Abstract
Context
There is increasing evidence for the role of altered metabolism in the pathogenesis of renal cancer.Objective
This review characterizes the metabolic effects of genes and signaling pathways commonly implicated in renal cancer.Evidence acquisition
A systematic review of the literature was performed using PubMed. The search strategy included the following terms: renal cancer, metabolism, HIF, VHL.Evidence synthesis
Significant progress has been made in the understanding of the metabolic derangements present in renal cancer. These findings have been derived through translational, in vitro, and in vivo studies. To date, the most well-characterized metabolic features of renal cancer are linked to von Hippel-Lindau (VHL) loss. VHL loss and the ensuing increase in the expression of hypoxia-inducible factor affect several metabolic pathways, including glycolysis and oxidative phosphorylation. Collectively, these changes promote a glycolytic metabolic phenotype in renal cancer. In addition, other histologic subtypes of renal cancer are also notable for metabolic derangements that are directly related to the causative genes.Conclusions
Current knowledge of the genetics of renal cancer has led to significant understanding of the metabolism of this malignancy. Further studies of the metabolic basis of renal cell carcinoma should provide the foundation for the development of new treatment approaches and development of novel biomarkers.Free full text

Metabolism of Kidney Cancer: From the Lab to Clinical Practice
Abstract
Context
There is increasing evidence for the role of altered metabolism in the pathogenesis of renal cancer.
Objective
This review characterizes the metabolic effects of genes and signaling pathways commonly implicated in renal cancer.
Evidence acquisition
A systematic review of the literature was performed using PubMed. The search strategy included the following terms: renal cancer, metabolism, HIF, VHL.
Evidence synthesis
Significant progress has been made in the understanding of the metabolic derangements present in renal cancer. These findings have been derived through translational, in vitro, and in vivo studies. To date, the most well-characterized metabolic features of renal cancer are linked to von Hippel-Lindau (VHL) loss. VHL loss and the ensuing increase in the expression of hypoxia-inducible factor affect several metabolic pathways, including glycolysis and oxidative phosphorylation. Collectively, these changes promote a glycolytic metabolic phenotype in renal cancer. In addition, other histologic subtypes of renal cancer are also notable for metabolic derangements that are directly related to the causative genes.
Conclusions
Current knowledge of the genetics of renal cancer has led to significant understanding of the metabolism of this malignancy. Further studies of the metabolic basis of renal cell carcinoma should provide the foundation for the development of new treatment approaches and development of novel biomarkers.
1. Introduction
Renal cell carcinoma (RCC) is among the top 10 most common malignancies in both men and women. In 2012, it is estimated that approximately 64 000 individuals will be diagnosed, with an estimated 13 570 dying from this disease in the United States [1]. Although the incidence has been increasing in the United States, the rates have leveled off or even declined in other parts of the world [2].
Is it is now clear that altered metabolism plays a key role in the pathogenesis of many disease states. Moreover, there has been increasing recognition of the role of metabolism in the pathogenesis of cancer, including RCC. To date, most studies have examined the risk of RCC in disease with known metabolic links. For example, the rising incidence of RCC has paralleled the increase in body mass index, particularly in Western countries [3]. This finding has been validated by several recent studies that reported increased incidence of RCC in disease states with known effects on metabolism, including obesity and diabetes [4]. Additionally, links between atherosclerosis and the risk of RCC have been identified [5].
Collectively, these data point to the emerging link between metabolism and cancer. Correspondingly, treatment of these disease processes, such as statins for atherosclerosis, may be associated with a reduced risk of RCC [6]. With continued examination of this link, identification of metabolic factors may point to new directions for therapeutics as well as biomarkers for diagnosis and prognosis. This review will focus on the metabolic alterations in the more common variants of RCC and the clinical relevance of these alterations.
2. Evidence acquisition
We performed a systematic search of PubMed through May 2012. Search terms included renal cancer, metabolism, HIF, and VHL. Searches were limited to articles written in English. All authors screened the potential references retrieved by the search and a final consensus on all publications included in the present review was reached.
3. Evidence synthesis
Evidence related to the review and that examined general aspects of cancer metabolism was acquired. Next, metabolic aspects of RCC were analyzed based on the histologic subtype. Metabolic aspects of RCC are described in the context of genes commonly alerted for each subtype.
3.1. Cancer metabolism: general considerations
One of the most basic physiologic functions of cells is to convert nutrients into energy equivalents such as adenosine triphosphate (ATP). The most basic starting nutrient is glucose, which is broken down by a series of enzymatic reactions, referred to as glycolysis, to the metabolite pyruvate [7,8]. Under conditions where oxygen is present (ie, aerobic conditions), pyruvate is transferred to the mitochondria where it is converted to acetyl coenzyme A (acetyl coA). In turn, acetyl coA is metabolized in the tricarboxylic acid (TCA) cycle (also referred to as the citric acid cycle). This process yields reducing equivalents that drive the process of oxidative phosphorylation that, in turn, leads to ATP generation. Oxidative phosphorylation requires oxygen. Therefore, under low oxygen (anaerobic) conditions, oxidative phosphorylation is suppressed and pyruvate is converted to lactate via lactate dehydrogenase (LDH). One of the most notable alterations in cancer metabolism was initially described by Otto Warburg [9]. This observation, now referred to as the Warburg effect, refers to the increased rate of glycolysis with reduced oxidative phosphorylation in tumor tissues despite the presence of normal levels of oxygen. An unresolved question is whether the Warburg effect is merely the result of metabolic adaptations that tumor cells make to survive or whether this phenotype contributes to tumorigenesis. The finding that the loss of several tumor suppressor genes may promote the Warburg effect suggests the latter [8].
3.2. Clear cell renal cancer: the role of VHL
RCC is actually composed of several tumor types linked by the site of origin. Current understanding of the molecular biology of RCC has benefited significantly from the study of hereditary forms of renal cancer. Most notable is von Hippel-Lindau (VHL) disease. Patients with VHL disease have a genetic predisposition to the development of clear cell RCC (ccRCC) among other tumor types affecting organ sites, including the brain, spinal cord, pancreas, and adrenal gland [10]. Through genetic linkage performed in VHL families, the VHL gene was identified on the short arm of chromosome 3 and was eventually cloned [11,12]. Subsequent studies led to the finding that VHL alterations are common genetic events in sporadic ccRCC, indicating that, from the standpoint of tumor-initiating events, ccRCCs are a relatively homogenous population relative to other tumor types [13]. It is estimated that 70% to 90% of ccRCCs demonstrate alterations of the VHL gene either via mutation or gene silencing (ie, hypermethylation) [14,15]. As such, current understanding of the basic biology of ccRCC has been largely derived through the study of the functions of VHL protein.
3.3. The VHL pathway
Through protein interactions studies, it was subsequently determined that VHL was a member of the E3 ubiquitin ligases. VHL associates with the proteins elongin B, elongin C, and cullin 2 [16,17]. Together, the VHL complex ubiquitinates proteins, which targets them for degradation by the proteosome. Among the most studied targets of the VHL complex are the hypoxia-inducible factor (HIF) proteins HIF-1α and HIF-2α [18,19]. The labile HIFα proteins form a complex with the constitutively expressed HIF-1β subunit [20]. Together, this complex binds to DNA motifs referred to as hypoxia response elements to regulate the expression of genes involved in angiogenesis, cell proliferation, and metabolism. Under normal oxygen-containing (aerobic) conditions, HIFα proteins are hydroxylated on conserved proline residues by a series of enzymes referred to as prolyl hydroxylases [18,21]. Hydroxylation of HIF permits recognition by the VHL complex, which therefore targets these transcription factors for proteosomal degradation. Under hypoxic conditions when oxygen is limiting, HIFs remain unhydroxylated, and therefore stable, due to lack of recognition by VHL. HIFs are stabilized in the absence of VHL despite the presence of oxygen (referred to as a pseudohypoxic state). Alternatively, HIF may be stabilized under conditions in which proline hydroxylation of HIF is inhibited. Interestingly, recent data demonstrate the in vitro and in vivo antitumor efficacy of an HIF inhibitor in RCC cells [22].
3.4. Hypoxia-inducible factor: a master regulator of renal cell carcinoma metabolism
As VHL loss is a common event in ccRCC, a significant proportion of the current understanding of this tumor type has been derived from the study of HIF biology. In addition to effects on angiogenesis and growth factor expression, HIF has now been shown to have profound effects on cellular metabolism (Fig. 1). HIF is known to regulate the expression of glucose transporters that regulate glucose uptake from the extracellular milieu. Increased expression of the glucose transporter 1, which promotes cellular glucose uptake, has been observed in VHL-deficient ccRCC [23]. Additionally, HIF has been shown to transcriptionally upregulate many of the genes that encode the enzymes of glycolysis, including hexokinase 1 and 2 (HK1 and HK2) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [24,25]. HIF shunts cellular metabolism away from the TCA cycle/oxidative phosphorylation through multiple mechanisms. As noted previously, the end product of glycolysis is pyruvate, which has one of two fates: (1) It may be converted to lactate via lactate dehydrogenase (LDH), or (2) it may be converted to acetyl CoA by the enzyme pyruvate dehydrogenase (PDH), which then proceeds through the reaction of the TCA cycle. HIF has been shown to upregulate LDH expression and thereby promote pyruvate’s conversion to lactate. In addition, HIF shunts cellular metabolism away from the TCA cycle through the regulation of PDH [26,27]. PDH is subject to regulation by the enzyme pyruvate dehydrogenase kinase 1 (PDK1). Phosphorylation of PDH by PDK1 inactivates PDH, thereby blocking the conversion of pyruvate to acetyl CoA. Multiple studies have now demonstrated that HIF upregulates the expression of PDK1 [26,27]. Moreover, PDK-1 expression is increased in RCC [28]. Recent investigation has focused on the role of microRNAs (miRNAs) in cancer. miRNAs are noncoding RNA molecules that can regulate gene expression. HIF has previously been shown to regulate the expression of several miRNAs, including mir-210 [29]. Several reports have established that miR-210 is overexpressed in ccRCC [30–32]. Among the targets of miR-210 are the iron–sulfur cluster assembly proteins ISCU1 and ISCU2 [33]. ISCU1 and ISCU2 facilitate the assembly of iron–sulfur cluster proteins, including components of the electron transport chain that drives mitochondrial oxidative phosphorylation. Accordingly, miR-210 has been shown to block mitochondrial respiration. In agreement with these findings, ICSU1/ISCU2 expression is decreased in ccRCC as well as in animal models with genetic disruption of VHL [33,34]. Furthermore, constitutive HIF activation has been shown to inhibit mitochondrial respiration in vivo [35]. Interestingly, several recent studies have examined the potential for miRNAs as biomarkers for RCC [36–38].
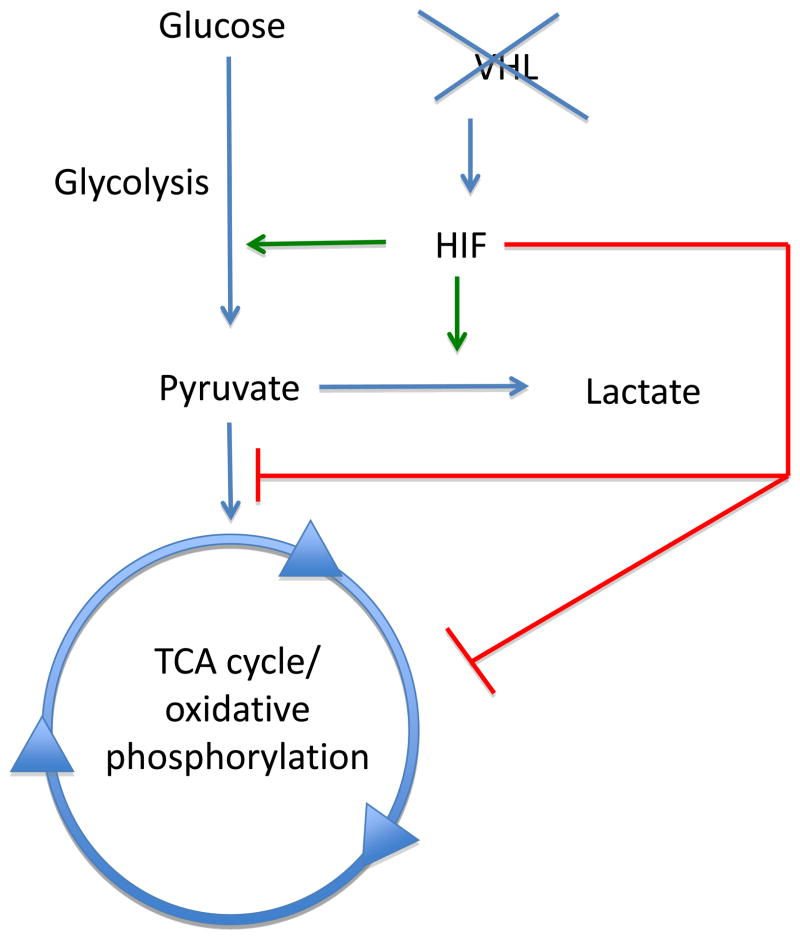
Hypoxia-inducible factor (HIF) accumulation resulting from von Hippel-Lindau (VHL) loss regulates several metabolic processes. HIF promotes the expression of many genes that encode glycolytic enzymes. HIF also prevents entry and metabolism through the tricarboxylic acid (TCA) cycle via several mechanisms including (1) promoting pyruvate conversion to lactate, (2) blocking pyruvate conversion to acetyl-coenzyme A, and (3) lowering the expression of enzymes involved in oxidative phosphorylation.
3.5. The potential role of alternate clear cell renal cell carcinoma genes in metabolism
The recent utilization of deep-sequencing technologies has led to the identification of new ccRCC genes, many of which are involved in chromatin biology. Among the more commonly mutated genes in ccRCC is polybromo 1 (PBRM1)[39]. PBRM1 is a member of the switch/sucrose nonfermentable (SWI/SNF) protein complex that remodels chromatin [39]. Interestingly, gene-expression profiling of ccRCCs with PBRM1 mutations falls into a hypoxic gene profiling. Given the connection between hypoxia and metabolism as demonstrated through the studies on the VHL/HIF axis, it would seem likely that PBRM1 may have effects on cellular metabolism. In addition, several other genes, such as SET domain containing 2 (SETD2), that regulate histone modifications are mutated in a subset of ccRCC [40,41]. Given the role of histones in the regulation of gene expression, the function of these genes in the regulation of ccRCC metabolism warrants further investigation.
3.6. The IGF/IGF-1R axis and mechanistic target of rapamycin signaling in clear cell renal cell carcinoma
The type 1 insulin-like growth factor receptor (IGF1R) is commonly overexpressed in several tumor types, including RCC. Recent studies have demonstrated a role for VHL in the regulation of IGF1R expression in a HIF-independent fashion [42]. Downstream effectors of IGF1R include phosphoinositide-3-kinase (PI3K), which in turn activates AKT signaling. Additionally, PI3K regulates signaling by mechanistic target of rapamycin (mTOR).
MTOR’s relevance to RCC has received much attention lately due to the emergence of mTOR inhibitors as US Food and Drug Administration–approved agents for the treatment of advanced renal cancer. The mTOR inhibitors used currently are analogues of the molecule rapamycin, which is an allosteric inhibitor of mTOR. MTOR is a key mediator of cellular physiology through effects on cellular metabolism. It serves as a nutrient sensor. Many tumor suppressor genes such as phosphatase and tensin homolog (PTEN), tuberous sclerosis 1 (TSC1), tuberous sclerosis 2 (TSC2), neurofibromin 1 (NF1), and liver kinase B1 (LKB1) are negative regulators of mTOR signaling [43]. Hence, loss or mutation of these genes may lead to aberrant mTOR activation. Hyperactivation of mTOR signaling has been observed in several cancer types, including RCC [44]. An enhanced understanding of the mTOR pathway has been derived from the study of patients with tuberous sclerosis (TS), who are at risk for the development of renal angiomyolipomas (AMLs). Patients with TS have germline mutations at either the TSC1 or TSC2 locus that encode for the proteins hamartin and tuberin, respectively. Hamartin and tuberin form a protein complex that blocks mTOR activation [43]. As such, loss of either TSC1 or TSC2 leads to mTOR activation. Consistent with this pathway are recent studies demonstrating the efficacy of sirolimus, a rapamycin analog and mTOR inhibitor, in the treatment of AMLs in patients with TS [45]. Of note are recent reports demonstrating mTOR and TSC1 mutations in ccRCC [46,47].
MTOR is known to regulate cell size and proliferation. mTOR can drive several metabolic processes, including fatty acid synthesis and the pentose phosphate pathway [48]. In addition, mTOR has been shown to promote glucose uptake and glycolysis, indicating that mTOR signaling promotes the Warburg phenotype in cancer. These latter effects are, in part, thought to be mediated by mTOR’s effects on HIF-1α expression through promotion of HIF-1α mRNA translation [48,49]. Given mTOR’s role in the regulation of metabolism, the efficacy of mTOR inhibition in renal cancer suggests the possibility of a metabolic Achilles heel of renal cancer, and that targeting the critical metabolic pathways may be of therapeutic benefit.
3.7. Metabolic aspects of papillary renal cancer
Papillary renal cancer accounts for approximately 15% of RCC and can be broadly classified as type 1 and type 2 papillary RCC (pRCC). As with ccRCC, our current understanding of pRCC has benefited from the study of families with hereditary papillary renal cancer (HPRC). Patients with HPRC harbor activating mutations of the met proto-oncogene (hepatocyte growth factor receptor) (c-MET) in the germline [50]. These individuals are predisposed to the development of type 1 pRCC. c-MET encodes for the receptor of hepatocyte growth factor [51]. MET is a receptor tyrosine kinase that may activate PI3K signaling. Given the emerging role of PI3K signaling in cellular metabolism, pRCC type 1 cancers would be expected to demonstrate metabolic derangements. c-MET appears to have a role in sporadic cancer as well as mutations, and amplifications of this gene have been identified in a subset of tumors from patients with sporadic type 1 pRCC [52]. Of note is that c-MET inhibitors are currently being evaluated for efficacy in the treatment of pRCC.
Until recently, very little was known about metabolic aspects of type 2 pRCC. However, the report of genetic predisposition to type 2 pRCC of patients with germline mutations of the fumarate hydratase (FH) gene led to further understanding of the metabolic aspect of this disease [53]. FH encodes for the enzyme fumarate hydratase. This enzyme is part of the TCA cycle that contributes to oxidative phosphorylation and is responsible for converting fumarate to malate. As expected, tumors that lack FH have elevated levels of fumarate [54]. Consistent with the role of FH in the TCA cycle, cells lacking FH enzymatic activity are heavily reliant on glucose (and glycolysis) for cell survival relative to cells with intact FH and they demonstrate reduced oxidative phosphorylation [55,56]. FH-null tumor cells have recently been reported to demonstrate a Warburg phenotype [57]. Hence, targeting how these cells take up glucose and use it to generate energy equivalents required for cellular homeostasis may have therapeutic potential, as recently attempted [58].
Another interesting aspect of FH-null tumors is that they also demonstrate elevated levels of HIF protein expression (particularly HIF-1α) [59]. As noted previously, HIF is usually very labile as it undergoes proline hydroxylation, which therefore targets it for proteosomal degradation. Fumarate has been shown to inhibit the proline hydroxylation of HIF and therefore lead to aberrant stabilization of HIF proteins [59]. Additionally, cellular models of FH loss demonstrated elevated levels of reactive oxygen species (ROS), which may also contribute to HIF stabilization [56]. The increase in ROS may be due to the highly glycolytic phenotype of these cells. Interestingly, two recent studies suggest that antioxidant pathways are activated in FH-null HPRC tumors as well as sporadic type 2 pRCC [60,61]. One possibility is that activation of antioxidant pathways may perhaps cope with the elevated levels of oxidative stress. From a therapeutic standpoint, ROS levels could be further raised above a certain threshold that would induce cell death in such tumor cells, as recently proposed by Sourbier et al[62].
3.8. Metabolic aspects of oncocytomas and chromophobe renal cell carcinoma
Recent evidence suggests that oncocytomas and chromophobe RCCs also have alterations in cellular metabolism relative to normal kidney. Gene-expression studies of these tumors demonstrate elevated levels of mitochondrial and oxidative phosphorylation gene expression in these tumors. These data are consistent with the observation that these tumor types have an abundance of mitochondria. In a similar fashion, tumors from patients with Birt-Hogg-Dubé (BHD) syndrome also demonstrate upregulation of mitochondrial/oxidative phosphorylation genes [63]. Patients with BHD syndrome are genetically predisposed to the development of chromophobe RCC, oncocytomas, and hybrid oncocytic tumors (tumors with a histologic mixture of both oncocytoma and chromophobe RCC) [64]. The implications of this finding as it pertains to tumor biology remains unknown at this time. However, it does underscore the finding that RCC is a spectrum of disease that varies at many levels, including metabolism.
Cellular studies of BHD syndrome demonstrate a potential link with 5′ adenosine monophosphate-activated protein kinase (AMPK) [65]. APMK is a major cellular regulator of energy homeostasis and therefore a key metabolic regulator. AMPK is a negative regulator of mTOR. Interestingly, animal models of BHD syndrome demonstrate activation of mTOR signaling [66]. As such, mTOR inhibitors may have therapeutic potential in these tumor types. Animal models of BHD syndrome demonstrate a renal cystic phenotype that can be abrogated by treatment with the mTOR inhibitor rapamycin.
3.9. Metabolomics in renal cancer
With an improved understanding of genetic alterations in RCC, particularly ccRCC, investigators are now in a position to exploit the ensuing metabolic alterations for clinical applications. Underlying gene defects such as VHL loss have profound impacts on the gene expression of many enzymes involved in key metabolic processes. As these enzymes interconvert small molecules, changes in the expression of these enzymes can affect levels of small molecules, that is, metabolites. The use of separation and identification technologies has now led to the development of metabolomics. Genomics, transcriptomics, and proteomics characterize a data set at the DNA, RNA, and protein levels, respectively. Metabolomics can characterize a biological sample at the level of small molecules (metabolites). Metabolomic analyses have generally applied two analytic approaches: nuclear magnetic resonance and mass spectrometry [67]. More recent approaches have used chromatography, either gas (GC) or liquid (LC), to separate metabolites in a biological sample from each other combined with mass spectrometry (MS) to allow for identification as well as quantification of molecules of interest. Given the relatively small number of molecules in the human metabolome (approximately 2200) as compared with the human genome and transcriptome, the potential for biomarker discovery exists for a variety of clinical applications [67].
The use of metabolomics for genitourinary malignancies has received considerable attention after the identification of the small molecule sarcosine as a potential mediator of prostate cancer progression. Using normal prostate tissue as well prostate cancer specimens (both localized and metastatic), Sreekumar et al were able to identify increasing levels of sarcosine as a function of cancer progression [68]. Moreover, they were able to identify sarcosine in urine samples of patients with prostate cancer. Metabolomics has begun to be applied to studies on renal cancer. Using GC-MS, Catchpole et al performed a metabolic profile of renal cancer tissues relative to normal tissue [69]. The primary histology analyzed in this data set was ccRCC. Consistent with a glycolytic phenotype representative of the Warburg effect, the levels of glucose were higher in RCC relative to normal tissue. In contrast, metabolites such as fumarate, malate, and succinate that participate in the mitochondrial TCA cycle/oxidative phosphorylation were reduced in RCC. These findings are consistent with the aforementioned metabolic biology of ccRCC. As most ccRCCs demonstrate loss of VHL, the enhanced HIF expression would be expected to promote glycolytic metabolism while downregulating mitochondrial metabolism. In addition, this study found markedly elevated levels (>5-fold) of α-tocopherol, a form of vitamin E, in RCC compared with normal tissue. As RCC cells are noted to have elevated levels of oxidative stress [70], the increased levels of α-tocopherol, which has antioxidant properties, may allow renal cancer cells to cope with increased levels of ROS. The potential for biomarker development was also investigated in this study. Using just α-tocopherol levels in a receiver operator curve analysis, this group was able to correctly classify almost 85% of tumors and >90% of normal tissues.
The potential for metabolomics in RCC biomarker development has also been applied to urine and serum analyses. Two recent analyses were able to distinguish patients with RCC from patients without based on the metabolic profile of urine specimens [71,72]. However, these studies did not identify the individual molecules that constituted this profile. Subsequent analysis by Kim et al was able to identify metabolites in the urine with differential levels in patients with RCC relative to unaffected controls [73]. Among the molecules identified with differential levels in patients with RCC were quinolinate, 4-hydroxybenzoate, and genistate. Not surprisingly, these molecules are involved in amino acid and energetic metabolism. Zira et al recently reported on serum metabolomic analysis using nuclear magnetic resonance [74]. Using principal component analysis, they were able to identify a metabolic profile that could distinguish between patients with RCC and those with benign urologic disease. More recently, Lin et al were able to generate a metabolic profile using LC-MS of serum samples to distinguish with 100% sensitivity and 100% specificity patients with RCC from healthy volunteers [75]. In addition, they were able to identify candidate metabolic biomarkers with differential levels in patients with early and advanced stage disease. More recent studies have furthered the potential for metabolic biomarkers of RCC. Ganti et al implanted ccRCC cells (Caki-1 cell line) into nude mice and performed concurrent metabolic analysis of tumor tissue, serum, and urine [76]. Interestingly, they found that many metabolites that could be detected in tumor tissue could also be identified in serum and urine. Moreover, they found a general concordance of metabolite levels in tumor tissue and serum. These data suggest that serum metabolomics analysis may serve as a proxy for tumor metabolite alterations.
3.10. Altered metabolism in renal cancer: clinical implications
With recognition of the changes in metabolism characteristic of RCC histologic subtypes, and in particular ccRCC, opportunities are emerging for biomarker development that can assist with prognosis, diagnosis, and therapeutics. In some cases, the level of metabolic alteration may correlate with prognosis. Many of the studies have focused on HIF expression in RCC as a prognostic biomarker [77]. Interestingly, VHL-null ccRCC can be broadly characterized into two groups based on the expression of HIF-1 and HIF-2: (1) tumors that express both HIF-1 and HIF-2 and (2) tumors that express HIF-2 alone [78]. Recent data by Shen et al suggest that HIF-1α may have a tumor suppressor function in ccRCC [79]. From a diagnostic standpoint, the gene expression of metabolic enzymes, or the metabolic profiles themselves, may assist in the diagnosis of renal cancer. A readily apparent application would be to assist in the interpretation of biopsy results of small renal masses. As up to 20% of biopsies in this setting may be nondiagnostic [80], the additional analysis of these metabolic parameters may improve the diagnostic yield of biopsy specimens.
Moreover, it may also help further stratify patients if additional metabolic prognostic factors could be assessed, once these are validated. The metabolic alterations found in ccRCC may also have therapeutic implications given recent evidence suggesting a correlation between tumor and serum metabolite levels in an animal model. Perhaps serum metabolite levels may serve as biomarker of tumor burden. This could be particularly helpful in the surveillance of patients who may be at high risk of recurrence. Alternatively, measurement of serum metabolite levels may be used to follow clinical response to therapy in patients with advanced/metastatic disease. For example, increases in serum cholesterol were recently reported to predict response to the mTOR inhibitor temsirolimus in patients with RCC [81]. At this time, serial imaging is the only reliable method of monitoring patients at high risk of recurrence and those on systemic therapy for advanced disease. Serum metabolite biomarkers theoretically could provide the advantage of assessing tumor burden in a more dynamic fashion. The technology currently exists to measure metabolites in serum. Hence, this could be easily incorporated into clinical practice once serum metabolites with differential levels in the presence or absence of RCC are identified and the measurements are standardized.
4. Conclusions
The last few decades have witnessed a marked advance in our understanding of the genetics of renal cancer, particularly ccRCC. ccRCC is remarkable for the fact that common genetic changes (ie, VHL loss) have dramatic effects on the metabolism that are biologically relevant to the growth and survival of renal tumor cells. As such, further studies into ccRCC will provide a unique opportunity for researchers to apply understanding of tumor metabolism to clinically relevant applications. These characteristic changes in metabolism will provide avenues for the development of novel therapeutic strategies, perhaps in synergy with agents currently in use. Additionally, further understanding of RCC metabolism will likely open the door to biomarker development that will assist with diagnosis, prognosis, and monitoring therapeutic response.
Acknowledgments
Funding/Support and role of the sponsor: None.
Footnotes
Author contributions: Sunil Sudarshan had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Thompson, Margulis, Brugarolas, Karam, Uzzo, Rini, Escudier, Patard, Sudarshan, Linehan.Acquisition of data: Sudarshan.
Analysis and interpretation of data: Thompson, Margulis, Brugarolas, Karam, Uzzo, Rini, Escudier, Patard, Sudarshan, Linehan.
Drafting of the manuscript: Thompson, Margulis, Brugarolas, Karam, Uzzo, Rini, Escudier, Patard, Sudarshan, Linehan.
Critical revision of the manuscript for important intellectual content: Thompson, Margulis, Brugarolas, Karam, Uzzo, Rini, Escudier, Patard, Sudarshan, Linehan.
Statistical analysis: None.
Obtaining funding: None.
Administrative, technical, or material support: None.
Supervision: Linehan.
Other (specify): None.
Financial disclosures: Sunil Sudarshan certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.eururo.2012.09.054
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3709870?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Enhancing breast cancer outcomes with machine learning-driven glutamine metabolic reprogramming signature.
Front Immunol, 15:1369289, 01 May 2024
Cited by: 0 articles | PMID: 38756785 | PMCID: PMC11097668
Single-cell landscape and spatial transcriptomic analysis reveals macrophage infiltration and glycolytic metabolism in kidney renal clear cell carcinoma.
Aging (Albany NY), 15(20):11298-11312, 16 Oct 2023
Cited by: 3 articles | PMID: 37847178 | PMCID: PMC10637799
PDK1 promotes breast cancer progression by enhancing the stability and transcriptional activity of HIF-1α.
Genes Dis, 11(4):101041, 15 Jul 2023
Cited by: 2 articles | PMID: 38560503 | PMCID: PMC10978537
Potential Clinical Value of Pretreatment De Ritis Ratio as a Prognostic Biomarker for Renal Cell Carcinoma.
Front Oncol, 11:780906, 21 Dec 2021
Cited by: 6 articles | PMID: 34993141 | PMCID: PMC8724044
Identification of a Novel Stem Cell Subtype for Clear Cell Renal Cell Carcinoma Based on Stem Cell Gene Profiling.
Front Oncol, 11:758989, 29 Nov 2021
Cited by: 1 article | PMID: 34912710 | PMCID: PMC8667732
Go to all (46) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
(1)H NMR metabolomics analysis of renal cell carcinoma cells: Effect of VHL inactivation on metabolism.
Int J Cancer, 138(10):2439-2449, 29 Jan 2016
Cited by: 13 articles | PMID: 26620126
The relationship of erythropoietin overexpression with von Hippel-Lindau tumour suppressor gene mutations between hypoxia-inducible factor-1α and -2α in sporadic clear cell renal carcinoma.
Int J Mol Med, 26(6):907-912, 01 Dec 2010
Cited by: 11 articles | PMID: 21042786
The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C.
Oncogene, 31(6):776-786, 04 Jul 2011
Cited by: 103 articles | PMID: 21725364 | PMCID: PMC4238297
The VHL tumor suppressor and HIF: insights from genetic studies in mice.
Cell Death Differ, 15(4):650-659, 25 Jan 2008
Cited by: 94 articles | PMID: 18219317 | PMCID: PMC3799983
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS
NCI NIH HHS (1)
Grant ID: K08 CA138774





