Abstract
Background
Mutations at arginine 132 of isocitrate dehydrogenase 1/2 (IDH1/2) have recently been demonstrated to be recurrent gene alterations in acute myeloid leukemia (AML). Subsequently, this mutation was also found in a variety of other hematologic malignancies, including myelodysplastic syndromes, myeloproliferative diseases, and non-Hodgkin lymphoma. Only a few cases were so far identified in acute lymphoblastic leukemia (ALL). To study the IDH status in ALL patients, we analyzed 54 adult and 34 pediatric ALL samples' IDH1/2 gene.Results
Three adult cases and no pediatric case with an isocitrate dehydrogenase 1 (IDH1) mutation were identified. No isocitrate dehydrogenase 2 (IDH2) mutation was identified in the total of 88 samples. The frequency of the IDH1 mutation in adult ALL was 5.5%. Among the three IDH1-mutated patients, two had normal karyotype and expressed the myeloid lineage markers. All three patients with an IDH1 mutation relapsed or died within 6 months.Conclusions
The results suggested that the IDH1 R132 mutation might be a recurrent gene alteration in ALL; patients carrying the mutation have a trend to aberrantly express myeloid antigen and the mutation may imply a dismal outcome.Free full text

Mutation Analysis of Isocitrate Dehydrogenase in Acute Lymphoblastic Leukemia
Abstract
Background: Mutations at arginine 132 of isocitrate dehydrogenase 1/2 (IDH1/2) have recently been demonstrated to be recurrent gene alterations in acute myeloid leukemia (AML). Subsequently, this mutation was also found in a variety of other hematologic malignancies, including myelodysplastic syndromes, myeloproliferative diseases, and non-Hodgkin lymphoma. Only a few cases were so far identified in acute lymphoblastic leukemia (ALL). To study the IDH status in ALL patients, we analyzed 54 adult and 34 pediatric ALL samples' IDH1/2 gene. Results" Three adult cases and no pediatric case with an isocitrate dehydrogenase 1 (IDH1) mutation were identified. No isocitrate dehydrogenase 2 (IDH2) mutation was identified in the total of 88 samples. The frequency of the IDH1 mutation in adult ALL was 5.5%. Among the three IDH1-mutated patients, two had normal karyotype and expressed the myeloid lineage markers. All three patients with an IDH1 mutation relapsed or died within 6 months. Conclusions: The results suggested that the IDH1 R132 mutation might be a recurrent gene alteration in ALL; patients carrying the mutation have a trend to aberrantly express myeloid antigen and the mutation may imply a dismal outcome.
Introduction
Acute lymphoblastic leukemia (ALL) is a neoplastic disorder that is rapidly fatal if untreated. In children, considerable advances in ALL have resulted in most being cured of their disease. By contrast, the majority of adult patients with ALL are not cured, in part, due to an increased frequency of unfavorable genetic alterations (Mullighan and Downing, 2009). This severe malignancy presents great heterogeneity in genetic profiles, clinical features, response to treatment, and prognosis. The diversity of ALL results mainly from mutant genes that are causally implicated in the pathogenesis of the disease. The full complement of genetic alterations responsible for the pathogenesis of ALL is not yet known and has turned into a hotspot interesting many researchers. In recent decades, progress in understanding the molecular basis of the disease not only provides more clues to clarify the mechanism of leukemogenesis, but also allows a more detailed subclassification and more exact prognostic predictions in many patients. In general, BCR/ABL fusion, MLL-AF4 fusion, and hypodiploidy (<44 chromosomes per leukemic cell) all confer a poor outcome, whereas hyperdiploidy (>50 chromosomes), TEL-AML1 fusion, and trisomy 4, 10, and 17 are associated with favorable prognosis (Pui et al., 2008). In addition to predicting prognosis, these gene alterations provide the basis for more targeted, risk-adapted therapeutic approaches as well as the development of novel therapies (Pui et al., 2008). For example, the bcr-abl fusion kinase produced by the BCR-ABL fusion gene is a good target of imatinib (also called Gleevec) in treatment (Druker, 2004). γ-Secretase, a multicomponent membrane-associated enzyme, is needed for NOTCH1 signaling through mutant NOTCH receptors in T-cell acute lymphoblastic leukemia, providing an attractive target for therapeutic intervention with a newly developed γ-secretase inhibitor (Paganin and Ferrando, 2011).
Recently, the newly unraveled IDH mutation in leukemia brings one more focus in this field. Isocitrate dehydrogenase 1 (IDH1) is a cytosolic protein that catalyzes the NADP+-dependent carboxylation of isocitrate into α-ketoglutatrate. Recently, mutations at arginine 132 of IDH1 have been revealed in acute myeloid leukemia (AML) by genome-wide screening (Mardis et al., 2009). The IDH1 mutation occurred with a frequency of 4.4–9.6% in AML (Mardis et al., 2009; Boissel et al., 2010; Green et al., 2010; Ho et al., 2010; Zhang et al., 2011) and up to 16% in cytogenetically normal AML (Paschka et al., 2010). The mutation was demonstrated to be very stable during disease development and might be a potential marker for residual disease monitoring (Chou et al., 2010). Some reports demonstrated that the mutations implicate unfavorable outcome in distinct cytogenetic or genetic subgroups (Mardis et al., 2009; Abbas et al., 2010; Boissel et al., 2010; Green et al., 2010; Marcucci et al., 2010; Paschka et al., 2010), whereas in other studies, IDH1 mutations had no impact on prognosis (Chou et al., 2010; Wagner et al., 2010).
Isocitrate dehydrogenase 2 (IDH2) gene encodes a mitochondrial protein homologous to IDH1 and also catalyzes isocitrate carboxylation. Subsequent to the detection of the IDH1 mutation, mutations at arginine 140 and 172 of IDH2 were also detected in AML (Boissel et al., 2010; Marcucci et al., 2010). IDH2 mutations occurred with a frequency of up to 11% in AML (Abbas et al., 2010; Paschka et al., 2010; Patel et al., 2011; Zou et al., 2010) and were mutually exclusive with mutations in IDH1 (Marcucci et al., 2010). Some studies showed that the IDH2 mutation had no impact on the prognosis (Thol et al., 2010), whereas some other studies provided evidence that the mutation had an unfavorable impact on the complete remission rate and overall survival (Boissel et al., 2010; Marcucci et al. 2010).
In addition to AML, the IDH mutation was also found in a variety of hematologic malignancies, including myelodysplastic syndromes (Kosmider et al., 2010), myeloproliferative diseases (Pardanani et al., 2010; Tefferi et al., 2010), and non-Hodgkin lymphoma (Zou et al., 2010). So far, only a few IDH-mutated cases have been identified in ALL. Kang et al. (2009) reported one B-acute lymphoblastic leukemia (B-ALL) case with an IDH1 R132C mutation (1/60; 1.7%). Another single IDH1 mutation, also R132C, was detected in an analysis of 288 pediatric ALL samples by Andersson et al. (2011). Abbas et al. (2010) reported an ALL case with an IDH2 R140Q mutation (1/96; 1.0%). It seemed that the mutation was rare in ALL. With the aim to study the IDH status in ALL patients, we analyzed the IDH1/2 gene in 54 adult and 34 pediatric ALL patients.
Materials and Methods
Patients
Bone marrow samples were obtained at diagnosis from 54 adult (32 men and 22 women) and 34 pediatric ALL patients (20 boys and 14 girls) enrolled in the Institute of Hematology and Blood Disease Hospital, Chinese Academy of Medical Sciences. The adult patients were aged 15–81 years with a median age of 28. The pediatric patients were aged 17 months to 14 years with a median age of 5 years. Informed consent was obtained from all patients and parents according to the Declaration of Helsinki. This study was approved by the Ethics Committee of our institution. Establishment of diagnosis was based on the WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues (Vardiman et al., 2009) and the European Group for the Immunological Classification of Leukaemias (EGIL) (Bene et al., 1995).
Nucleic acid isolation
Bone marrow mononuclear cells (BMMNCs) were isolated by density gradient centrifugation over a Ficoll solution (Invitrogen) from 5 mL fresh bone marrow following the instructions of the manufacturer. Genomic DNA was extracted from BMMNCs by using the TIANamp Genomic DNA Kit (Tiangen Biotech). The concentration of DNA was determined by spectrophotometry (NanoDrop Technologies, Thermo Scientific) at 260
mL fresh bone marrow following the instructions of the manufacturer. Genomic DNA was extracted from BMMNCs by using the TIANamp Genomic DNA Kit (Tiangen Biotech). The concentration of DNA was determined by spectrophotometry (NanoDrop Technologies, Thermo Scientific) at 260 nm.
nm.
Mutation analysis
The hotspot mutation at codon 132 of IDH1 was prescreened by polymerase chain reaction (PCR) and restriction endonuclease-based detection as previously described (Meyer et al., 2010; Zhang et al., 2011) and identified by sequencing confirmation. PCR amplification of a 166-bp fragment overriding codon 132 was carried out using forward primer 5′-TGGGTAAAACCTATCATCATCGT-3′ and reverse primer 5′-TTCATACCTTGCTTAATGGGTGT-3′. The last four nucleotides (CGAT) of the forward primer made up one part of the PvuI restriction site (CGATCG), and the first two nucleotides (CG) of the wild-type sequence of IDH1 R132 made up the other part of the PvuI restriction site. Consequently, the PCR product of the wild-type IDH1 carried a PvuI restriction site. If substitution mutation occurred in either of the first two nucleotides of IDH1 R132, then the PvuI restriction site would be destroyed. About 2 μL of the PCR products was digested for 4
μL of the PCR products was digested for 4 h at 37°C using restriction endonuclease PvuI and appropriate buffer (New England Biolabs) in a total volume of 20
h at 37°C using restriction endonuclease PvuI and appropriate buffer (New England Biolabs) in a total volume of 20 μL. The sizes of the cleaved fragments were 142 and 24
μL. The sizes of the cleaved fragments were 142 and 24 bp. Separation on 3% agarose gels allowed detection of the noncleaved fragment of 166
bp. Separation on 3% agarose gels allowed detection of the noncleaved fragment of 166 bp and the larger cleaved fragment of 142
bp and the larger cleaved fragment of 142 bp, whereas the smaller cleaved fragment of 24
bp, whereas the smaller cleaved fragment of 24 bp was invisible. Complete restriction of the PCR product resulted in a single band of 142
bp was invisible. Complete restriction of the PCR product resulted in a single band of 142 bp. Detection of only one signal of 142
bp. Detection of only one signal of 142 bp indicates that the patient carried two wild-type alleles of IDH1 (Fig. 1). Detection of two signals of 142 and 166
bp indicates that the patient carried two wild-type alleles of IDH1 (Fig. 1). Detection of two signals of 142 and 166 bp indicates a heterozygote with one each of mutated and wild-type allele (Fig. 1). All mutations were determined consequently by cloning into a pMD18-T vector and sequencing (Fig. 2).
bp indicates a heterozygote with one each of mutated and wild-type allele (Fig. 1). All mutations were determined consequently by cloning into a pMD18-T vector and sequencing (Fig. 2).
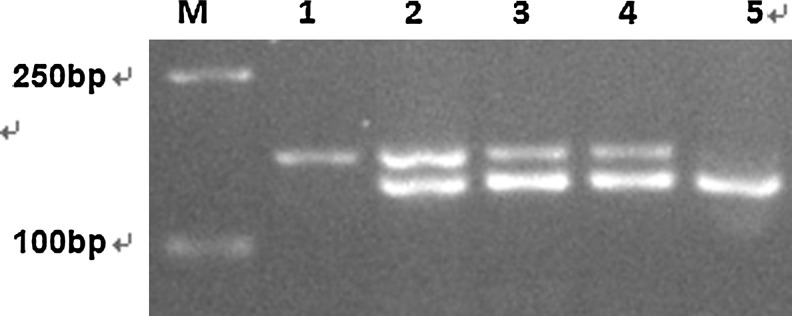
Prescreening of IDH1 R132 mutant by endonuclease digestion. M: DL2000; 1: polymerase chain reaction product without digestion; 2: patient 1; 3: patient 2; 4: patient 3 (the three patients were all heterozygous mutant); 5: wild type.
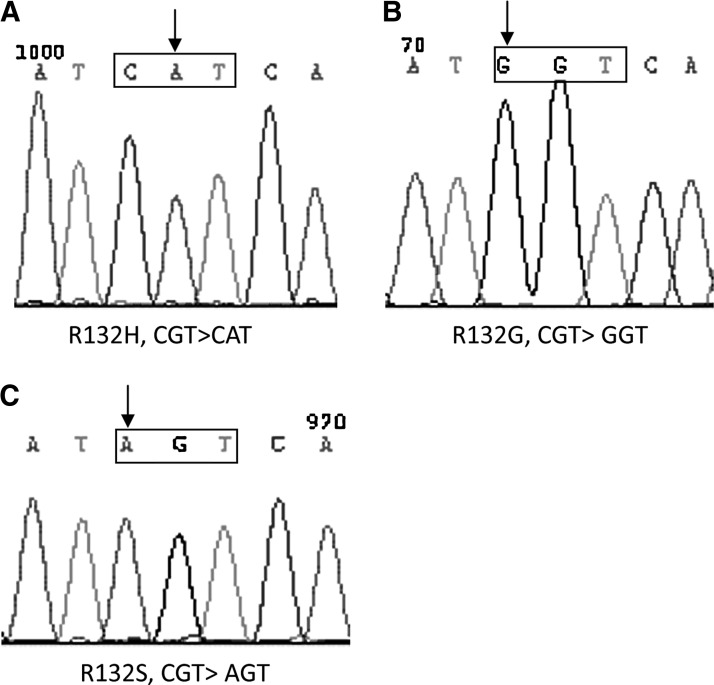
Sequences of IDH1 mutations. (A–C) The mutation types of patient 1, 2, and 3. The sequences in the frames were the mutated codon 132 of IDH1. The arrow indicates the single-mutated nucleotide.
For IDH2 mutational analysis, DNA fragments covering exon 4 were amplified using the primer pair IDH2F: AATTTTAGGACCCCCGTCTG/IDH2R: CTGCAGAGACAAGAGGATGG. The PCR product was directly sequenced as previously described (Marcucci et al., 2010).
Results
Three cases of IDH1-mutated ALL were identified in the total of 54 (5.5%) adult patients and no IDH1 mutation was detected in pediatric patients. No IDH2 mutation was identified in either the adult or the pediatric cases. The IDH1 mutation occurred in 5.3% (2/38) of adult B-ALL and 6.25% (1/16) of adult T-acute lymphoblastic leukemia (T-ALL). All the three mutants were heterozygous (Fig. 1). The mutation types were R132H (CGT>CAT; Fig. 2A), R132G (CGT>GGT; Fig. 2B), and R132S (CGT>AGT; Fig. 2C). The clinical characteristics of the three patients were described in Table 1.
Table 1.
Clinical Characteristics of the IDH1-Mutated Acute Lymphoblastic Leukemia Patients
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Sex | Female | Male | Male |
| Age (years) | 44 | 15 | 53 |
| Symptoms and signs | Metromenorrhagia, dizziness, fatigue, petechia, splenomegaly | Sore throat, fever, adenopathy of cervical lymph nodes, swelling of tonsils, hepatomegaly | Pale, fatigue, chest pain following exercise, paroxysmal throbbing headache |
| Diagnosis | Pro-B-ALL | Pre-T-ALL | Common-B-ALL |
| IDH1 R132 mutation type | CGT>CAT | CGT>GGT | CGT>AGT |
| WBC | 325, 380/μL | 1370/μL | 2920/μL |
| Hg | 80 g/dL g/dL | 84 g/dL g/dL | 58 g/L g/L |
| Plt | 23,000/μL | 117,000/μL | 92,000/μL |
| Blast % in bone marrow | 88% | 80% | 75% |
| Immunophenotype | CD19, CD22, CD38, cytTdT | cytCD3, CD7, CD8, cTdT, CD13, CD33, CD34, CD38, CD11b | CD19, CD10, CD79a, CD13, CD33, CD65, and HLA-DR |
| Karyotype | t(4;11) | Normal | Normal |
| Gene alteration | MLL/AF4 | None | None |
LDH (reference range 15,220 U/L) U/L) | 1134 U/L U/L | 204 U/L U/L | 286 U/L U/L |
| DFS (months) | 6 | 3 | 4 |
| OS (months) | 18 | 4 | 7 |
IDH1, isocitrate dehydrogenase 1; WBC, white blood cell; Hg, hemoglobin; Plt, platelet; LDH, lactate dehydrogenase; DFS, disease-free survival; OS, overall survival; B-ALL, B-acute lymphoblastic leukemia; T-ALL, T-acute lymphoblastic leukemia.
Discussion
Researchers are focusing on IDH mutation since it has been demonstrated to be recurrent gene alteration in AML (Mardis et al., 2009). Subsequently, the mutation was also detected in myeloproliferative neoplasm (Pardanani et al., 2010; Tefferi et al., 2010), myelodysplastic syndrome (Kosmider et al., 2010), and non-Hodgkin lymphoma (Zou et al., 2010). Only three IDH-mutated cases were so far identified in ALL (Kang et al., 2009; Abbas et al., 2010; Andersson et al., 2011). Our observation confirmed the occurrence of IDH1 mutation in ALL. Based on this observation, we predict that additional IDH-mutated ALL cases will be reported in further studies and IDH mutation may be a recurrent gene alteration in ALL.
As for the method to detect the mutation, there are a multitude of means. The most popular method is direct sequencing (Abbas et al., 2010; Andersson et al., 2011). In addition, single-strand conformation polymorphism-based assays (Kang et al., 2009; Zou et al., 2010) and real-time PCR with post-PCR fluorescence melting curve analysis assays (Lin et al., 2011; Noordermeer et al., 2011) were also used. The immunohistochemistry with the anti-IDH1 R132H antibody (Kato et al., 2009; Byers et al., 2012) allows the observation of IDH1 R132H-positive cells in the clinical samples and leads to diffused staining by the antibody in almost every single cell of IDH1 R132H-positive tumor. The restriction digestion method is less expensive and simple. Direct sequencing is simple and fast. However, the limitation of these two methods is that DNA needs to be extracted from samples containing a high proportion of tumor cells, so that if the IDH1-mutated tumor cell load was low, then the mutated sample could not be screened out. Therefore, the sensitivity of these two methods is not satisfied. In comparison to DNA sequencing, the restriction digestion method is not applicable to detect other IDH1 mutations in exon 4 in addition to R132.
In our observation, two IDH1-mutated patients (patient 2 and 3) had normal karyotype and expressed the myeloid antigens CD13 and CD33. Interestingly, the pediatric IDH1-mutated case reported by Andersson et al. (2011) also had a normal karyotype and aberrant myeloid antigen expression of CD13 and CD33. The expression of the myeloid antigens in the IDH1 mutation carriers suggested that the leukemia clone of IDH1-mutated ALL may come from an early stage of hematopoiesis and IDH1 mutation may be involved in myeloid differentiation. In previous reports, the IDH1 mutation was demonstrated to be related to an increase of reactive oxygen species (ROS) (Kolker et al., 2002; Ward et al., 2010). It was speculated that the increase of ROS may disturb the quiescence of the hematopoietic stem cells (HSCs) and act as an intracellular trigger for HSC differentiation to the myeloid lineage fate (Tothova and Gilliland, 2007). The expression of myeloid antigens in IDH1-mutated ALL observed in our study supported this speculation, which could partially explain how the IDH1 mutation contributes to the transformation of HSCs.
In our observation, all of the three IDH1-mutated patients relapsed or died within 6 months. Although the limited number of mutation carriers allows no final conclusions regarding the prognostic impact of ALL, we speculated that the mutation may imply a dismal outcome. Large scope studies are needed to verify the prognostic significance of the mutation in ALL.
In summary, the IDH1 mutations may be a recurrent gene alteration in adult ALL (3/54, 5.5%). The mutations occur in both B-ALL (2/38, 5.3%) and T-ALL (1/16, 6.25%). The IDH1-mutated ALL patients may have a trend to carry normal karyotype and express myeloid antigen aberrantly. ALL patients harboring the mutation may have unfavorable prognosis. However, the limited number of the mutation carriers identified in this study warranted strong conclusions, and we are expecting further studies to clarify the role of IDH in ALL.
Acknowledgments
Y.Z. performed the experiments and drafted the article; J.W. contributed to the concept and design as well as to the approval of the final article; K.T., L.W., D.L., C.W., and C.Z. contributed essential reagents or tools. M.W., H.W., and Y.M. contributed to the data interpretation and helped revise the article. This work was supported by National Natural Science Foundation (Grant No. 81070427, 30971290), Special Scientific Fund for Public Welfare Industry (Health) (Grant No. 201002024), and Tianjin Applied Fundamental Research Planning Key Project (Grant No. 10JCZDJC19600).
References
- Abbas S. Lugthart S. Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. [Abstract] [Google Scholar]
- Andersson AK. Miller DW. Lynch JA, et al. IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia. 2011;25:1570–1577. [Abstract] [Google Scholar]
- Bene MC. Castoldi G. Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9:1783–1786. [Abstract] [Google Scholar]
- Boissel N. Nibourel O. Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. [Abstract] [Google Scholar]
- Byers R. Hornick JL. Tholouli E, et al. Detection of IDH1 R132H mutation in acute myeloid leukemia by mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol. 2012;20:37–40. [Abstract] [Google Scholar]
- Chou WC. Huang HH. Hou HA, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116:4086–4094. [Abstract] [Google Scholar]
- Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. [Abstract] [Google Scholar]
- Green CL. Evans CM. Hills RK, et al. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116:2779–2782. [Abstract] [Google Scholar]
- Ho PA. Alonzo TA. Kopecky KJ, et al. Molecular alterations of the IDH1 gene in AML: a Children's Oncology Group and Southwest Oncology Group study. Leukemia. 2010;24:909–913. [Europe PMC free article] [Abstract] [Google Scholar]
- Kang MR. Kim MS. Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. [Abstract] [Google Scholar]
- Kato Y. Jin G. Kuan CT, et al. A monoclonal antibody IMab-1 specifically recognizes IDH1R132H, the most common glioma-derived mutation. Biochem Biophys Res Commun. 2009;390:547–551. [Europe PMC free article] [Abstract] [Google Scholar]
- Kolker S. Pawlak V. Ahlemeyer B, et al. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci. 2002;16:21–28. [Abstract] [Google Scholar]
- Kosmider O. Gelsi-Boyer V. Slama L, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. [Abstract] [Google Scholar]
- Lin J. Qian J. Yao DM, et al. Rapid and reliable detection of IDH1 R132 mutations in acute myeloid leukemia using high-resolution melting curve analysis. Clin Biochem. 2011;44:779–783. [Abstract] [Google Scholar]
- Marcucci G. Maharry K. Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. [Europe PMC free article] [Abstract] [Google Scholar]
- Mardis ER. Ding L. Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. [Europe PMC free article] [Abstract] [Google Scholar]
- Meyer J. Pusch S. Balss J, et al. PCR- and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol. 2010;20:298–300. [Abstract] [Google Scholar]
- Mullighan CG. Downing JR. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009;23:1209–1218. [Abstract] [Google Scholar]
- Noordermeer SM. Tonnissen E. Vissers I, et al. Rapid identification of IDH1 and IDH2 mutations in acute myeloid leukaemia using high resolution melting curve analysis. Br J Haematol. 2011;152:493–496. [Abstract] [Google Scholar]
- Paganin M. Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2011;25:83–90. [Europe PMC free article] [Abstract] [Google Scholar]
- Pardanani A. Lasho TL. Finke CM, et al. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia. 2010;24:1146–1151. [Abstract] [Google Scholar]
- Paschka P. Schlenk RF. Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. [Abstract] [Google Scholar]
- Patel KP. Ravandi F. Ma D, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am J Clin Pathol. 2011;135:35–45. [Abstract] [Google Scholar]
- Pui CH, et al. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. [Abstract] [Google Scholar]
- Tefferi A. Lasho TL. Abdel-Wahab O, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. [Europe PMC free article] [Abstract] [Google Scholar]
- Thol F. Damm F. Wagner K, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–616. [Abstract] [Google Scholar]
- Tothova Z. Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–152. [Abstract] [Google Scholar]
- Vardiman JW. Thiele J. Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. [Abstract] [Google Scholar]
- Wagner K. Damm F. Gohring G, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. [Abstract] [Google Scholar]
- Ward PS. Patel J. Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Y. Wei H. Wang M, et al. Some novel features of IDH1-mutated acute myeloid leukemia revealed in Chinese patients. Leuk Res. 2011;35:1301–1306. [Abstract] [Google Scholar]
- Zou Y. Zeng Y. Zhang DF, et al. IDH1 and IDH2 mutations are frequent in Chinese patients with acute myeloid leukemia but rare in other types of hematological disorders. Biochem Biophys Res Commun. 2010;402:378–383. [Abstract] [Google Scholar]
Articles from Genetic Testing and Molecular Biomarkers are provided here courtesy of Mary Ann Liebert, Inc.
Full text links
Read article at publisher's site: https://doi.org/10.1089/gtmb.2011.0323
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3422558?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1089/gtmb.2011.0323
Article citations
Biosynthesis and Significance of Fatty Acids, Glycerophospholipids, and Triacylglycerol in the Processes of Glioblastoma Tumorigenesis.
Cancers (Basel), 15(7):2183, 06 Apr 2023
Cited by: 4 articles | PMID: 37046844 | PMCID: PMC10093493
Review Free full text in Europe PMC
Occurrence of non-central nervous system cancers during postoperative follow-up of patients who underwent surgery for a WHO grade II glioma: implications for therapeutic management.
J Neurooncol, 162(1):237-244, 13 Mar 2023
Cited by: 0 articles | PMID: 36913047
2-Hydroxyglutarate in Cancer Cells.
Antioxid Redox Signal, 33(13):903-926, 22 Jan 2020
Cited by: 62 articles | PMID: 31847543 | PMCID: PMC7533892
Review Free full text in Europe PMC
Recent Advances in the Targeting of Epigenetic Regulators in B-Cell Non-Hodgkin Lymphoma.
Front Genet, 10:986, 16 Oct 2019
Cited by: 15 articles | PMID: 31681423 | PMCID: PMC6807552
Review Free full text in Europe PMC
The Role of Immunohistochemistry and Molecular Analysis of Succinate Dehydrogenase in the Diagnosis of Endocrine and Non-Endocrine Tumors and Related Syndromes.
Endocr Pathol, 30(1):64-73, 01 Mar 2019
Cited by: 15 articles | PMID: 30421319
Review
Go to all (17) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
IDH1 and IDH2 mutations in pediatric acute leukemia.
Leukemia, 25(10):1570-1577, 07 Jun 2011
Cited by: 60 articles | PMID: 21647154 | PMCID: PMC3883450
IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome.
Ann Hematol, 91(4):519-525, 14 Oct 2011
Cited by: 77 articles | PMID: 21997850
Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value.
Blood, 116(12):2122-2126, 10 Jun 2010
Cited by: 240 articles | PMID: 20538800
Isocitrate dehydrogenase mutations in myeloid malignancies.
Leukemia, 31(2):272-281, 10 Oct 2016
Cited by: 181 articles | PMID: 27721426 | PMCID: PMC5292675
Review Free full text in Europe PMC
 1,2
1,2



