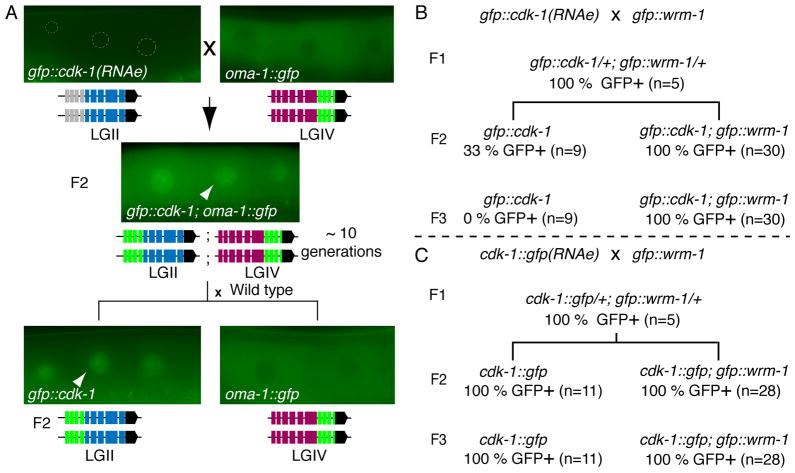
(A) Schematic illustrating the cross between neSi11 gfp::cdk-1(RNAe) and teIs1 oma-1::gfp. The micrographs show the expression status of GFP::CDK-1 in oocyte nuclei (arrowhead) when expressed and OMA-1::GFP in the oocyte cytoplasm. The dashed circles (top left) show the position of GFP-negative oocyte nuclei in the neSi11 gfp::cdk-1(RNAe) strain. The cartoon below each micrograph indicates whether the transgene is expressed (green) or silent (gray).
(B–C) Schematics illustrating crosses between neIs2 gfp::wrm-1 males and (B) neSi11 gfp::cdk-1(RNAe) or (C) neSi12 cdk-1::gfp(RNAe) hermaphrodites. After each cross the two transgenes were either maintained together or allowed to segregate away from each other. The GFP::WRM-1 signal is very weak and was scored periodically during the analysis. The percentage of GFP+ worms indicates the expression of the CDK-1 fusion proteins.
DISCUSSION
Recognition of self and non-self nucleic acids
Organisms employ an array of mechanisms that afford some control over the expression of foreign sequences (Hornung and Latz, 2010; Murray, 2002). However, little is known about how foreign sequences are distinguished from self, and how silencing is initiated and maintained. Using homologous recombination to introduce single-copy transgenes at a defined locus in the C. elegans genome, we have shown that otherwise identical transgenes can adopt either active or silent epigenetic states. Our analysis of this phenomenon has identified distinct AGO systems that function in the initiation and maintenance phases of silencing. The initiation of silencing requires the Piwi family member, PRG-1, while maintenance depends on multiple members of the WAGO clade. Interestingly, silencing was only observed when foreign sequences were included in the transgene, and was more prevalent when foreign sequences were 5′ relative to endogenous sequences in the transgene construct. In genetic crosses, strains bearing a silent transgene exhibited a trans-acting signal that induced heritable silencing on a second newly-generated active transgene. Surprisingly, however, at least two independently-generated, actively-expressed transgenes that were made several years ago exhibited a potent trans-acting signal that could activate newly generated silent transgenes. These findings indicate that competing epigenetic pathways generate trans-acting signals that can silence or activate homologous genes.
We propose a model in which three AGO pathways may function together in a system that allows the animal to maintain an inventory of expressed mRNAs and to recognize and silence foreign sequences (Figure 6B). In this system, PRG-1 uses genomically encoded piRNA co-factors to scan, via imperfect base pairing interactions, for foreign RNAs expressed in the germ line. Upon targeting, PRG-1 recruits RdRP to produce anti-sense 22G-RNAs, which are loaded onto WAGO Argonautes. In turn, WAGOs mediate silencing and establish a memory of non-self RNA. A third, as yet unidentified pathway provides a memory of self and is capable of acting as an anti-silencing signal. Although our studies have not yet identified the anti-silencing (self-recognition) mechanism, the CSR-1 22G-RNA pathway provides an attractive candidate for this activity (See further discussion below). We propose that the self-recognition pathway can prevent PRG-1 from recruiting the WAGO pathway, providing a function that helps expressed transgenes to maintain their expression and helps endogenous genes to recover from WAGO-mediated silencing in RNAi. The initial decision to silence or express the transgene represents a stochastic outcome of competition between establishment of these epigenetic self- or non-self memories.
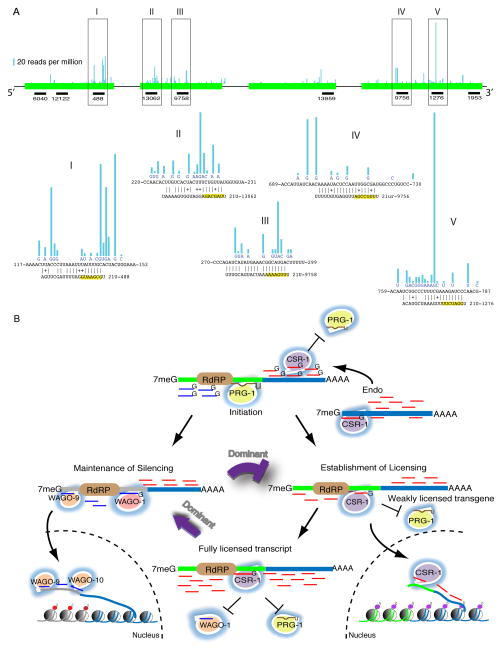
(A) Schematic showing the density of 22G-RNAs targeting GFP in neSi8 gfp::csr-1(RNAe) worms, as described in the legend of Figure 3J. Scale bar indicates 20 reads per million. The positions of several 21U-RNAs that could base pair with mismatches to the gfp sequence are indicated below the gene diagram. Five major 22G hotspots (numbered boxes) are enlarged to show the base pairing between the candidate 21U-RNA and gfp, as well as the density of 22G-RNAs at single-nucleotide resolution. Each 21U-RNA has at most two G:U pairs within the seed region (nts 2–8th, yellow highlight), and at most 3 non-seed mismatches (nts 9–21st).
(B) Model for the allelic interactions between transgenes observed in this study.
Repetitive and single copy transgenes exhibit distinct but overlapping silencing mechanisms
The silencing of high-copy and single-copy transgenes share several features, including chromatin and WAGO 22G pathway requirements. However, several observations suggest that high-copy transgenes are subject to distinct modes of recognition and silencing. First, high-copy transgenes were at best only partially de-silenced in the mutant contexts described above (data not shown) whereas single copy transgenes were fully desilenced and in some cases even maintained their expression after outcrossing to wild type. Second, high-copy transgenes were fully and rapidly silenced in the germ-line of prg-1 mutant animals (data not shown), indicating that a distinct initiation step is involved in silencing. Third, high-copy number silencing was observed even when only the native germ-line gene sequences were present in the transgene (data not shown), whereas silencing of the single-copy transgene was correlated with the presence of foreign sequences within the germ-line-expressed portion of the transgene construct. Finally, unlike the single-copy silencing described here, where trans-silencing remains focused on foreign sequences, high-copy transgenes were found to elicit co-suppression of the endogenous gene (Dernburg et al., 2000; Ketting and Plasterk, 2000). Taken together these observations are consistent with the existence of at least two distinct modes of silencing that act on transgenes, one that depends on high-copy number and can spread throughout the transgene, and a second that requires PRG-1 and is restricted to portions of the transgene composed of foreign sequences.
21U-RNAs complementary to gfp are correlated with 22G biogenesis
Our findings suggest that transgene silencing is initiated by PRG-1 and depends on the presence of foreign gfp sequences in the transgene. In a parallel study, PRG-1 was shown to initiate silencing of synthetic reporters containing sites perfectly complementary to 21U-RNAs (Lee et al., co-submitted and Miska E., personal communication). Mismatched pairing was also correlated with silencing both on transgenes (Miska E., personal communication) and on presumptive endogenous targets (Lee et al., co-submitted and Miska E., personal communication). We have not identified 21U-RNAs that are perfectly complementary to gfp, however, there are potentially dozens of high affinity 21U-RNA-GFP target sites (data not shown). Our recent studies (Lee et al., co-submitted) suggest that PRG-1/21U-RNA targeting initiates 22G-RNA biogenesis within a +/− 40 nt window around the site of 21U-RNA complementarity on the target RNA. We found 8 regions on gfp where 22G-RNAs were detected at greater than 75 reads per million in a silent strain (Figure 6A). We identified potential high-affinity 21U-RNA interactions in all 8 regions. These base-pairing interactions and the proximal 22G-RNAs found in a silent transgenic strain are shown at single-nucleotide resolution in Figure 6A (also see EXPERIMENTAL PROCEDURE). Validation of these candidate 21U-RNA target sites and the general rules that govern piRNA targeting remain to be elucidated in the future.
CSR-1 as an anti-silencing Argonaute
At least three mechanisms must work together to explain the all-or-none nature (expressed or silent) of the epigenetic states observed, and the stable heritability of these states once established (Figure 6B). The genetic studies, thus far, have implicated PRG-1 in the initiation of silencing and the WAGO pathway in the maintenance of silencing. The third pathway required is a “maintenance of expression” or “anti-silencing pathway”. Such a pathway is necessary to explain why, once established, active transgenes are stably transmitted from one generation to the next without undergoing spontaneous silencing. An anti-silencing pathway could also explain how certain active transgenes are able to dominantly activate silent transgenes (Figure 6B).
The CSR-1 22G pathway targets endogenous germ-line-expressed mRNAs (Claycomb et al., 2009), and is an ideal candidate for an anti-silencing pathway. In vitro, CSR-1 is catalytically active and capable of cleaving a target (Aoki et al., 2007), whereas the all WAGOs lack key catalytic residues (Yigit et al., 2006). Perhaps CSR-1 can compete by selectively destroying RNAs on which RdRP is bound, thus preventing or attenuating the production of WAGO 22G-RNAs. It is not known how CSR-1 targeting is first established. However, all of the transgenes that we analyzed contain endogenous germ-line expressed sequences known to be targeted by CSR-1 22Gs. Perhaps CSR-1 22Gs can spread in trans along a target transcript as has been shown for the transitive RNAi mediated by WAGOs after dsRNA targeting (Pak and Fire, 2007; Sijen et al., 2007; Yigit et al., 2006). If so, then stable expression of a transgene may reflect the spread of CSR-1 targeting to the foreign portion of the transgene prior to PRG-1 recognition.
Interestingly, although the anti-silencing signal initially appears to be sufficient to prevent PRG-1 driven silencing, it is not sufficient to prevent silencing initiated in crosses with a silent transgene or when dsRNA is used to stimulate gene silencing. If CSR-1 22G-RNAs represent the anti-silencing signal, then it will be interesting to explore whether the levels of CSR-1 22G-RNAs build up over generations. If so then, the older transgenes, which were able to activate a silent transgene, may show relatively high levels of CSR-1 22G-RNAs targeting gfp when compared to newly established lines. However, it is also possible that as yet unknown features of the chromatin environments of the different transgenes drives their different sensitivity to trans-silencing and their differing abilities to trans-activate or to recover from silencing spontaneously.
Finally, it is worth noting that PRG-1 may function upstream of RdRP recruitment for both the CSR-1 and WAGO pathways. If so, then the decision to express or silence a new transgene may represent the result of a competition between the CSR-1 and WAGO pathways for RdRP loading, downstream of this initial recruitment. An expectation for such a model would be that both the maintenance-of-silencing (non-self) and maintenance-of-expression (self) pathways should fail to initiate when PRG-1 is absent. To further explore this question it will be important to analyze the behavior of additional transgenes established in the prg-1 mutant background.
RNAe, RNA-induced epigenetic inheritance
While RNAe likely serves as a defense against transposons and other invasive sequences, it is also possible that it could have a more general role with significant potential impact on evolution. For example, RNAe could accelerate evolutionary change by heritably modulating the expression of unpaired parental alleles to allow the phenotypic expression of recessive traits among F1 progeny. Consistent with this idea a recent report has shown that a paternally derived allele with no homolog in the hermaphrodite genome is subject to dominant silencing, and that silencing was prevented by injecting single stranded RNAs matching the coding region of the absent gene into hermaphrodite gonads prior to the cross (Johnson and Spence, 2011). These observations are consistent with a mechanism for the licensing of gene expression by maternal RNA and, along with the present study, support the existence of an epigenetic switch that is sensitive to prior expression of a gene. These phenomena are also similar to a form of allelic interaction known as paramutation that has been described in organisms ranging from mice to corn (Erhard and Hollick, 2011). Thus, it appears likely that diverse organisms can both track and respond epigenetically to the history of gene expression. In C. elegans, this process overlaps mechanistically with RNAi, but involves a distinct triggering mechanism that requires the genomically encoded piRNAs.
EXPERIMENTAL PROCEDURES
Genetics
All C. elegans strains were derived from the Bristol N2 strain and cultured as described (Brenner, 1974). The strains used in this study are listed in Supplemental Information.
MosSCI by direct injection
MosSCI lines were generated by the direct insertion method using strain EG4322 and EG5003 as described (Frokjaer-Jensen et al., 2008). Targeting vectors are described in Supplemental Information.
MosSCI by heat-shock and ivermectin selection
Strain WM189 was injected with a DNA mixture containing 50 ng/μl each of pRF4::rol-6(su1006), pCCM416::Pmyo-2::avr-15, and pJL44::Phsp-16.48::MosTase::glh-2utr (Frokjaer-Jensen et al., 2008), and either 1 ng/μl or 50 ng/μl of targeting vector. MosSCI was performed using the heat-shock method (Frokjaer-Jensen et al., 2008) and single-copy insertion lines were selected on ivermectin to select against animals carrying the extrachromosomal array. Additional details are provided in Supplemental Information.
Small RNA cloning from isolated germ lines
Ten gonads from each strain were dissected in 1x PBS containing 0.1 mM EDTA, 1 mM Aurin tricarboxylate, 0.1% Tween 20, and 0.2 mM levamisole (Wang et al., 2009). Total RNAs were extracted with 5 volumes of TRI Reagent (MRC). Small RNAs were gel-purified and cloned as described (Gu et al., 2009). gfp::csr-1 small RNAs were pre-treated with Tobacco Acid Phosphatase (TAP, Epicenter Biotechnologies). gfp::cdk-1 and cdk-1::gfp small RNAs were pre-treated with CIP/PNK (NEB). Libraries were sequenced in the UMass Deep Sequencing Core using an Illumina HiSeq instrument.
Small RNA cloning from FLAG::WAGO-9 immune complexes
Synchronous adult flag::wago-9 worms were dounced in a stainless steel homogenizer. FLAG::WAGO-9 was immunoprecipitated from 20 mg of lysate essentially as described (Gu et al., 2009). Small RNAs were extracted from WAGO-9 immune complexes as well as a portion of the input lysate, gel-purified, pre-treated with TAP, cloned and sequenced as above.
Computational analysis of small RNAs
Deep sequencing data were processed and analyzed using custom Perl scripts (Gu et al., 2009). Definition of WAGO and CSR-1 22Gs are described in (Gu et al., 2009). Candidate 21U-RNAs that target gfp were identified by searching for seed sequences (nts 2–8th) that base-pair with at most two G:U wobbles, and at most 3 unpaired non-seed residues (nts 9–21st). Additional details are provided in Supplemental Information. Perl scripts are available on request.
Chromatin Immunoprecipitation
ChIP was performed essentially as described (Claycomb et al., 2009) except that synchronized adult neSi8 gfp::csr-1 (RNAe) and neSi9 gfp::csr-1 worms were dounced in a stainless steel homogenizer (30 strokes) prior to cross-linking with 2.6 % formaldehyde. Immunoprecipitations were performed in a total volume of 1 mL (5 mg) with 10 μg of anti-Histone H3 (ab1791, Abcam) or anti-H3K9me3 (ab8898, Abcam) antibodies. Immune complexes were recovered with 50 μL of Protein A Dynabeads (Invitrogen). Three independent ChIP experiments were performed and analyzed by quantitative PCR.
Quantitative PCR
Quantitative PCR was performed as described (Claycomb et al., 2009) using an ABI 7500 Fast Real-Time PCR instrument. For RNA analysis, cDNA was generated from 1 μg of total RNA using random hexamers and Superscript III Reverse Transcriptase (Invitrogen). gfp::csr-1 expression was measured relative to clp-3 mRNA levels. H3K9me3 ChIP was first normalized to Histone H3 ChIP, and fold enrichment was then determined relative to an H3K9me3 negative control locus. Primer sequences are provided in Supplemental Information.
Transgenerational RNAi phenotype
A single neSi9 gfp::csr-1, neSi12 cdk-1::gfp, tsIs1 oma-1::gfp or neIs2 gfp::wrm-1 adult worm was placed onto each of 10 plates seeded with gfp(RNAi) food. A single F1 worm from each plate was transferred to OP50 (control) or gfp(RNAi) food and each line was maintained for 10 generations by transferring a single worm from each plate to the corresponding food source, OP50 or gfp(RNAi). In each generation, 10 progeny from each plate were scored for gfp expression (100 total for each condition).
Western blot analysis
Antibodies used for Western blotting are anti-CSR-1 (Claycomb et al., 2009), GFP (A01704, Genscript) and α-Tubulin (MCA78A, Serotec) antibodies.
Microscopy
Transgenic worms were mounted in dH2O on RITE-ON glass slides (Beckton Dickinson). Epi-fluorescence and differential interference contrast (DIC) microscopy were performed using an Axioplan2 Microscope (Zeiss). Images were captured with an ORCA-ER digital camera (Hamamatsu) and AxioVision (Zeiss) software.
Supplementary Material
Supplementary Figure 1
Supplementary Legend and Experimental Procedures
Supplementary Table 1
Acknowledgments
We thank Kirsten Hagstrom and Anna Zinovyeva for primer information and technical advice on ChIP experiment and helpful discussion. We are grateful to the members of the Mello lab for their input and discussion, especially Sandra Vergara for her help with genetic studies, William Stanney III for his technical help, and Elaine Youngman and Colin Conine for their advices on figure presentation. Some strains used in this study were obtained from the Caenorhabditis Genetic Center. H.C.L. is supported by Ruth L. Kirschstein National Research Service Awards fellowship (GM099372-2). This work is supported by NIH grant (GM058800) to C. C. M. and Howard Hughes Medical Institute.
References
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. [Europe PMC free article] [Abstract] [Google Scholar]
- Antonovics J, Boots M, Abbate J, Baker C, McFrederick Q, Panjeti V. Biology and evolution of sexual transmission. Ann N Y Acad Sci. 2011;1230:12–24. [Abstract] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. Embo J. 2007;26:5007–5019. [Europe PMC free article] [Abstract] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. [Abstract] [Google Scholar]
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. [Abstract] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. [Europe PMC free article] [Abstract] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. [Abstract] [Google Scholar]
- Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 2010;6:e1000830. [Europe PMC free article] [Abstract] [Google Scholar]
- Birchler J, Pal-Bhadra M, Bhadra U. Transgene cosuppression in animals. In: Hannon G, editor. RNAi: A guide to gene silencing. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. pp. 23–42. [Google Scholar]
- Bosher JM, Labouesse M. RNA interference: genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–36. [Abstract] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. [Europe PMC free article] [Abstract] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. [Abstract] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19683–19688. [Europe PMC free article] [Abstract] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. [Abstract] [Google Scholar]
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol. 2005;15:378–383. [Abstract] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. [Europe PMC free article] [Abstract] [Google Scholar]
- Couteau F, Guerry F, Muller F, Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 2002;3:235–241. [Europe PMC free article] [Abstract] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. [Europe PMC free article] [Abstract] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. [Europe PMC free article] [Abstract] [Google Scholar]
- Dernburg AF, Zalevsky J, Colaiacovo MP, Villeneuve AM. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 2000;14:1578–1583. [Europe PMC free article] [Abstract] [Google Scholar]
- Erhard KF, Jr, Hollick JB. Paramutation: a process for acquiring trans-generational regulatory states. Curr Opin Plant Biol. 2011;14:210–216. [Abstract] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [Abstract] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. [Europe PMC free article] [Abstract] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. [Abstract] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. [Europe PMC free article] [Abstract] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. [Europe PMC free article] [Abstract] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. [Europe PMC free article] [Abstract] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. [Europe PMC free article] [Abstract] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. [Abstract] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. [Abstract] [Google Scholar]
- Johnson CL, Spence AM. Epigenetic licensing of germline gene expression by maternal RNA in C. elegans. Science. 2011;333:1311–1314. [Abstract] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. [Europe PMC free article] [Abstract] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. [Europe PMC free article] [Abstract] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. [Europe PMC free article] [Abstract] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. [Europe PMC free article] [Abstract] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. [Abstract] [Google Scholar]
- Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. [Abstract] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. [Abstract] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. [Abstract] [Google Scholar]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev Biol. 2003;258:226–239. [Abstract] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. [Abstract] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. [Abstract] [Google Scholar]
- Murray NE. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. [Abstract] [Google Scholar]
- Nakamura K, Kim S, Ishidate T, Bei Y, Pang K, Shirayama M, Trzepacz C, Brownell DR, Mello CC. Wnt signaling drives WRM-1/beta-catenin asymmetries in early C. elegans embryos. Genes Dev. 2005;19:1749–1754. [Europe PMC free article] [Abstract] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. [Abstract] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. Rna. 2001;7:1397–1402. [Europe PMC free article] [Abstract] [Google Scholar]
- Praitis V. Creation of transgenic lines using microparticle bombardment methods. Methods Mol Biol. 2006;351:93–107. [Abstract] [Google Scholar]
- Rieckher M, Kourtis N, Pasparaki A, Tavernarakis N. Transgenesis in Caenorhabditis elegans. Methods Mol Biol. 2009;561:21–39. [Abstract] [Google Scholar]
- Robert VJ, Vastenhouw NL, Plasterk RH. RNA interference, transposon silencing, and cosuppression in the Caenorhabditis elegans germ line: similarities and differences. Cold Spring Harb Symp Quant Biol. 2004;69:397–402. [Abstract] [Google Scholar]
- Rulicke T, Hubscher U. Germ line transformation of mammals by pronuclear microinjection. Exp Physiol. 2000;85:589–601. [Abstract] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. [Abstract] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. [Abstract] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. [Abstract] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. [Abstract] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. [Abstract] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. [Abstract] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. [Europe PMC free article] [Abstract] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. [Abstract] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2012.06.015
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867412007647/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cell.2012.06.015
Article citations
Small RNA-mediated genetic switches coordinate ALG-3/4 small RNA pathway function.
Nucleic Acids Res, 52(16):9431-9449, 01 Sep 2024
Cited by: 0 articles | PMID: 38967024 | PMCID: PMC11381353
RNAi-dependent expression of sperm genes in ADL chemosensory neurons is required for olfactory responses in <i>Caenorhabditis elegans</i>.
Front Mol Biosci, 11:1396587, 11 Jul 2024
Cited by: 0 articles | PMID: 39055986 | PMCID: PMC11269235
Germ granule compartments coordinate specialized small RNA production.
Nat Commun, 15(1):5799, 10 Jul 2024
Cited by: 0 articles | PMID: 38987544 | PMCID: PMC11236994
Paramutation at the maize pl1 locus is associated with RdDM activity at distal tandem repeats.
PLoS Genet, 20(5):e1011296, 30 May 2024
Cited by: 0 articles | PMID: 38814980
Heritable epigenetic changes are constrained by the dynamics of regulatory architectures.
Elife, 12:RP92093, 08 May 2024
Cited by: 2 articles | PMID: 38717010 | PMCID: PMC11078544
Go to all (373) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression.
Dev Cell, 27(6):656-663, 01 Dec 2013
Cited by: 147 articles | PMID: 24360782 | PMCID: PMC3954781
C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts.
Cell, 150(1):78-87, 25 Jun 2012
Cited by: 260 articles | PMID: 22738724 | PMCID: PMC3410639
piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans.
Cell, 150(1):88-99, 25 Jun 2012
Cited by: 442 articles | PMID: 22738725 | PMCID: PMC3464430
Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.
Adv Genet, 83:1-69, 01 Jan 2013
Cited by: 53 articles | PMID: 23890211
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIGMS NIH HHS (5)
Grant ID: F32 GM099372
Grant ID: R37 GM058800
Grant ID: GM058800
Grant ID: R01 GM058800
Grant ID: GM099372





